Abstract
Background
Adipose-derived stem cells (ASC) and its exosomes are gaining utmost importance in the field of regenerative medicine. The ASCs tested for their potential in wound healing are predominantly derived from the subcutaneous depot of lean donors. However, it is important to characterize the ASC derived from different adipose depots as these depots have clinically distinct roles.
Methods
We characterized the ASC derived from subcutaneous and omental depots from a lean donor (sc-ASCn and om-ASCn) and compared it to the ASC derived from an obese donor (sc-ASCo and om-ASCo) using flow cytometry and real time qPCR.
Results
We show that stem cell markers Oct4, Sal4, Sox15, KLF4 and BMI1 have distinct expression patterns in each ASC. We evaluated the secretome of the ASC and characterized their secreted exosomes. We show long noncoding RNAs (lncRNAs) are secreted by ASC and their expression varied between the ASC’s derived from different depots. Protein kinase C delta (PKCδ) regulates the mitogenic signals in stem cells. We evaluated the effect of silencing PKCδ in sc-ASCn, om-ASCn, sc-ASCo and om-ASCo. Using β-galactosidase staining, we evaluated the percentage of senescent cells in sc-ASCn, om-ASCn, sc-ASCo and om-ASCo. Our results also indicated that silencing PKCδ increases the percentage of senescent cells.
Conclusions
Our case-specific study demonstrates a role of PKCδ in maintaining the adipose stem cell niche and importantly demonstrates depot-specific differences in adipose stem cells and their exosome content.
Keywords: Adipose-derived stem cells (ASCs), adipose stem cell markers, long noncoding RNAs (lncRNA), exosome, senescence, obesity, Protein kinase C delta (PKCδ)
Introduction
Human adult stem cells are capable of regenerating the cellular population via self-renewal and their ability to differentiate and maintain asymmetric cellular division. The hematopoietic and mesenchymal stem cells were predominantly characterized and studied for stem cell markers and differentiation into specific lineages. Since then, it is now recognized that stem cells reside within stem cell niches of the adult organ where the number of stem cells are regulated. Amongst these, the adipose-derived stem cells (ASCs) show great therapeutic potential in regenerative medicine as they are relatively easy to isolate and are obtained at higher yields.
Human ASCs reside within the stromal vascular fraction (SVF) of the adipose tissue. ASC maintain a quiescent stage in vivo and can undergo differentiation into adipocytes, osteoblasts or chondrocytes (1,2). ASC maintain their self-renewal ability by expressing transcriptional factors that regulate its ability to differentiate. Tissue damage and diseases often cause the ASC to undergo hyper-proliferation leading to dysfunctional cells. We previously showed that the ASC obtained from obese patients have distinct differences in their genetic profiles during differentiation to adipocytes compared to lean (1). In addition, it is known that depot-specific differences are prominent in white adipose tissue. For example, leptin is produced predominantly by subcutaneous adipose tissue; adiponectin is expressed higher in omental adipose tissue. Excess omental fat is central to increased risk factor for cardiovascular diseases and diabetes mellitus.
Protein kinase C is a family of serine/threonine kinases with 11 isoforms. The primary amino acid structure of PKCs can be divided into conserved regions (C1-C4) separated by the variable regions (V1-V5). All PKCs have an N-terminal regulatory domain and a C-terminal catalytic domain separated by the V3 hinge region. The protein kinase C family is subdivided into three groups based upon their activation by calcium, phosphatidyl serine, diacyl glycerol or phorbol esters: classical or conventional PKCs (α, βI, βII and γ), novel PKCs (δ, ε, η and θ) and atypical PKCs (ζ, λ/ι). PKCs are also activated by proteolytic cleavage at the V3 hinge region by calpain I, II or caspase-3 to generate a constitutively active catalytic domain of PKC. PKCδ, a novel PKC, plays an important role in cellular differentiation, proliferation and apoptosis. In addition, several reports have indicated the role of PKCδ in stem cell differentiation (3,4). PKCδ is alternatively spliced to generate PKCδI and PKCδVIII variants in humans (5,6). We have previously shown the expression of PKCδ splice variants in the ASC as well as the role of PKCδVIII in regulating hTERT in senescence (7).
Here we investigated the differences in stem cells derived from subcutaneous and omental adipose depots from a lean and an obese donor. We compared stem cell markers, exosomes and cellular senescence in the ASC isolated from different depots and evaluated the role of PKCδ in ASC niche.
Methods
Adipose samples
White adipose tissue was obtained as discarded tissue from surgeries performed at Tampa General Hospital by Dr. Murr. Donors consented to their waste tissue to be used in research. The subcutaneous and omental depots were collected from the same subject. The lean adipose tissue samples were obtained from a female donor with BMI of 21.3 and the obese adipose tissue samples were from a female donor with BMI of 54.6. Both subjects were non-diabetic, non-smokers and did not have any form of cancer. The de-identified samples were obtained under an Institutional Review Board approved protocol (University of South Florida IRB #20295) with a “not human research activities determination” and was transported to the laboratory and processed within 24 h of collection.
Adipose-derived stem cells (ASCs)
ASC were isolated as previously described by our lab (2). Briefly, adipose tissue was cut up into small pieces and digested with 0.075% collagenase type 1 (Worthington) in modified PBS for 2 h at 37 °C. The digestion was stopped by adding α-MEM +20% heat-inactivated FBS. The suspension was filtered and centrifuged at 400 g at room temperature. The pellet contains the SVF. The pellet was resuspended in 1 mL of the erythrocyte lysis buffer (stem cell technologies) for 10 min and washed in 20 mL of PBS with 2% P/S/A before centrifugation, 300–500 g, 5 min. The supernatant was aspirated and the cell pellet resuspended in a 3 mL stromal medium (α-MEM; Mediatech, Mannassas, VA) with 20% FBS, 1% l-glutamine (Mediatech), 1% P/S/A. Following three rinses in the stromal medium, SVF cells were plated for initial cell culture at 37 °C with 5% CO2 in ASC medium from ZenBioTM (Cat# PM-1). Subconfluent cells were passaged by trypsinization. Experiments were conducted within passages 2–3.
In vitro differentiation of adipose-derived stem cells (ASC) to adipocytes
The ASC lines were tested in culture to differentiate into mature adipocytes and show accumulation of lipid and secrete adiponectin and leptin. At the start of all experiments, cells were grown to confluency such that all cells are synchronized and then differentiated. The cells were cultured as follows. ASC were passaged with preadipocyte medium (PM-1; DMEM/Ham’s F-12 medium, HEPES, FBS, penicillin, streptomycin, amphotericin B; ZenBioTM) and then plated at 50,000 cells/cm2 with PM-1. Cells were fed every other day with PM-1 until confluent. To induce differentiation, PM-1 medium was replaced with differentiation medium (DM2; ZenBioTM) which included biotin, pantothenate, human insulin, dexamethasone, isobutylmethylxanthine and a PPARγ agonist (days 0–7). After 7 days, DM-2 medium was replaced with Adipocyte Medium (AM1; ZenBioTM; days 7–14), which included PM-1, biotin, pantothenate, human insulin and dexamethasone. By day 14, cells contained large lipid droplets and were considered mature adipocytes. Cells were maintained at 37 °C in a humidified 5% CO2 atmosphere.
Exosome isolation
The four adipose stem cells sc-ASCn, om-ASCn, sc-ASCo and om-ASCo were grown to confluency and the medium was replaced with serum-free mesenchymal stem cell basal medium (MSC-BM-CD from Lonza #00190620). Conditioned media (CM) was collected from the ASC after 48 h and centrifuged at 3,000 g for 15 min to remove dead cells. ExoQuickTM (SBI) reagent was added to the CM and incubated overnight at 4 °C. Following centrifugation at 1,500 g for 30 min, the pellet was further processed. ExoCapTM (JSR Life Sciences) composite reagent containing magnetic beads for CD9, CD63 and CD81 was used to purify exosomes. Exosomes were eluted from beads using the manufacturer’s elution buffer and used in western blot analysis or in qPCR.
Western blot analysis: Protein lysates were obtained from the ASC using lysis buffer containing protease inhibitors. The lysates (40 µg) were separated by SDS-PAGE with 10% gels, electrophoretically transferred to nitrocellulose membranes, blocked with Tris-buffered saline containing 0.1% Tween 20 and 5% nonfat dried milk, washed, and incubated with antibody specific for CD9, CD63 or CD81 (cell signaling). After incubation with anti-rabbit IgG-HRP, enhanced chemiluminescence (Pierce) was used for detection. The blots were analyzed on ProteinSimple Fluor M and Alpha viewTM software was used for densitometric analysis.
SiRNA transfection
PRKCD siRNA (ID: 103702) and scrambled siRNA were purchased from Thermo Fisher Scientific. These siRNA were previously validated for specificity and off-target gene effects were eliminated. The 50 nM of PRKCD siRNA or scrambled siRNA was transfected in ASC for 48hours using siPORT NeoFX® transfection agent. Total RNA was harvested and used as described in experiments.
Quantitative real-time qPCR
Total RNA was isolated from ASC using RNAzol according to the manufacturer’s protocol (TelTest Inc., Friendswood, TX). The 2 µg RNA was reverse-transcribed with Omniscript R kit (Qiagen) using oligo dT primers. QPCR was performed using 1.0 µL cDNA and Maxima SYBR Green/Rox qPCR master mix (Thermo Scientific). The primers for the stem cell transcription factors are: Oct4 sense: 5'-TCCCATGCATTCAAACTGAGG-3' and antisense 5'-CCAAAACCCTGGCACAAACT-3'; Sal4 sense: 5'-GCCCAGATATCCTGGAAACCA-3' and antisense 5'-TTCTCGGAGCTCTCTTGCTTTG-3'; Sox15 sense: 5'-GAACAGGTTGGAAGCAAAGGC-3' and antisense 5'-GAACAGGTTGGAAGCAAAGGC-3'; KLF4 sense: 5'-CTGCGGCAAAACCTACACAA-3' and antisense 5'-GGTCGCATTTTTGGCACTG-3'; BMI1 sense: 5'-AATGTCTTTTCCGCCCGCT-3' and antisense 5'-ACCCTCCACAAAGCACACCACAT-3'; PKCδVIII sense: 5'-TGGGTCCATTGCCCCATTAC-3' and antisense 5'-CGTAGGTCCCACTGTTGTCC-3'; PKCδI sense: 5'-ACATCCTAGGTACAACAACGGGAC-3' and antisense 5'-ACCACGTCCTTCTTCAGACAC-3'. Amplification was performed on the ViiaA 7 (Applied Biosystems). Real-time PCR was then performed in triplicate on samples and standards. The plate setup included a standard series, no template control, no RNA control, no reverse transcriptase control, and no amplification control. After primer concentrations were optimized to give the desired standard curve and a single melt curve, relative quotient (RQ) was determined using the ∆∆CT method with U1RNA as the endogenous control and sc-ASCn as the calibrator sample. The long noncoding RNAs (lncRNAs) array was purchased from SBI (catalog # RA910-A1) and includes cDNA master mix and SYBR Green master mix for real time qPCR. Experiments were repeated four times.
Flow cytometry
Immunophenotypical analysis of cultured cells was performed using the FITC-, PE-, or APC conjugated monoclonal antibodies against CD31, CD34, CD44, CD45, CD73, CD90, CD105, CD106, and CD117. Cells were detached using the EDTA buffer, washed, and resuspended at a concentration of 106 cells/mL. Cells were incubated at 4 °C for 10 min in PBS with 10% FBS and centrifuged for 5 min at 200 g. The cell pellet was resuspended in the binding buffer (PBS/2% FBS/0.01% sodium azide) followed by incubation with the specific antibodies at 4 °C for 30 min, then washed with the binding buffer, and resuspended in 0.5 mL of the same buffer and analyzed by flow cytometry (BD Accuri C6).
Senescence
ASC were grown in 60 mm plates. Cells were washed with 1X PBS and fixed with 1X fixative solution (20% formaldehyde, 2% glutaraldehyde in 10X PBS) for 15 min at room temperature. Cells were rinsed and 1 mL of β-galactosidase staining solution (400 mM citric acid/sodium phosphate, 1.5 M NaCl, 20 mM MgCl2, 500 mM potassium ferrocyanide and 20 mg/mL X-gal in DMF) was added to cells and incubated overnight in a dry incubator. The images were captured brightfield on a Nikon Eclipse fluorescent microscope using 4× or 10× objective with 0.38/µM pixel.
Statistical analysis
Analyses were performed using PRISMTM software and analyzed using two-tailed Student’s t-test. P<0.05 was significant; P<0.01 was highly significant; P<0.001 was extremely significant. Analysis was performed either within group or between groups as determined by the experiment.
Results
ASCs from subcutaneous and omental depot show differences in expression of stem cell transcription factors: ASCs from the subcutaneous and omental depot were isolated from either a lean or an obese donor (methods). To validate the pluripotency of the ASC, we evaluated the stem cell surface antigens on the ASC from subcutaneous (sc-ASCn) and omental depot (om-ASCn) from lean subjects and ASC from subcutaneous and omental depots from obese subjects (sc-ASCo and om-ASCo) (Table 1). Our results indicate that ASC from all depots were negative for CD34; CD45 was absent in the omental ASC from lean and obese donors (om-ASCn and om-ASCo) while CD105 was 10-fold higher in ASC from omental depots (om-ASCn and om-ASCo) compared to ASC derived from subcutaneous depots (sc-ASCn and sc-ASCo) of either lean or obese. Interestingly, CD106 was detected only in sc-ASCo.
Table 1. Comparison of stem cell antigens and markers.
| Maker | Other names | sc-ASCn (%) | Om-ASCn (%) | Sc-ASCo (%) | Om-ASCo (%) |
|---|---|---|---|---|---|
| CD31 | PECAM-1 | 1.10 | 0.00 | 8.30 | 0.20 |
| CD34 | L-selectin ligand | Negative | Negative | Negative | Negative |
| CD44 | Pgp-1 | 89.10 | 91.60 | 87.90 | 96.60 |
| CD45 | LCA | 0.10 | Negative | 1.80 | Negative |
| CD73 | eNT | 88.10 | 99.70 | 89.10 | 99.50 |
| CD90 | Thy-1 | 99.40 | 99.50 | 98.50 | 99.00 |
| CD105 | Endoglin | 18.00 | 98.00 | 19.00 | 99.00 |
| CD106 | VCAM-1 | Negative | Negative | 1.10 | Negative |
| CD117 | c-Kit | 0.30 | 0.20 | 0.30 | 0.20 |
Representative scatter patterns for ASC populations using flow cytometry. Table summarizes data from flow cytometry for stem cell antigens and markers. sc-ASCn, om-ASCn, sc-ASCo and om-ASCo were analyzed using flow cytometry with antibodies as indicated for stem cell antigens. Results represent five separate experiments.
We evaluated transcription factors that define the stem cell niche. Our results (Figure 1A) indicated that sc-ASCn had significantly higher expression of Oct4, Sal4, Sox15, KLF4 and BMI1 compared to om-ASCn. We next compared the stem cell markers between the ASC derived from lean and obese donors. Our results (Figure 1B) comparing the subcutaneous depots show that sc-ASCn had higher expression levels of Oct4, Sal4, Sox15, KLF4 while sc-ASCo had higher expression of BMI1. Our results (Figure 1C) comparing the omental depots show that om-ASCo had higher expression of Oct4, Sal4, Sox15, KLF4 and BMI1 compared to om-ASCn. These results indicate that obesity changes the adipose stem cell niche.
Figure 1.
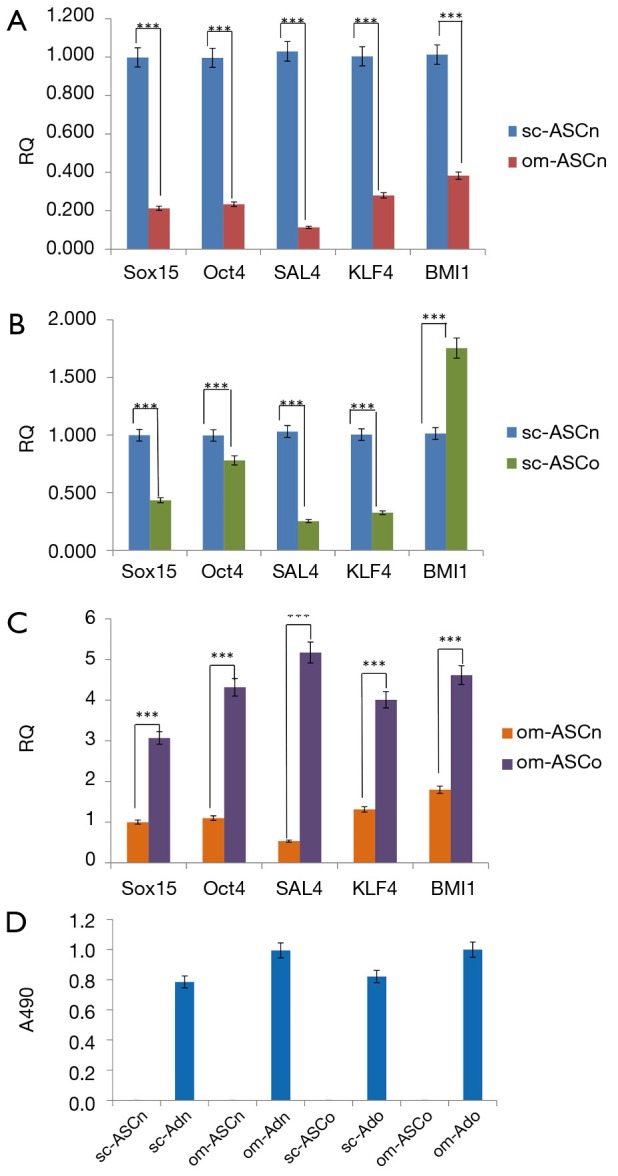
Comparison of stem cell transcription factors. Expression of stem cell transcription factors in sc-ASCn, om-ASCn, sc-ASCo and om-ASCo were determined by real time qPCR. Total RNA was collected and real time qPCR was used to measure expression of Sox15, Oct4, SAL4, KLF4, BMI1. (A-C) shows relative quantification (RQ). The experiment was independently repeated 5 times with similar results. Statistical analysis performed by 2-way analysis of variance. (D) sc-ASCn, om-ASCn, sc-ASCo and om-ASCo were differentiated in vitro to mature adipocytes. The lipid droplets were stained with Oil Red O. The dye was extracted using isopropanol and absorbance was read at 490 nM. The ASCs do not have lipid accumulation while the mature adipocytes (Ad) derived from the respective ASCs show lipid staining. The experiments were independently repeated five times with similar results. ***, P<0.001.
We have previously demonstrated the multipotency of ASC derived from lean and obese subjects in vitro (1,2). Here, we verified the stem cell potential of sc-ASCn, om-ASCn, sc-ASCo and om-ASCo by differentiating them to adipocytes in vitro. Oil Red O was added to stain the lipid droplets in mature adipocytes, followed by extraction of Oil Red O by isopropanol and quantification of absorbance at 490 nM. Our results (Figure 1D) show robust lipid droplet accumulation in sc-ASCn, om-ASCn, sc-ASCo and om-ASCo adipocytes while the ASC do not show lipid accumulation.
Adipose-derived stem cells (ASC) secrete exosomes
Factors secreted by stem cells also referred to as its secretome often influence the neighboring organs. The protein and RNA factors in the secretome are packaged into extracellular vesicles and exosomes. Exosomes are the smallest vesicles in diameter and have a high therapeutic potential as its cargo can be transferred to the recipient cells where it affects gene expression. The four adipose stem cells sc-ASCn, om-ASCn, sc-ASCo and om-ASCo were grown to confluency and medium replaced with serum-free defined medium. To analyze the secretome, CM from the ASC was collected after 48 h. The extracellular vesicles were isolated from CM using ExoQuickTM. Since ExoQuickTM also precipitates lipo-proteins and larger vesicles, ExoCapTM was then applied to the samples to obtain pure exosomes. ExoCapTM uses magnetic beads coated with exosome specific markers CD9, CD63 and CD81 to purify exosomes. We validated our exosome preparation using western blot analysis performed with antibodies against the exosome-specific markers (Figure 2).
Figure 2.
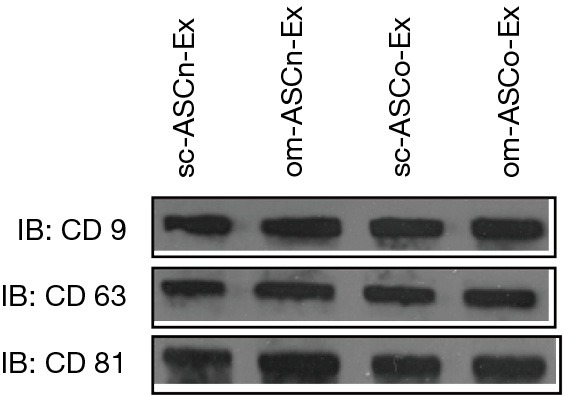
Isolation of exosomes. (A) Exosomes were isolated and purified from sc-ASCn, om-ASCn, sc-ASCo and om-ASCo as described in methods. Western blot analysis was performed using exosome surface protein specific antibodies CD9, CD63 and CD81. Experiments were repeated four times with similar results.
We evaluated the concentration and sizes of the exosomes secreted from subcutaneous and omental (sc-ASCn and om-ASCn) and compared them to subcutaneous and omental ASC derived from obese donor (sc-ASCo and om-ASCo). Using nanoparticle tracking analysis from NanoSight (NTA 3.1, Build 3.1.46), we observed that the size particle was physically homogeneous for each ASC exosome subtype with the sizes in the range of [90–100] ±5 nm as summarized in Table 2. The exosome yield per 106 ASCs per day varied between sc-ASCn, om-ASCn, sc-ASCo and om-ASCo as shown in Table 2. The omental depots secreted larger exosomes at higher concentrations compared to the subcutaneous depots. These results indicate that exosomes, which may function as intracellular messengers, are secreted differentially into the CM by sc-ASCn, om-ASCn, sc-ASCo and om-ASCo.
Table 2. Nanoparticle tracking analysis from NanoSight (NTA 3.1, Build 3.1.46).
| Items | Peak diameter (nm) | Concentration (×108 mL) |
|---|---|---|
| sc-ASCn | 89±7 | 1.10±0.08 |
| om-ASCn | 101±4 | 1.41±0.04 |
| sc-ASCo | 91±8 | 1.24±0.09 |
| om-ASCo | 104±7 | 1.49±0.02 |
The Nanoparticle tracking analysis was used to analyze peak diameter (nm) and concentration of exosomes obtained from 106 cells from sc-ASCn, sc-ASCo, om-ASCn and om-ASCo. Analysis repeated thrice with similar results.
LncRNA content of adipose-derived stem cells (ASC) exosomes
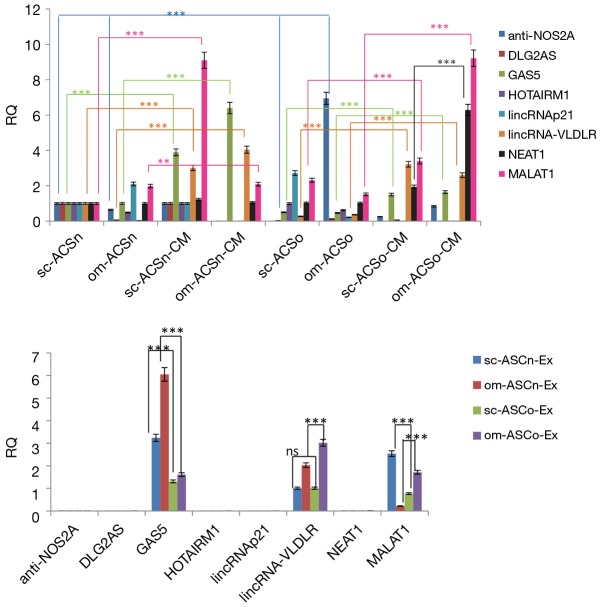
LncRNAs are important regulators of gene expression and epigenetic regulation. LncRNAs are packaged in exosomes to prevent degradation. We sought to evaluate the expression of lncRNAs within the ASC and compare it to the expression levels in its secretome. Using an lncRNA array (SBITM) we evaluated a spectrum of lncRNAs within the ASC and its secretome measured in the CM. We isolated total RNA from ASC, CM or from exosomes from sc-ASCn, om-ASCn, sc-ASCo and om-ASCo. Our analysis included only those lncRNAs which were consistently observed in the sc-ASCn, om-ASCn, sc-ASCo and om-ASCo and were statistically significant. This set included anti-NOS2a, DLG2A5, GAS5, HOTAIRM1, lincRNAp21, lincRNA-VLDL, NEAT1, MALAT1. Our results (Figure 3A) indicated that the lncRNAs expression levels varied between sc-ASCn, om-ASCn, sc-ASCo and om-ASCo. Comparing the levels of lncRNAs within the ASC, we observe that the expression of anti-NOS2a was seven-fold higher in om-ASCo.
Figure 3.
Identification of lncRNAs in cell, conditioned media and exosomes. (A) Total RNA was isolated form sc-ASCn, om-ASCn, sc-ASCo and om-ASCo and its respective conditioned media. Real time qPCR was used to measure expression of anti-NOS2A, DLG2AS, HOTAIRM1, lincRNAp21, lincRNA-VLDLR and MALAT1. GAPDH and 18S RNA were used to normalize the data. The graph shows relative quantification (RQ) with sc-ASCn sample set as reference. Statistical analysis performed by two-way analysis of variance. The experiments were independently repeated five times with similar results. (B) LncRNAs in exosomes. Exosomes were purified from conditioned media and RNA was used in real time qPCR analysis to measure expression of anti-NOS2A, DLG2AS, GAS5, HOTAIRM1, lincRNAp21, lincRNA-VLDLR and MALAT1. The graph shows RQ. The experiment was independently repeated five times with similar results. Statistical analysis performed by two-way analysis of variance between samples as indicated in graphs. **, P<0.01; ***, P<0.001.
Next, we measured the expression of lncRNAs in the secretome (measured in CM) and compared their levels to the lncRNA expression in their respective ASC. Our results show that anti-NOS2a, DLG2A5, GAS5, HOTAIRM1, lincRNAp21, lincRNA-VLDLR, NEAT1, MALAT1 are present at higher concentration in the CM. Interestingly, we detected significantly higher amounts of GAS5, lincRNA-VLDLR, NEAT1 and MALAT1 in the conditioned medium (CM) compared to the corresponding ASC. GAS5 levels were predominantly increased in CM from both subcutaneous and omental depots of lean ASC; while NEAT1 and MALAT1 were significantly higher in CM from omental depot of obese ASC.
We evaluated the expression of these lncRNAs within the exosomes. Our results (Figure 3B) indicate that GAS5, lincRNA-VLDLR and MALAT1 are enriched in the exosomes while anti-NOS2a, DLG2A5, HOTAIRM1, lincRNAp21, NEAT1 are not detected in the exosomes from CM of sc-ASCn, om-ASCn, sc-ASCo and om-ASCo. Our results further show that lincRNA-VLDLR expression is highest in exosomes isolated from CM of om-ASCo and MALAT1 expression is highest in exosomes isolated from CM of sc-ASCn.
Senescence in adipose-derived stem cells (ASC)
Adipose stem cells tightly regulate their self-renewal capacity and ability to differentiate and commitment duality. Obesity is often associated with senescence. Normal cells remain quiescent and obesity or other disease states amplify the ASC to differentiate and exit the senescence stage. We determined the senescence-associated (SA) β-galactosidase activity by visualizing it as a blue stain in cells. We evaluated the senescent state of subcutaneous and omental ASC from lean and obese donors. Our results show that senescence is decreased in both sc-ASCo and om-ASCo obese cells (Figure 4) compared to sc-ASCn and om-ASCn.
Figure 4.
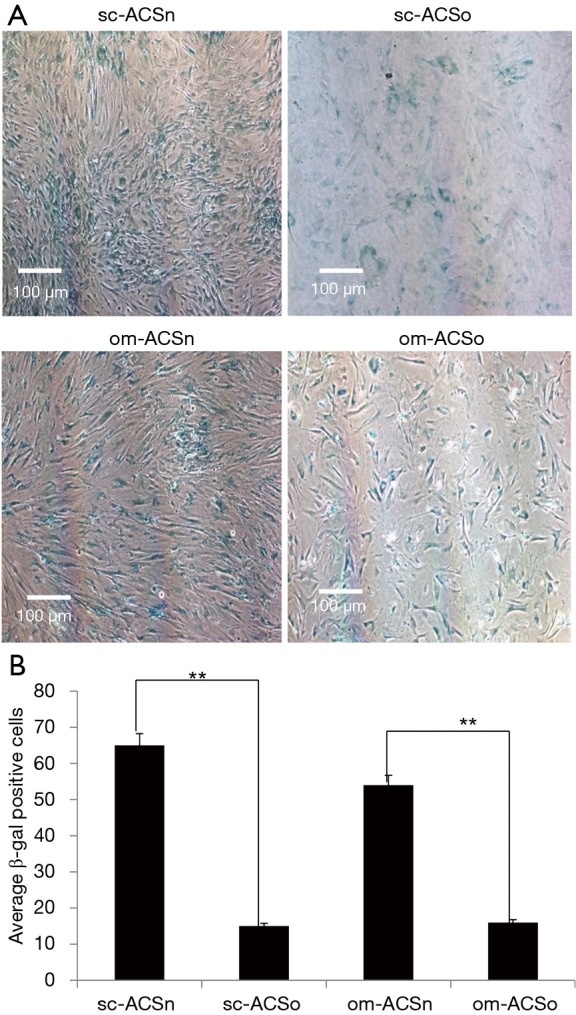
Senescence in ASC. (A) sc-ASCn, om-ASCn, sc-ASCo and om-ASCo were stained for β-galactosidase to detect senescence. Cells were then visualized under 4X light microscopy using Nikon Eclipse microscope. The percent of cells staining positive was calculated by counting five areas of the same size from each field per sample using NIS elements advance research image tool software. (B) The graphs show average of β-gal positive cells. Statistical analysis performed by two-tail Student’s t-test. **, P<0.01.
PKCδ expression in adipose-derived stem cells (ASC)
PKCδ is implicated in the regulation of transcription factors which maintain the stem cell niche (3,4). Hence, we evaluated the expression of PKCδ in sc-ASCn, om-ASCn, sc-ASCo and om-ASCo. Our results showed that both alternatively spliced products of human PKCδ: PKCδI and PKCδVIII were increased in sc-ASCo and om-ASCo derived from obese donor compared to sc-ASCn and om-ASCn (Figure 5) derived from lean donor.
Figure 5.
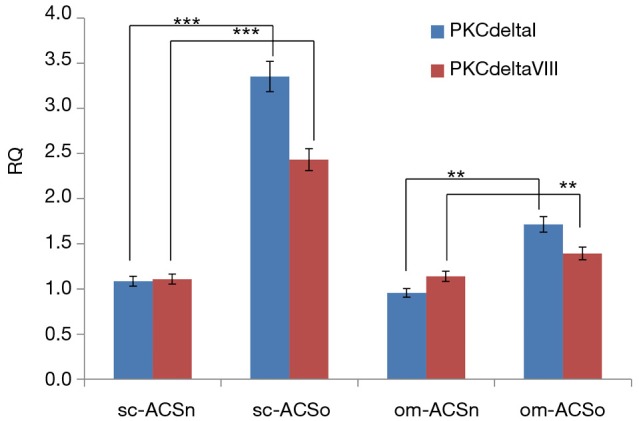
Expression of PKCδI and PKCδVIII in ASC. Total RNA was isolated form sc-ASCn, om-ASCn, sc-ASCo and om-ASCo. Real time qPCR was used to measure expression of PKCδ alternatively spliced variants PKCδI and PKCδVIII. The graph shows relative quantification (RQ). The experiment was independently repeated five times with similar results. Statistical analysis performed by two-way analysis of variance; **, P<0.01; ***, P<0.001.
Effect of PRKCD silencing on adipose-derived stem cells (ASC) markers and senescence
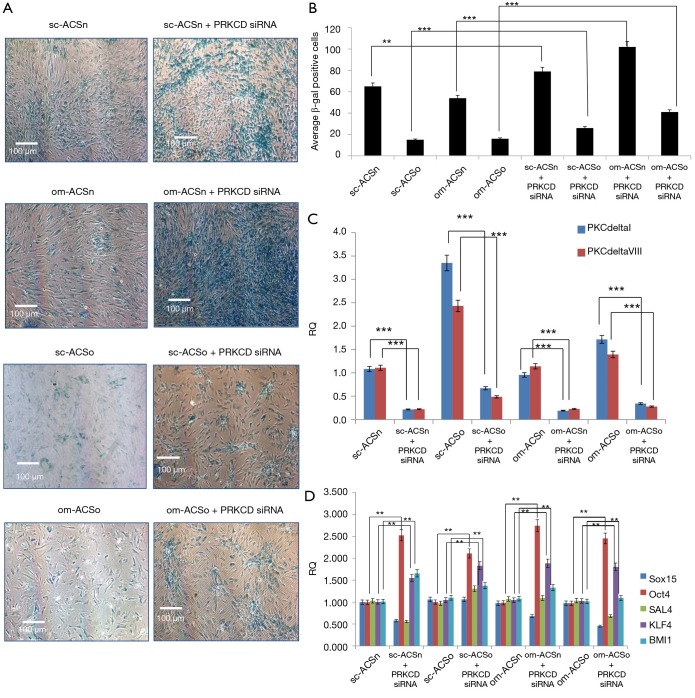
PKCδ regulates mitogenic signals to regulate a cascade of molecular events underlying senescence (8). Our results above showed that PKCδ was increased in obese ASC. To evaluate its effect on senescence in ASC, PKCδ expression was knocked down using PRKCD siRNA which silences both PKCδI and PKCδVIII. Our results indicated an increased number of senescent cells in both sc-ASCo and om-ASCo. Further, our results demonstrate that in om-ASCn transfected with PRKCD siRNA, the percent of senescent cells observed doubled compared to om-ASCn (Figure 6A-C).
Figure 6.
Senescence in ASC. (A) sc-ASCn, om-ASCn, sc-ASCo and om-ASCo were transfected with 50 nM PRKCDsiRNA for 48 h. Cells were stained for β-galactosidase to detect senescence. Cells were then visualized under 4× light microscopy using Nikon Eclipse microscope. The percent of cells staining positive was calculated by counting five areas of the same size from each field per sample using NIS elements advance research image tool software. (B) The graphs show average of β-gal positive cells. Statistical analysis performed by two-tail Student’s t-test. (C) Total RNA was isolated and real time qPCR was performed using PKCδI and PKCδVIII primers. The graph shows relative quotient (RQ). Statistical analysis performed by two-tail Student’s t-test. (D) Total RNA was isolated and real time qPCR was performed using transcription factor Oct4, Sal4, Sox15, KLF4 and BMI1 primers. The graph shows relative quotient (RQ). Statistical analysis performed by two-tail Student’s t-test. **, P<0.01; ***, P<0.001.
We further evaluated the expression of stem cell markers after PRKCD siRNA treatment. Our results (Figure 6D) show that Oct4, KLF4 and BMI1 were up-regulated with PRKCD knockdown in sc-ASCn, om-ASCn, sc-ASCo and om-ASCo. This suggested that PKCδ may regulate the expression of transcription factors which are important in maintaining the stem cell niche.
Discussion
ASCs from white adipose tissue have tremendous potential in regenerative medicine and hence we sought to characterize ASC from subcutaneous and omental adipose depots. The two depots are highly clinically relevant and have distinct functions in adipose biology. Subcutaneous adipose tissue secretes higher levels of leptin while omental fat has higher expression of lipogenic and lipolytic genes (9,10). Due to the inherent differences in the adipose tissue depot (11,12), it is important to evaluate the genetic profiles of the stem cells isolated from different adipose depots. In addition, we sought to evaluate stem cell markers in obesity as our work (2) and others (13) have shown that obesity changes the stem cell niche. Our results demonstrate that stem cell markers Oct4, Sal4, Sox15, KLF4 and BMI1 had expression patterns specific to the ASC obtained from either subcutaneous or omental depots as well as between lean or obese donors. The ASC derived from the subcutaneous depot of obese donor had lower expression of Oct4, Sal4, Sox15, KLF4 but higher expression of BMI1 compared to subcutaneous depot of the lean donor. In contrast, the ASC derived from omental depot of obese donor had higher expression of Oct4, Sal4, Sox15, KLF4 and BMI1 compared to omental depot of the lean donor. These results further attest to the finding that obesity changes the adipose stem cell niche and is dependent on the depot-specific source of the ASC.
Exosomes serve as intracellular transmitters of RNA and proteins. The therapeutic potential of the ASC also lies in its ability to secrete factors that influence other cells. We used a combination of ExoQuickTM and ExoCapTM to purify the exosomes. This enabled us to distinguish between the lncRNAs in the secretome which may be present in larger vesicles and those which are present in the smaller exosomes (sizes vary between 30–150 nm depending on the source). Ultracentrifugation may also be used to isolate exosomes (14); however we chose to utilize the ExoCapTM method for higher yield of pure exosomes for our studies. Other studies have also verified that the reagent method provides higher yields and purity compared to the ultracentrifugation method (15). Here, we report that the size and concentration of exosomes secreted from omental depots of lean or obese ASC are higher compared to those secreted from subcutaneous depots of lean or obese ASC. Importantly, we show that their lncRNA content varies significantly between ASC, its secretome and the cargo contained within the exosomes.
Our results indicated that lincRNA-VLDLR, GAS5 and MALAT1 are enriched in the exosomes and this may suggest that they function as intra-cellular messengers. The very low expression of anti-Nos2a, DLG2A5, HOTAIRM1, NEAT1 and lincRNAp21 in our exosome preparations may suggest that these lncRNAs are packaged in larger secretory vesicles. Our results indicate that exosomes from sc-ASCn, the predominately used ASC source in regenerative medicine, is enriched in MALAT1. Interestingly, NEAT1 expression is significantly elevated in om-ASCo-CM compared to om-ASCo. Our results suggest that extracellular vesicles from the secretome of obese ASC could also be used in therapeutic applications or to decipher disease state mechanisms. Karastergiou et al. (16) showed expression of homeobox genes from abdomen or gluteal depot in the adipose tissue as well as in the stromal cells. The differential expression of these genes contributed to the distinct phenotypic characteristics of peripheral fat. In this study, our report is limited to evaluation of lncRNAs observed consistently in sc-ASCn, om-ASCn, sc-ASCo and om-ASCo. Additional analysis using RNAseq will provide an exhaustive list of all RNA including miRNAs, mRNAs, lncRNAs and other non-coding RNAs present in the exosomes derived from sc-ASCn, om-ASCn, sc-ASCo and om-ASCo.
In obesity, the omental fat is infiltrated by immune-inflammatory cells and is implicated in metabolic syndrome to a higher extent compared to subcutaneous fat. Cellular senescence evolved as a means to prevent hyperproliferation of dysfunctional cells. We previously showed that the stem cell niche is altered in obesity and expression of PKCδ, a kinase mediating cellular proliferation, differentiation and apoptosis, is higher in obese adipocytes compared to lean adipocytes (1). Senescence in mature adipocytes and adipose tissue has been studied earlier with reports indicating that adipose tissue upon aging accumulates senescent cells in WAT and causes age-associated morbidity (17-20). Here, we sought to evaluate senescence in ASC and compare the number of senescent cells in ASC from lean and obese. Our results demonstrated that lean ASC from subcutaneous and omental depots have higher senescent cells compared to obese ASC depots. This result suggests that the obese cells have undergone an epigenetic modification to promote hyperproliferation. It is reported that SA-RNA-1 lncRNA is associated with senescence and levels of lncRNAs change with senescence (21). Obesity is characterized by hyperplasia and hypertrophy which may be influenced by the altered stem cells niche. PKCδ modulates cellular senescence by mediating a cascade of mitogenic signals (8,22). Our experiments with PKCδ silencing in the ASC showed an increase in cellular senescence in obese ASC in both subcutaneous and omental depots. These results suggest that PKCδ regulates cell cycle arrest in senescent cells and modulating its expression may influence the stem cell niche in adipose tissue and obesity. We previously showed that PKCδ regulates cell cycle in differentiating mouse pre-adipocyte 3T3L1 cell line (23).
In conclusion, we have characterized the stem cell markers in ASCs isolated from different depots from a lean and an obese donor. We evaluated the secretome and exosomes secreted by ASC and show that specific lncRNAs are enriched in the exosomes which may be transferred to recipient cells to regulate gene expression. Importantly, we show that the exosome content varies between depots and between lean and obese phenotype. Finally, we demonstrate that levels of senescent cells are higher in sc-ASCn and om-ASCn. PKCδ modulates senescence in ASC and it may be through the phosphorylation of transcription factors Oct4, KLF4 or BMI1. Additional molecular manipulations are needed to decipher the mechanisms underlying phosphorylation by PKCδ in ASC. These results have significant implications in understanding the adipose stem cell niche in the field of obesity. In addition, our results shed light on understanding the impact of obesity on diseases such as cancer and endocrine disorders via their exosome content which serve as intra-cellular messengers.
Acknowledgements
This work was supported by the Department of Veterans Affairs Medical Research grant.
Disclaimer: The contents do not represent the views of the Department of Veterans Affairs or the United States Government.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Carter G, Apostolatos A, Patel R, et al. Dysregulated Alternative Splicing Pattern of PKCδ during Differentiation of Human Preadipocytes Represents Distinct Differences between Lean and Obese Adipocytes. ISRN Obes 2013;2013:161345. [DOI] [PMC free article] [PubMed]
- 2.Watson JE, Patel NA, Carter G, et al. Comparison of Markers and Functional Attributes of Human Adipose-Derived Stem Cells and Dedifferentiated Adipocyte Cells from Subcutaneous Fat of an Obese Diabetic Donor. Adv Wound Care (New Rochelle) 2014;3:219-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hamdorf M, Berger A, Schüle S, et al. PKCδ-induced PU.1 phosphorylation promotes hematopoietic stem cell differentiation to dendritic cells. Stem Cells 2011;29:297-306. [DOI] [PubMed] [Google Scholar]
- 4.Lee HJ, Jeong CH, Cha JH, et al. PKC-delta inhibitors sustain self-renewal of mouse embryonic stem cells under hypoxia in vitro. Exp Mol Med 2010;42:294-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Patel NA, Song SS, Cooper DR. PKCdelta alternatively spliced isoforms modulate cellular apoptosis in retinoic acid-induced differentiation of human NT2 cells and mouse embryonic stem cells. Gene Expr 2006;13:73-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang K, Apostolatos AH, Ghansah T, et al. Identification of a novel antiapoptotic human protein kinase C delta isoform, PKCdeltaVIII in NT2 cells. Biochemistry 2008;47:787-97. [DOI] [PubMed] [Google Scholar]
- 7.Carter G, Patel R, Apostolatos A, et al. Protein kinase C delta (PKCδ) splice variant modulates senescence via hTERT in adipose-derived stem cells. Stem Cell Investig 2014;1:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Katakura Y, Udono M, Katsuki K, et al. Protein kinase C delta plays a key role in cellular senescence programs of human normal diploid cells. J Biochem 2009;146:87-93. [DOI] [PubMed] [Google Scholar]
- 9.Bortolotto JW, Reis C, Ferreira A, et al. Higher content of trans fatty acids in abdominal visceral fat of morbidly obese individuals undergoing bariatric surgery compared to non-obese subjects. Obes Surg 2005;15:1265-70. [DOI] [PubMed] [Google Scholar]
- 10.Caserta F, Tchkonia T, Civelek VN, et al. Fat depot origin affects fatty acid handling in cultured rat and human preadipocytes. Am J Physiol Endocrinol Metab 2001;280:E238-47. [DOI] [PubMed] [Google Scholar]
- 11.Baglioni S, Cantini G, Poli G, et al. Functional differences in visceral and subcutaneous fat pads originate from differences in the adipose stem cell. PLoS One 2012;7:e36569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dusserre E, Moulin P, Vidal H. Differences in mRNA expression of the proteins secreted by the adipocytes in human subcutaneous and visceral adipose tissues. Biochim Biophys Acta 2000;1500:88-96. [DOI] [PubMed] [Google Scholar]
- 13.Oñate B, Vilahur G, Camino-López S, et al. Stem cells isolated from adipose tissue of obese patients show changes in their transcriptomic profile that indicate loss in stemcellness and increased commitment to an adipocyte-like phenotype. BMC Genomics 2013;14:625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zubiri I, Vivanco F, Alvarez-Llamas G. Proteomic analysis of urinary exosomes in cardiovascular and associated kidney diseases by two-dimensional electrophoresis and LC-MS/MS. Methods Mol Biol 2013;1000:209-20. [DOI] [PubMed] [Google Scholar]
- 15.Schageman J, Zeringer E, Li M, et al. The complete exosome workflow solution: from isolation to characterization of RNA cargo. Biomed Res Int 2013;2013:253957. [DOI] [PMC free article] [PubMed]
- 16.Karastergiou K, Fried SK, Xie H, et al. Distinct developmental signatures of human abdominal and gluteal subcutaneous adipose tissue depots. J Clin Endocrinol Metab 2013;98:362-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shen X, Du Y, Shen W, et al. Adipose-derived stem cells promote human dermal fibroblast function and increase senescence-associated β-galactosidase mRNA expression through paracrine effects. Mol Med Rep 2014;10:3068-72. [DOI] [PubMed] [Google Scholar]
- 18.Mantovani C, Terenghi G, Magnaghi V. Senescence in adipose-derived stem cells and its implications in nerve regeneration. Neural Regen Res 2014;9:10-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jun HS, Dao LT, Pyun JC, et al. Effect of cell senescence on the impedance measurement of adipose tissue-derived stem cells. Enzyme Microb Technol 2013;53:302-6. [DOI] [PubMed] [Google Scholar]
- 20.Markowski DN, Thies HW, Gottlieb A, et al. HMGA2 expression in white adipose tissue linking cellular senescence with diabetes. Genes Nutr 2013;8:449-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abdelmohsen K, Panda A, Kang MJ, et al. Senescence-associated lncRNAs: senescence-associated long noncoding RNAs. Aging Cell 2013;12:890-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Byun HO, Jung HJ, Kim MJ, et al. PKCδ phosphorylation is an upstream event of GSK3 inactivation-mediated ROS generation in TGF-β1-induced senescence. Free Radic Res 2014;48:1100-8. [DOI] [PubMed] [Google Scholar]
- 23.Patel RS, Carter G, Cooper DR, et al. Transformer 2β homolog (Drosophila) (TRA2B) regulates protein kinase C δI (PKCδI) splice variant expression during 3T3L1 preadipocyte cell cycle. J Biol Chem 2014;289:31662-72. [DOI] [PMC free article] [PubMed] [Google Scholar]