Abstract
Denaturing gradient gel electrophoresis (DGGE) has become a widely used tool to examine microbial diversity and community structure, but no systematic comparison has been made of the DGGE profiles obtained when different hypervariable (V) regions are amplified from the same community DNA samples. We report here a study to make such comparisons and establish a preferred choice of V region(s) to examine by DGGE, when community DNA extracted from samples of digesta is used. When the members of the phylogenetically representative set of 218 rrs genes archived in the RDP II database were compared, the V1 region was found to be the most variable, followed by the V9 and V3 regions. The temperature of the lowest-melting-temperature (Tm(L)) domain for each V region was also calculated for these rrs genes, and the V1 to V4 region was found to be most heterogeneous with respect to Tm(L). The average Tm(L) values and their standard deviations for each V region were then used to devise the denaturing gradients suitable for separating 95% of all the sequences, and the PCR-DGGE profiles produced from the same community DNA samples with these conditions were compared. The resulting DGGE profiles were substantially different in terms of the number, resolution, and relative intensity of the amplification products. The DGGE profiles of the V3 region were best, and the V3 to V5 and V6 to V8 regions produced better DGGE profiles than did other multiple V-region amplicons. Introduction of degenerate bases in the primers used to amplify the V1 or V3 region alone did not improve DGGE banding profiles. Our results show that DGGE analysis of gastrointestinal microbiomes is best accomplished by the amplification of either the V3 or V1 region of rrs genes, but if a longer amplification product is desired, then the V3 to V5 or V6 to V8 region should be targeted.
The inherent limitations associated with cultivation-based approaches to characterizing microbial communities are widely recognized, and a number of cultivation-independent, molecular methods have emerged in recent years to improve our understanding of this aspect of microbial ecology. Techniques such as denaturing gradient gel electrophoresis (DGGE) (8, 20, 21, 27), terminal restriction fragment length polymorphism (15, 22, 28, 38), length heterogeneity PCR (29, 34), automated rRNA intergenic spacer analysis (13), and ribosomal intergenic spacer length polymorphism (9, 40) are now widely used and reported in the literature. In the in silico analyses of 41 completely sequenced bacterial genomes, DGGE appeared to be one of the best molecular community fingerprinting techniques in terms of predicting the actual Shannon-Wiener diversity index, richness, and evenness (3). Additionally, DGGE supports the identification of community members because the amplification products can be recovered and sequenced (4, 6, 20, 31). This may explain why DGGE has become the most frequently used method of molecular community fingerprinting.
In PCR-DGGE, either a single hypervariable (V) region or a combination of two or three V regions in rrs genes is amplified (1, 2, 7, 12, 14, 16, 17, 19, 31-33, 35-37). In studies of rumen and gastrointestinal microbiomes, the V1, V3, V1 to V3, and V6 to V8 regions have all been examined (5, 11, 24, 25, 31). Although it is well recognized that the quality of information produced by PCR-DGGE is dependent on both the number and resolution of the amplicons in denaturing gradient gels, few authors have explained their justification of primer choice and DGGE conditions. Indeed, there appears to have been little or no systematic evaluation of how the choice of primers or denaturing conditions influence data quality and veracity of the analysis.
In this context, we have compared the PCR-DGGE profiles arising from a common set of community DNA samples, using primer sets that have been previously reported in the literature. We have also used the phylogenetically representative set of rrs genes archived in the RDP II database to calculate melting temperature (Tm) values of the lowest-melting-temperature (Tm(L)) domains for each V region and to formulate DGGE conditions for each set of amplification products. We expect that our results can be used to standardize the primer sets and DGGE conditions needed to produce the most comprehensive information about the microbial diversity present in rumen and gastrointestinal microbiomes.
MATERIALS AND METHODS
Microbial community samples and DNA extraction.
Microbial community samples were collected from four sheep. Two were fed a mixture of hay and grain (60:40 on a dry-matter basis; sheep are referred to as C1 and C2), and the other two were fed a mixture of hay, grain, and tallow (60:20:20 on a dry-matter basis; sheep are referred to as F1 and F2). Samples of rumen digesta were collected via rumen cannulae 14 and 23 days after the sheep were started on those diets. The digesta were strained through two layers of cheesecloth, and community DNA from these eight rumen fluid samples was extracted using the RBB + C method (41). The DNA recovered from each sample was quantified using the PicoGreen double-stranded DNA quantitation kit (Molecular Probes, Inc., Eugene, Oreg.) and diluted to a final concentration of 50 ng/μl with Tris-EDTA.
In silico analysis of rrs gene V regions.
The phylogenetically representative set of 218 rrs genes was downloaded from the RDP II database (release 8.1). The V regions were delimited by the primer sequences flanking them, and the respective sequence identities were calculated using BioEdit (http://www.mbio.ncsu.edu/BioEdit/bioedit.html). The average (± standard deviation) of the percent sequence identities for each individual V region, or combination thereof, was calculated using Microsoft Excel.
The Tm(L) value of each V region was also calculated for the same sequences with Primo Melt 3.4 software (http://www.changbioscience.com/primo/primomel.html), and the frequencies of Tm(L) values for the V1, V3, and V5 regions were graphed using Microsoft Excel. It was assumed that a 40-bp GC clamp was present at one end of each amplicon. The average Tm(L) for each V region and its standard deviation were calculated, and these values were used to predict the denaturing gradients needed to effectively resolve 95% of all the amplicons produced by a specific primer set, with the following equations:
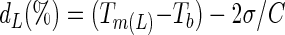 |
(1) |
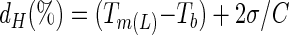 |
(2) |
where dL and dH are the low and high denaturant concentrations, respectively; Tm(L) is the average Tm value of the lowest-melting-temperature domains of each amplicon; Tb is the temperature of the running buffer (typically 60°C); σ is the standard deviation corresponding to the average Tm(L) of that amplicon; and C is a constant, relating chemical denaturant concentration to melting temperature of double-stranded DNA (in DGGE, C = 0.3°C/1% denaturant as described in reference 10).
PCR and DGGE.
The primers, annealing temperatures, and DGGE conditions used in this study are listed in Table 1. The following degenerate primer sets were used to evaluate the effects of primer degeneracy on DGGE profiles: Deg-63f (5′-GCY TAA BVC ATG CRA GTC-3′), Deg-109r (5′-AYG YRT TAC TSA SCC KT-3′); Deg-357f (5′-ACW CCT ACG GGD SGC WGC A-3′), and Deg-518r (5′-GTA TTA CCG CGG CKG CTG-3′). Degenerate bases (equimolar ratios) were introduced at positions where two or more nucleotides occur at relatively high frequency within the phylogenetically representative set of rrs genes. Inosine-containing primers with the underlined bases replaced with inosine were also tested. All PCR amplifications were performed using a PTC-100 thermocycler (MJ Research, Waltham, Mass.) in 50-μl volumes containing 1× PCR buffer (20 mM Tris-HCl [pH 8.4] and 50 mM KCl), 200 μM deoxynucleoside triphosphates, 500 nM (each) primer, 1.75 mM MgCl2, 670 ng of bovine serum albumin/μl, and 1.25 U of Platinum Taq DNA polymerase (Invitrogen Corporation, Carlsbad, Calif.), which allows for hot-start PCR. After an initial denaturation at 94°C for 5 min, 10 cycles of touchdown PCR were performed (denaturation at 94°C for 30 s, annealing for 30 s with an 0.5°C/cycle decrement at a temperature 5°C above the respective annealing temperatures indicated in Table 1, and extension at 72°C for 1 min), followed by 25 cycles of regular PCR (94°C for 30 s, 30 s at the respective annealing temperature, and 72°C for 1 min [0.5 min for PCR amplification of the V1 region]) and a final extension step for 7 min at 72°C. Negative controls, containing all the components except DNA templates, were included in parallel.
TABLE 1.
PCR primers, targeted hypervariable regions, PCR, and DGGE conditions used in this study
| Primer | Sequence (5′→3′) | Annealing positionsb | Target | Annealing temp | Amplicon length (bp)c | DGGE conditions | Reference(s) |
|---|---|---|---|---|---|---|---|
| −63fa | GCC TAA CAC ATG CAA GTC | 46-63 | V1 | 58→53°C, −0.5°C/cycle | 80 | 8%, 30-50%, 620 V · h | 2,31 |
| 109r | ACG TGT TAC TCA CCC GT | 109-125 | |||||
| −63fa | GCC TAA CAC ATG CAA GTC | 46-63 | V1-V3 | 58→53°C, −0.5°C/cycle | 489 | 6.5%, 20-70%, 1,230 V · h | 5 |
| 518r | ATT ACC GCG GCT GCT GG | 518-534 | |||||
| −357fa | CCT ACG GGA GGC AGC AG | 341-357 | V3 | 61→56°C, −0.5°C/cycle | 194 | 6.5%, 40-60%, 1,230 V · h | 7,35 |
| 518r | ATT ACC GCG GCT GCT GG | 518-534 | |||||
| 533f | GTG CCA GCA GCC GCG GTA A | 515-533 | V4-V5 | 56→51°C, −0.5°C/cycle | 412 | 6.5%, 20-70%, 1,230 V · h | 16 |
| 907ra | CCG TCA ATT CCT TTG AGT TT | 907-926 | |||||
| −357fa | CCT ACG GGA GGC AGC AG | 341-357 | V3-V5 | 56→51°C, −0.5°C/cycle | 586 | 6.5%, 30-60%, 1,230 V · h | 1,37 |
| 907r | CCG TCA ATT CCT TTG AGT TT | 907-926 | |||||
| −954fa | GCA CAA GCG GTG GAG CAT GTG G | 933-954 | V6-V8 | 61→56°C, −0.5°C/cycle | 456 | 6.5%, 35-60%, 1,230 V · h | 18,19 |
| 1369r | GCC CGG GAA CGT ATT CAC CG | 1369-1388 | |||||
| F-968a | AAC GCG AAG AAC CTT AC | 968-984 | V6-V8 | 61→56°C, −0.5°C/cycle | 434 | 6.5%, 35-60%, 1,230 V · h | 25,42 |
| R-1401 | CGG TGT GTA CAA GAC CC | 1385-1401 | |||||
| 1070f | ATG GCT GTC GTC AGC T | 1055-1070 | V8 | 53→48°C, −0.5°C/cycle | 352 | 6.5%, 40-60%, 1,230 V · h | 12,14 |
| −1392ra | ACG GGC GGT GTG TAC | 1392-1406 |
Primers with a 40-bp GC clamp at the 5′ end.
Numbering according to the rrs gene of Escherichia coli.
Calculated from the rrs gene of E. coli; the primers were included.
After PCR, 5-μl aliquots were subjected to agarose gel electrophoresis with 1.5% (wt/vol) agarose gels (2% [wt/vol] for the V1-region amplicons). Then, 15-μl aliquots were resolved on polyacrylamide gels (37.5:1) containing a gradient of denaturants (100% denaturants consisting of 40% [vol/vol] formamide and 7 M urea) as indicated in Table 1. All the DGGE gels were run at 60°C and 82 V to reach the volts-hours indicated in Table 1, with a Dcode Universal Mutation detection system (Bio-Rad Laboratories, Hercules, Calif.). The DGGE gels were stained with GelStar (BioWhittaker Molecular Applications, Rockland, Maine) according to the manufacturer's specifications, and the images were captured using a FluorChem Imager (Alpha Innotech Corp., San Leandro, Calif.).
DGGE gel analysis.
The DGGE bands were detected using the band-searching algorithm of BioNumerics software (BioSystematica, Tavistock, Devon, United Kingdom). After normalization of the gels, only those bands with a peak height intensity exceeding 2.0% of the strongest band in each lane were included in further analyses. Diversity indices were also calculated: richness (S) was determined from the number of bands in each lane, and the Shannon-Wiener index (H′) was calculated from H′ = −ΣPilnPi (30), where Pi is the importance probability of the bands in a lane, calculated from ni/N, where ni is the peak height of a band and N is the sum of all peak heights in the densitometric curve. Evenness (E) was calculated as E = H′/H′max, where H′max = ln S (26).
RESULTS
Sequence divergence within various V regions and their Tm(L) heterogeneity.
With the use of the phylogenetically representative set of rrs genes archived at RDP II, the V1 region was found to be the most divergent, as indicated by the low average sequence identities and the high standard deviation (Table 2). The V3 and V9 regions are also more divergent than the remaining V regions examined. Accordingly, the V1, V2, and V3 regions combined were found to possess the highest sequence divergence compared to the other multiple V regions.
TABLE 2.
Sequence identities of the PCR amplicons calculated from the phylogenetically representative set of 218 rrs genes in RDP II and the melting temperatures of the lowest-melting-temperature domains (Tm(L))
| Hypervariable region | % Sequence identitya | No. of Tm(L) groupsb | No. of sequences sharing the same Tm(L) | Avg Tm(L) (°C)a | Denaturant gradient (%)c |
|---|---|---|---|---|---|
| V1 | 34.4 (12.1) | 120 | 1.83 | 77.6 (4.9) | 26-91 |
| V2 | 62.9 (8.0) | 108 | 2.03 | 74.1 (3.3) | 25-69 |
| V3 | 60.9 (15.4) | 89 | 2.46 | 76.3 (2.6) | 37-72 |
| V4 | 67.4 (8.4) | 93 | 2.35 | 76.3 (2.7) | 36-72 |
| V5 | 73.5 (7.1) | 77 | 2.84 | 76.3 (2.1) | 40-68 |
| V6 | 69.3 (15.3) | 64 | 3.42 | 77.1 (1.6) | 46-68 |
| V8 | 76.5 (7.6) | 73 | 3.00 | 75.7 (1.9) | 40-65 |
| V9 | 48.3 (16.2) | 84 | 2.61 | 77.6 (2.5) | 42-75 |
| V1-V3 | 52.7 (20.7) | 101 | 2.17 | 74.5 (3.2) | 27-70 |
| V3-V5 | 69.2 (9.0) | 91 | 2.41 | 74.7 (2.7) | 31-67 |
| V4-V5 | 71.7 (7.2) | 89 | 2.46 | 74.8 (2.5) | 33-66 |
| V6-V8d | 70.6 (10.5) | 80 | 2.74 | 73.6 (2.2) | 31-60 |
| V6-V8e | 71.8 (10.3) | 78 | 2.80 | 75.1 (2.2) | 36-65 |
| V7-V8 | 74.2 (7.5) | 75 | 2.92 | 75.3 (2.2) | 36-66 |
Values in parentheses are standard deviations.
Difference of >0.1°C.
Assuming that DGGE gels are run at 60°C.
Amplicons delimited with GC-954f and 1368r.
Amplicons delimited with F-968-GC and R-1401.
The average Tm(L) values for the V regions ranged from 73.6 to 77.6°C (Table 2), and the V1, V2, and V1 to V3 regions possess the greatest Tm(L) heterogeneity, while the V5, V6, V8, and V7-V8 regions have the lowest Tm(L) heterogeneity. Moreover, the frequency of calculated Tm(L) values for the V1, V3, and V5 regions all appeared to be normally distributed about the average value (Fig. 1). Equations 1 and 2 described in Materials and Methods were therefore devised to predict the denaturing gradient necessary to resolve 95% of the amplification products arising for each V region. Table 2 shows that the V1 region requires the largest denaturing gradient (26 to 91%), while the V6 region requires the smallest (46 to 68%). With the use of these equations, the denaturing gradients listed for the V1, V3, and V5 regions should effectively resolve 95.0, 95.4, and 92.7% of the 218 phylogenetically representative rrs sequences, respectively.
FIG. 1.
Frequency distribution of Tm(L) values for the V1 region (A), V3 region (B), and V5 region (C), derived from the phylogenetically representative set of 218 rrs gene sequences archived in RDP II (release 8.1). The arrows indicate the average Tm(L) value calculated for each V region, and the Tm(L) values appear to be normally distributed.
DGGE profiles of rumen microbial community DNA samples.
The DGGE profiles produced using the different primer sets and conditions listed in Table 1 are shown in Fig. 2, and the diversity indices calculated from the PCR-DGGE banding profiles are shown in Table 3. Amplification of either the V1 or V3 region alone yielded more intense bands, and the V3 region produced the largest number of bands (and richness score), followed by the V1 and V8 regions. However the V1, V3, and V8 DGGE profiles all produced relatively low evenness scores, apparently due to the existence of a relatively small number of intense bands. When multiple V regions were amplified, the richness scores were all lower than that for the V3 region alone, because of the reduced number of discernible bands, and the V1 to V3 region was not amplified as efficiently as were other V regions, as judged by the intensity of the bands on the DGGE gel (Fig. 2). In contrast, the evenness scores were all higher for the multiple-V-region profiles, suggesting that band intensity was more uniform in these DGGE profiles. Another interesting finding was the differences arising in DGGE profiles and diversity indices when the different V6 to V8 primer sets were used (Fig. 2 and Table 3). Overall, the primer set consisting of GC-954f and 1369r resulted in higher values than those obtained with F-968-GC and R-1401, although both primer sets were found to produce lower richness, Shannon-Wiener, and evenness values than those for the V3 to V5 region (Table 3). Based on these results, it appears that amplification of the V3 region alone produced the most informative DGGE profiles, and if a longer amplification product is required, then either the V3 to V5 region or the V6 to V8 region amplified with the GC-954f-1369r primer set should be selected.
FIG. 2.
DGGE banding profiles of V regions produced from the community DNA extracted from eight different samples of ruminal digesta. The nondegenerate primers and DGGE conditions described in Table 1 were used in the PCR and DGGE. The lanes labeled C1 and C2 represent samples of digesta collected from animals fed a mixture of hay and grain, and the lanes labeled F1 and F2 represent samples of digesta collected from animals fed a mixture of hay, grain, and beef tallow. D14 and D23, samples collected 14 and 23 days, respectively, after the animals were started on the diet; Marker, electrophoresis marker; V6 to V8a, DGGE profiles generated using primers GC-954f and 1369r; V6 to V8b, DGGE profiles generated using primers F-968-GC and R-1401.
TABLE 3.
Diversity indices calculated from the DGGE banding profiles generated from various hypervariable regions
Impact of degenerate PCR primers on DGGE profile.
The introduction of degenerate bases into the V1-specific primer set substantially decreased the number of bands (Fig. 3A), and the replacement with inosine resulted in unsuccessful PCR amplification at the various annealing temperatures (42 to 64°C) and MgCl2 concentrations (1.5 to 2.75 mM) tested. The degenerate primers specific for the V3 region increased the number of bands at the upper portion of the DGGE gel, but there was a decrease in the number of bands appearing at the lower portion of the gel (Fig. 3B). Although the use of inosine-containing primers resulted in a more even band distribution in the DGGE gel (Fig. 3C), there was little influence on the diversity indices (data not shown). Based on these results, it appears that the introduction of degenerate bases and/or inosine in the V1 and V3 primer sequences does not improve the PCR-DGGE profiles.
FIG. 3.
DGGE gel banding profiles of the V1 and V3 regions produced with V1-specific inosine-containing primers (A), V3-specific degenerate primers (B), or V3-specific inosine-containing primers (C). The labeling of lanes follows the same protocol as described for Fig. 2.
DISCUSSION
Although there are numerous published reports of using PCR-DGGE to examine microbial diversity (e.g., references 1, 12, 14, 16, 18, and 19), how the quality of the information obtained is impacted by the choice of V region(s) amplified has not been previously evaluated. The results presented here with eight different community DNA samples clearly show that the V region(s) chosen for amplification can greatly influence the PCR-DGGE profiles and diversity indices produced from community DNA samples, and even subtle differences in primer sequences can result in substantially different profiles and assessment of microbial diversity. The denaturing gradient used will also affect the results obtained, but this, too, has received little attention. The gradient estimation approach described here was very effective in producing well-resolved banding profiles, with all the primer sets used here. By using the set of 218 rrs gene sequences archived in RDP II to calculate average Tm(L) values (and standard deviations), we avoided bias towards those taxa that have more sequence representations in RDP II. However, our approach to choosing denaturing gradients should also be applicable to guild-, genus-, or species-specific PCR-DGGE analyses, where closely related sequences require a narrow denaturing gradient to maximize the resolving power of DGGE.
Theoretically the V1 region, with the highest sequence divergence and Tm(L) heterogeneity, should have produced DGGE profiles with the greatest number of resolved bands. However, the DGGE profiles of the V1 region alone were inferior to those obtained with V3-specific primers (Table 3 and Fig. 2). This may be attributed to the limited length of the V1 region, rather than the limited universality of the primer set used. This explanation is supported by the observation that degenerate primers did not improve DGGE profiles of the V1 region (Fig. 3A). For the V3 region, although inosine replacements resulted in an increase in the number of bands at the lower portion of the DGGE gel (Fig. 3C), the use of degenerate primers did not improve the assessment of microbial diversity in the samples. For these reasons, we propose that PCR-DGGE targeting the V1 region be avoided in future studies of gut microbiomes and that degenerate primers be used with caution in PCR-DGGE analyses.
A necessary requirement of all DGGE-based studies of microbial ecology is the reamplification and sequencing of excised amplicons, to provide a more detailed characterization of these microbial communities. For such purposes, longer amplicons would be preferable to facilitate identification to species level with a higher degree of probability. Most automated DNA sequencers now produce in excess of 500 bp of unambiguous sequence, which can span at least two V regions. Based on the results obtained here, amplification of the V3 to V5 region produced superior DGGE profiles. Given the results obtained when either the V1 or V3 region was amplified alone, we were also surprised by the relatively poor DGGE profiles when the V1 to V3 region was amplified. An in silico analysis of the primer set consisting of GC-63f and 518r performed using Primer Designer (Scientific & Educational Software, Durham, N.C.) did not show primer self-annealing or hairpin formation. Combined with the results obtained when the V1 region alone was amplified, a different forward primer (e.g., 27f, 5′-AGA GTT TGA TCM TGG CTC AG-3′ [23]) may improve the amplification and DGGE profiles of the V1 to V3 region.
It is interesting to notice the difference in results obtained when two different primer sets were used to amplify the V6 to V8 region (Fig. 2 and Table 3). The primer set consisting of F-968-GC and R-1401 set has been frequently used for PCR-DGGE analysis of community DNA samples from human and animal gastrointestinal tracts (11, 21, 39, 42), while the primer set consisting of GC-954f and 1369r has been used with other types of environmental samples (18, 19). The annealing sites for these primers are in high proximity to each other: the F-968-GC and GC-954f primers are 30 nucleotides apart, and the R-1401 and 1369r primers are 16 nucleotides apart. However, the primer set consisting of GC-954f and 1369r is more universal than that consisting of F-968-GC and R1401, because when compared to all 16S rrs sequences in RDP II (release 8.1), 1369r matches many more (8,595 perfect matches and 4,482 nearly perfect matches [0.9 < Sab < 1.0]) than does R-1401 (2,049 perfect matches and 495 nearly perfect matches). The improved DGGE profiles derived with GC-954f and 1369r are qualitatively consistent with the above in silico analysis, and the different DGGE profiles generated from these two primer sets suggest detection of different populations in the samples. For these reasons, we propose that, if the V6 to V8 region is chosen for PCR-DGGE analyses, a more informative analysis of gastrointestinal microbiomes will be produced by using GC-954 and 1369r rather than F-968-GC and R-1401.
In conclusion, given the similarities among the microbial communities present in the gastrointestinal tracts of humans and other herbivores, in terms of the dominance of these communities by members of the phylum Bacteroidetes and class Clostridia, we recommend that the V3 region be routinely used in PCR-DGGE analyses with such samples. Alternatively, the V3 to V5 or V6 to V8 (with the use of GC-954f and 1369r) region can be chosen if a longer amplicon is preferred.
Acknowledgments
This work was supported by funds available to the authors through an Ohio Board of Regents Academic Enrichment Award and funds from the Ohio Agricultural Research and Development Center (Hatch Research Subsidy 530189 and OHOG0592-500486).
We also thank Burk Dehority, Ohio Agricultural Research and Development Center, Wooster, for the collection of the samples of rumen digesta.
REFERENCES
- 1.Casamayor, E. O., H. Schafer, L. Baneras, C. Pedros-Alio, and G. Muyzer. 2000. Identification of and spatio-temporal differences between microbial assemblages from two neighboring sulfurous lakes: comparison by microscopy and denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 66:499-508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cocolin, L., M. Manzano, C. Cantoni, and G. Comi. 2001. Denaturing gradient gel electrophoresis analysis of the 16S rRNA gene V1 region to monitor dynamic changes in the bacterial population during fermentation of Italian sausages. Appl. Environ. Microbiol. 67:5113-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crosby, L. D., and C. S. Criddle. 2003. Understanding bias in microbial community analysis techniques due to rrn operon copy number heterogeneity. BioTechniques 34:790-802. [DOI] [PubMed] [Google Scholar]
- 4.Curtis, T. P. 1998. The comparison of the diversity of activated sludge plants. Water Sci. Technol. 37:71-78. [Google Scholar]
- 5.De Boever, P., R. Wouters, V. Vermeirssen, N. Boon, and W. Verstraete. 2001. Development of a six-stage culture system for simulating the gastrointestinal microbiota of weaned infants. Microbiol. Ecol. Health Dis. 13:111-123. [Google Scholar]
- 6.Diez, B., C. Pedros-Alio, T. L. Marsh, and R. Massana. 2001. Application of denaturing gradient gel electrophoresis (DGGE) to study the diversity of marine picoeukaryotic assemblages and comparison of DGGE with other molecular techniques. Appl. Environ. Microbiol. 67:2942-2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dilly, O., J. Bloem, A. Vos, and J. C. Munch. 2004. Bacterial diversity in agricultural soils during litter decomposition. Appl. Environ. Microbiol. 70:468-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donskey, C. J., A. M. Hujer, S. M. Das, N. J. Pultz, R. A. Bonomo, and L. B. Rice. 2003. Use of denaturing gradient gel electrophoresis for analysis of the stool microbiota of hospitalized patients. J. Microbiol. Methods 54:249-256. [DOI] [PubMed] [Google Scholar]
- 9.Eriksson, M., E. Sodersten, Z. Yu, G. Dalhammar, and W. W. Mohn. 2003. Degradation of polycyclic aromatic hydrocarbons at low temperature under aerobic and nitrate-reducing conditions in enrichment cultures from northern soils. Appl. Environ. Microbiol. 69:275-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ezra, S., S. Abrams, and V. Stanton. 1992. Use of denaturant gradient gel electrophoresis to study conformational transitions in nucleic acids. Methods Enzymol. 212:71-104. [DOI] [PubMed] [Google Scholar]
- 11.Favier, C. F., E. E. Vaughan, W. M. de Vos, and A. D. L. Akkermans. 2002. Molecular monitoring of succession of bacterial communities in human neonates. Appl. Environ. Microbiol. 68:219-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferris, M., G. Muyzer, and D. Ward. 1996. Denaturing gradient gel electrophoresis profiles of 16S rRNA-defined populations inhabiting a hot spring microbial mat community. Appl. Environ. Microbiol. 62:340-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fisher, M. M., and E. W. Triplett. 1999. Automated approach for ribosomal intergenic spacer analysis of microbial diversity and its application to freshwater bacterial communities. Appl. Environ. Microbiol. 65:4630-4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friedrich, M., R. J. Grosser, E. A. Kern, W. P. Inskeep, and D. M. Ward. 2000. Effect of model sorptive phases on phenanthrene biodegradation: molecular analysis of enrichments and isolates suggests selection based on bioavailability. Appl. Environ. Microbiol. 66:2703-2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gong, J., R. J. Forster, H. Yu, J. R. Chambers, R. Wheatcroft, P. M. Sabour, and S. Chen. 2002. Molecular analysis of bacterial populations in the ileum of broiler chickens and comparison with bacteria in the cecum. FEMS Microbiol. Ecol. 41:171-179. [DOI] [PubMed] [Google Scholar]
- 16.Henckel, T., M. Friedrich, and R. Conrad. 1999. Molecular analyses of the methane-oxidizing microbial community in rice field soil by targeting the genes of the 16S rRNA, particulate methane monooxygenase, and methanol dehydrogenase. Appl. Environ. Microbiol. 65:1980-1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hespell, R. B., D. E. Akin, and B. A. Dehority. 1997. Bacteria, fungi, and protozoa of the rumen, p. 59-141. In R. I. Mackie, B. A. White, and R. E. Isaacson (ed.), Gastrointestinal microbiology, vol. 2. Chapman and Hall, New York, N.Y. [Google Scholar]
- 18.Iwamoto, T., K. Tani, K. Nakamura, Y. Suzuki, M. Kitagawa, M. Eguchi, and M. Nasu. 2000. Monitoring impact of in situ biostimulation treatment on groundwater bacterial community by DGGE. FEMS Microbiol. Ecol. 32:129-141. [DOI] [PubMed] [Google Scholar]
- 19.Kawai, M., E. Matsutera, H. Kanda, N. Yamaguchi, K. Tani, and M. Nasu. 2002. 16S ribosomal DNA-based analysis of bacterial diversity in purified water used in pharmaceutical manufacturing processes by PCR and denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 68:699-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kocherginskaya, S. A., R. I. Aminov, and B. A. White. 2001. Analysis of the rumen bacterial diversity under two different diet conditions using denaturing gradient gel electrophoresis, random sequencing, and statistical ecology approaches. Anaerobe 7:119-134. [Google Scholar]
- 21.Konstantinov, S. R., W.-Y. Zhu, B. A. Williams, S. Tamminga, W. M. de Vos, and A. D. L. Akkermans. 2003. Effect of fermentable carbohydrates on piglet faecal bacterial communities as revealed by denaturing gradient gel electrophoresis analysis of 16S ribosomal DNA. FEMS Microbiol. Ecol. 43:225-235. [DOI] [PubMed] [Google Scholar]
- 22.Kuske, C. R., L. O. Ticknor, M. E. Miller, J. M. Dunbar, J. A. Davis, S. M. Barns, and J. Belnap. 2002. Comparison of soil bacterial communities in rhizospheres of three plant species and the interspaces in an arid grassland. Appl. Environ. Microbiol. 68:1854-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. D. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Inc., New York, N.Y.
- 24.Mai, V., H. A. Katki, H. Harmsen, D. Gallaher, A. Schatzkin, D. J. Baer, and B. Clevidence. 2004. Effects of a controlled diet and black tea drinking on the fecal microflora composition and the fecal bile acid profile of human volunteers in a double-blinded randomized feeding study. J. Nutr. 134:473-478. [DOI] [PubMed] [Google Scholar]
- 25.Nübel, U., B. Engelen, A. Felske, J. Snaidr, A. Wieshuber, R. Amann, W. Ludwig, and H. Backhaus. 1996. Sequence heterogeneities of genes encoding 16S rRNAs in Paenibacillus polymyxa detected by temperature gradient gel electrophoresis. J. Bacteriol. 178:5636-5643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pielou, E. C. 1969. An introduction to mathematical ecology. Wiley, New York, N.Y.
- 27.Plant, L., C. Lam, P. L. Conway, and K. O'Riordan. 2003. Gastrointestinal microbial community shifts observed following oral administration of a Lactobacillus fermentum strain to mice. FEMS Microbiol. Ecol. 43:133-140. [DOI] [PubMed] [Google Scholar]
- 28.Ranjard, L., F. Poly, and S. Nazaret. 2000. Monitoring complex bacterial communities using culture-independent molecular techniques: Application to soil environment. Res. Microbiol. 151:167-177. [DOI] [PubMed] [Google Scholar]
- 29.Ritchie, N. J., M. E. Schutter, R. P. Dick, and D. D. Myrold. 2000. Use of length heterogeneity PCR and fatty acid methyl ester profiles to characterize microbial communities in soil. Appl. Environ. Microbiol. 66:1668-1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shannon, C. E., and W. Weaver. 1949. The mathematical theory of communication. University of Illinois Press, Urbana.
- 31.Simpson, J. M., V. J. McCracken, H. R. Gaskins, and R. I. Mackie. 2000. Denaturing gradient gel electrophoresis analysis of 16S ribosomal DNA amplicons to monitor changes in fecal bacterial populations of weaning pigs after introduction of Lactobacillus reuteri strain MM53. Appl. Environ. Microbiol. 66:4705-4714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stewart, C., H. Flint, and M. Bryant. 1997. The rumen bacteria, p. 10-72. In P. N. Hobson and C. S. Stewart (ed.), The rumen microbial ecosystem, 2nd ed. Chapman and Hall, New York, N.Y.
- 33.Suau, A., R. Bonnet, M. Sutren, J.-J. Godon, G. R. Gibson, M. D. Collins, and J. Dore. 1999. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl. Environ. Microbiol. 65:4799-4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suzuki, M., M. S. Rappe, and S. J. Giovannoni. 1998. Kinetic bias in estimates of coastal picoplankton community structure obtained by measurements of small-subunit rRNA gene PCR amplicon length heterogeneity. Appl. Environ. Microbiol. 64:4522-4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tannock, G. W., K. Munro, H. J. M. Harmsen, G. W. Welling, J. Smart, and P. K. Gopal. 2000. Analysis of the fecal microflora of human subjects consuming a probiotic product containing Lactobacillus rhamnosus DR20. Appl. Environ. Microbiol. 66:2578-2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Temmerman, R., L. Masco, T. Vanhoutte, G. Huys, and J. Swings. 2003. Development and validation of a nested-PCR-denaturing gradient gel electrophoresis method for taxonomic characterization of bifidobacterial communities. Appl. Environ. Microbiol. 69:6380-6385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Teske, A., C. Wawer, G. Muyzer, and N. Ramsing. 1996. Distribution of sulfate-reducing bacteria in a stratified fjord (Mariager Fjord, Denmark) as evaluated by most-probable-number counts and denaturing gradient gel electrophoresis of PCR-amplified ribosomal DNA fragments. Appl. Environ. Microbiol. 62:1405-1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tiedje, J. M., S. Asuming-Brempong, K. Nüsslein, T. L. Marsh, and S. J. Flynn. 1999. Opening the black box of soil microbial diversity. Appl. Soil Ecol. 13:109-122. [Google Scholar]
- 39.van de Wielen, P. W. J. J., D. A. Keuzenkamp, L. J. A. Lipman, F. Knapen, and S. Biesterveld. 2002. Spatial and temporal variation of the intestinal bacterial community in commercially raised broiler chickens during growth. Microb. Ecol. 44:286-293. [DOI] [PubMed] [Google Scholar]
- 40.Yu, Z., and W. W. Mohn. 2001. Bioaugmentation with resin-acid-degrading bacteria enhances resin acid removal in sequencing batch reactors treating pulp mill effluents. Water Res. 35:883-890. [DOI] [PubMed] [Google Scholar]
- 41.Yu, Z., and M. Morrison. 2004. Improved extraction of PCR-quality community DNA from digesta and fecal samples. BioTechniques 36:808-812. [DOI] [PubMed] [Google Scholar]
- 42.Zoetendal, E. G., A. von Wright, T. Vilpponen-Salmela, K. Ben-Amor, A. D. L. Akkermans, and W. M. de Vos. 2002. Mucosa-associated bacteria in the human gastrointestinal tract are uniformly distributed along the colon and differ from the community recovered from feces. Appl. Environ. Microbiol. 68:3401-3407. [DOI] [PMC free article] [PubMed] [Google Scholar]





