Abstract
A transcription map was developed for the virulent Streptococcus thermophilus phage Sfi19 on the basis of systematic Northern blot hybridizations. All deduced 5′ ends were confirmed by primer extension experiments. Three classes of transcripts were detected based on the different times of appearance. Early transcripts were identified in three genome regions; middle transcripts covered cro-like, DNA replication, and transcriptional regulation genes; and late genes consisted of structural and lysis genes. Chloramphenicol treatment suppressed the translation of a putative transcriptional factor necessary for the production of late transcripts and shifted middle transcripts to early transcription times.
Data from phages of lactic acid bacteria have challenged some concepts developed with phages from Escherichia coli (3, 4, 5). One is the distinction of two fundamental classes of bacteriophages. Phages from Streptococcus thermophilus, a major starter bacterium used in the dairy industry, do not fit into that scheme: virulent and temperate phages are closely related at the genome level (4, 10). During serial passage, the temperate cos site phage Sfi21 spontaneously gave rise to virulent phages by a site-specific deletion process (6). Upon dual serial infections in the lytic mode, the virulent derivative phage outcompeted the parental temperate phage. The vast majority of the dairy streptococcal phages are in fact virulent phages, and only a few S. thermophilus strains are lysogenic (2). We also identified virulent phages, e.g., Sfi19, that could superinfect an Sfi21 lysogen, suggesting that phage-phage competition is a major selective force shaping the S. thermophilus phage genomes. To shed some light on the genetic relationship between temperate and virulent streptococcal phages, we sequenced the genomes from phages Sfi21 and Sfi19 (11). Except for point mutations (concentrated over DNA packaging, head, and tail genes), the two phages differed mainly in two regions. One was, as expected, the lysogeny module. The corresponding Sfi19 region could theoretically be derived from the lysogeny module of the temperate S. thermophilus pac-site phages O1205 by deletion/insertion and DNA rearrangement events (10). Sfi19, like many other virulent S. thermophilus phages, maintained an O1205-like cro repressor gene. Another area of difference between the DNA replication module and the cos site was identified (10). Over this region, transcription mapping in Sfi21-infected cells identified early genes possibly involved in transcriptional regulation (11). Conspicuous differences were detected in this region between Sfi21 and Sfi19. As in a number of other virulent streptococcal phages, an anonymous open reading frame (ORF) followed by a second origin of DNA replication replaced two Sfi21-specific genes in Sfi19 (10, 11). In addition, a candidate for early transcriptional regulator protein from Sfi21 showed a distinct DNA-binding domain in Sfi19. Since phages Sfi21 and Sfi19 differed in all three repressor/transcriptional regulator genes, we suspected a distinct transcription pattern in both phages.
Transcription mapping in phage Sfi19.
Total RNA was isolated from the Sfi19-infected indicator cell Sfi1 at 0 and 2 min postinfection (p.i.) and then every 5 min until cell lysis occurred (about 32 min p.i.). RNA extraction, probe preparation, Northern hybridization, and primer extension analysis were done as described previously (13, 15). In order to localize the transcripts on the genome map, we performed Northern blot hybridization using 17 different DNA probes (Fig. 1, probes a to m). The precise localization of the phage mRNAs on the phage genome map is limited by the number of DNA probes used in Northern blots and their spatial resolution. The probes were selected on the basis of a previous study (15). We focused our attention on gene expression of genome regions of phage Sfi19 that depicted sequence variability compared to those of phage Sfi21. Consequently, PCR-generated probes covering these genome regions were used in this study. In order to increase the reliability of our transcription map for the different times p.i., many probes spanning the same region were employed. Selected 5′ positions of the mRNA were experimentally determined by primer extension experiments (Table 1). Rho-independent terminators were predicted by in silico analysis. A summary of the results is presented in the transcription map shown in Fig. 1.
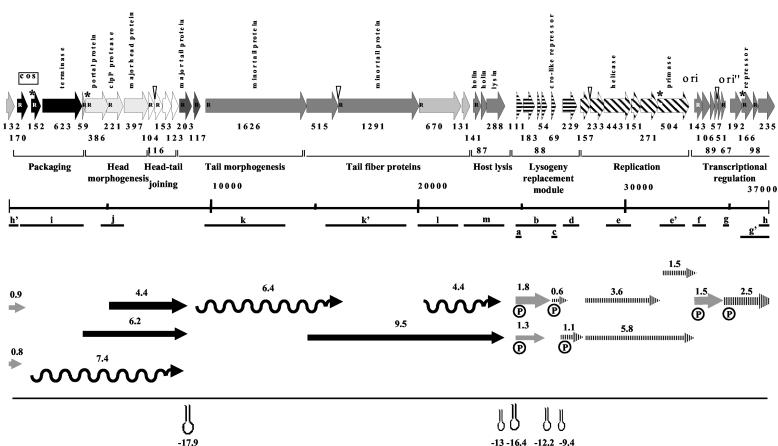
FIG. 1.
Genome and transcription map for the virulent S. thermophilus phage Sfi19. (Top) Gene map of phage Sfi19. ORFs are marked with their codon lengths. Putative gene functions and phage modules were identified by comparative genomics (10). Genes predicted to belong to the same module share same shading or pattern (arrows labeled “transcriptional regulation” show a lack of information). ORFs preceded by a potential RBS are marked with an R inside the arrow; ORFs with an asterisk have an unconventional initiation codon; overlap of a start and stop codon is indicated with a triangle. (Middle) The PCR products used for probing of the Northern blots are located on the gene map (a to m; scale in base pairs). (Bottom) The transcripts are located on the Sfi19 gene map; the arrows point to the 3′ end of the mRNA. The arrows indicate early (gray), middle (hatched), and late (black) transcripts. The length of the arrow is proportional to the length of the deduced mRNAs; the size of the mRNA is in kilobases. The width of the arrows indicates the relative abundance of the mRNA species. The wavy lines indicate mRNAs that did not yield defined bands. The 5′ ends determined in primer extension experiments are marked by a circle with a P. Hairpins indicate predicted rho-independent terminators, with their stability given in kilocalories per mole.
TABLE 1.
Primer sequences used in primer extension experiments
| Oligo- nucleotide | Position (bp)a | Sequence (5′→3′) |
|---|---|---|
| 111 | 25047-25074 | CCTTTGCATCAGACCAAGCCATTTTC |
| 69 | 26891-26919 | GTTCCTTTCTCATCTTTAGCTAAAATCC |
| 229 | 27776-27803 | CAGTTCTCCTTCTTCGACATTG |
| 143 | 33629-23657 | GCGTTGCATTGCACATCTATAAC |
| 67 | 34908-34929 | CGTGAGGACTTATTCAGACAG |
| 192 | 35411-35429 | CCTAAACCTGTAGTGCCAC |
| 166 | 36011-36036 | GCTGAATACTTCAACGTATCAGTTCC |
Positions refer to the Sfi19 sequence reported in AF115102.
Lysogeny replacement module.
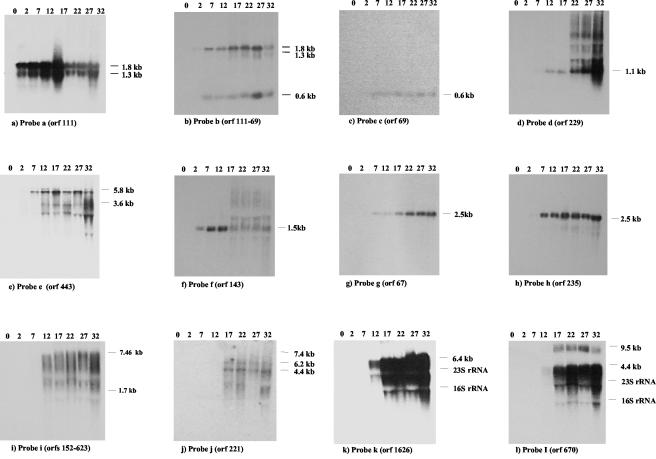
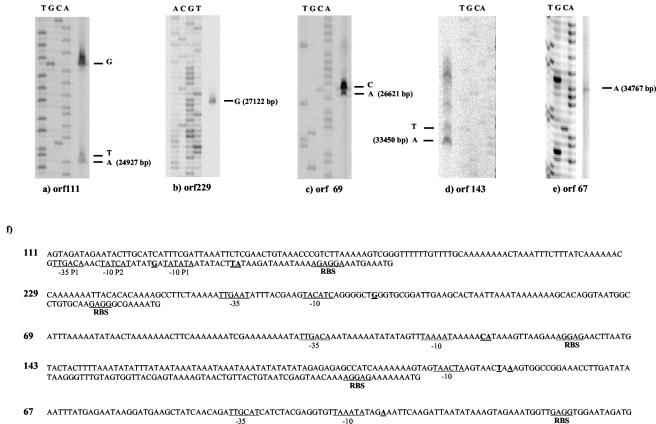
An ORF 111-specific probe revealed a major 1.8-kb and a less abundant 1.3-kb mRNA (Fig. 2a). ORF 111 shares sequence identity with a gene from the temperate S. thermophilus phage O1205 located in the lysogeny module between the phage lysin and integrase genes (14). It is one of the few prophage genes transcribed in the lysogenic state (16). ORF 111 transcription was already detectable at 2 min p.i. (Fig. 2a). Multiple promoter structures were found preceding ORF 111. In fact, two transcription start sites were identified at −25 bp (putative promoter P1) and at −42 bp (putative promoter P2) relative to the start site of the coding sequence (Fig. 3a). Putative promoter P1 had conventional −10 and −35 promoter regions and an extended −10 site (TG motif), while putative promoter P2 had a canonical −10 box but no consensus −35 sequences (Fig. 3f). On the basis of the mRNA size and the location of the 5′ ends, the longer transcript covers the left part of the lysogeny replacement module in phage Sfi19 (ORF 111 to ORF 54) (Fig. 1). In support of this diagnosis, a rho-independent terminator was placed between ORF 54 and ORF 69.
FIG. 2.
Northern blot analysis. Northern blots of total RNA isolated from Sfi19-infected cells at the indicated time after infection in minutes (top of the Northern blots) were hybridized with the indicated probes. The estimated sizes of the transcripts are given.
FIG. 3.
5′ ends of the mRNAs. (a through e) Primer extension experiments for the indicated ORFs. (f) Upstream sequences of selected ORFs. RBSs, −10 and −35 promoter sites, and the experimental 5′ ends of the mRNAs (bold and underlined) are indicated. Base pair numbers refer to entry AF115102.
When the Northern blot was tested with a probe covering ORF 111 to ORF 69 (ORF 69 is a predicted cro-like repressor gene), an additional signal of 0.6 kb was detected by autoradiography (Fig. 2b). The signal was detected from 7 min p.i. until the end of the infection cycle. When Northern blots were probed with ORF 69, a single 0.6-kb hybridization signal was detected by autoradiography (Fig. 2c). Primer extension experiments located the 5′ end 24 bp upstream of the start codon of ORF 69 (Fig. 3b). ORF 69 showed a conventional promoter and a good ribosome binding site (RBS) (Fig. 3f). A rho-independent terminator was found in the intergenic region between ORF 69 and ORF 229.
An ORF 229-specific probe detected a 1.1-kb mRNA signal at 12 min p.i., which increased substantially in intensity at later infection times (Fig. 2d). The protein encoded by ORF 229 lacks database matches, except for an anonymous Haemophilus protein, and consists of many small loosely conserved peptide repeats. Primer extension experiments located the 5′ end 65 bp upstream of the start codon of ORF 229 (Fig. 3c). The ORF 229 start codon is preceded by a good RBS and a perfect −10 consensus promoter site, but the ORF 229 −35 sequence differed from the consensus sequence (Fig. 3f).
From DNA replication module to the cos site.
Probes individually covering ORF 157, ORF 233 (nucleoside triphosphate binding protein with a Walker A motif), and ORF 443 (helicase) revealed mRNAs of 5.8 and 3.6 kb (Fig. 2e and data not shown). The signals became visible on a background of cross-hybridization with rRNA at 7 min p.i. As in the case of phage Sfi21, a probe covering ORF 504 (primase) hybridized with the 5.8- and a 1.5-kb transcript (data not shown).
An ORF 143-specific probe revealed an mRNA species of approximately 1.5 kb. ORF 143 lacks database matches. This mRNA was clearly detected at 2 min p.i., reached its highest intensity at 12 min p.i., and was undetectable at the later infection stages (Fig. 2f). Primer extension identified a series of putative transcription start sites upstream of ORF 143 (Fig. 3d).
ORF 67- and ORF 235-specific probes both revealed an mRNA species of approximately 2.5 kb which was detected at 7 min p.i., reached maximal transcription at 17 min p.i., and was maintained at that level until the end of the infection cycle (Fig. 2g and h). Primers placed ahead of ORF 192 and ORF 166 (transcriptional regulator of the CI/Cro family) gave no extension products, while ORF 67 yielded a weak extension, which placed the 5′ end 43 bp ahead of ORF 67 (Fig. 3e and 3f). ORF 67 resembled closely Streptococcus pyogenes, Listeria, and Lactococcus prophage genes located at comparable genome map positions; ORF 235-like proteins were not observed outside S. thermophilus.
Two transcripts of 0.8 and 0.9 kb were identified when we used a probe corresponding to ORF 132, which encodes a hypothetical protein (Fig. 4a). Both transcripts were visible within the first 2 min of infection. The 0.9-kb mRNA gradually decreased in abundance at later time points and was replaced by the 0.8-kb transcript at 17 p.i. ORF 132-like genes are also known from S. pyogenes prophages.
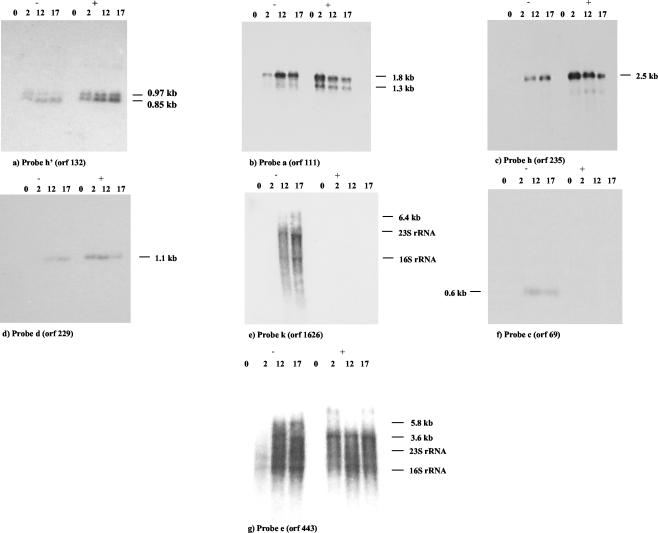
FIG. 4.
Effect of chloramphenicol on phage transcription (10 μg of chloramphenicol/ml). Northern blots of RNA isolated from Sfi19-infected cells at the indicated times p.i. (in minutes) in the absence (−) or presence (+) of chloramphenicol are shown. The blots were probed with early gene ORF 132 (a), early gene ORF 111 (b), middle gene ORF 235 (c), middle gene ORF 229 (d), late gene 1626 (e), middle gene ORF 69 (f), and middle gene ORF 443 (g).
From the cos site to the structural genes.
Northern blots probed with DNA covering the DNA packaging genes ORF 170, ORF 152, and ORF 623 revealed hybridization signals from 12 min p.i. onwards, reaching a maximum level 32 min after infection. The peak distribution of the hybridization smear was between 7.4 and 1.7 kb (Fig. 2i). Northern hybridization using a probe corresponding to ORF 221, a ClpP protease gene from the head morphogenesis module, showed two major transcripts of 6.2 and 4.4 kb from 17 min p.i. onwards (Fig. 2j). A rho-independent terminator was identified downstream of ORF 203, encoding the major tail gene.
Probes which independently covered the tail morphogenesis and host lysis genes yielded intensive hybridization signals consisting of multiple bands. The signals were first seen at 12 min p.i. and increased in intensity until the end of the infection cycle. The largest transcript was 6.4 kb when an ORF 1626-specific probe (tail tape measure gene) was used (Fig. 2k). When individual probes corresponding to ORF 1291 (antireceptor gene), ORF 670, ORF 131, and ORF 288 (lysin) were used (Fig. 2l and data not shown), a 9.5-kb mRNA and a trail of smearing starting at 4.4 kb towards lower molecular weights was seen. These transcripts became visible at 17 min after infection and were maintained at high level of expression until the end of the infection cycle. Two rho-independent terminators were identified downstream from ORF 288.
A phage protein(s) promotes the transcription of late-expressed genes.
As already described for phage Sfi21 (15), chloramphenicol (CA) treatment at 10 μg/ml prevents synthesis of transcriptional regulator and consequently suppressed the transcription of late phage genes in Sfi19-infected cells (Fig. 4e) but did not inhibit the transcription of early transcripts (Fig. 4a and b). In fact, CA increased the intensity of the hybridization signals markedly at the earliest time point. This stimulatory effect was even stronger on middle transcripts (Fig. 4c, d, and g). Under CA treatment the transcript revealed by probe ORF 235 appeared at 2 min p.i. with strongest intensity, while in untreated cells this transcript was detected only at 7 min p.i. (Fig. 4c). Likewise, transcription of the 3.6-kb mRNA from the DNA replication module was not suppressed by CA treatment as in Sfi21-infected cells but was already detectable at 2 min p.i. (Fig. 4g). CA affected the adjacent genes ORF 69 and ORF 229 differently. The 0.6-kb cro repressor transcript was no longer detected (Fig. 4f), while the ORF 229-specific transcript increased in intensity and became detectable at 2 min p.i. (Fig. 4d).
Conclusion.
Three different classes of mRNAs, early, middle, and late, were detected on the basis of the time dependence of appearance, but transcript initiation kinetics may be obscured by differences in mRNA stability. In fact, translation inhibitors have been described to affect mRNA stability (9). Overall the transcription map of the virulent phage Sfi19 resembles that of the temperate phage Sfi21 in the lytic infection mode. For example, the left part of the lysogeny replacement module in Sfi19 (ORF 111 to ORF 54) gave rise to early transcripts. These Sfi19 genes are partially derived from the lysogeny module of temperate S. thermophilus phages (10). Notably, the corresponding lysogeny genes in phage Sfi21 are also transcribed as early genes in the lytic infection mode of phage Sfi21 (15). These genes thus seem to play an important role in the early lytic infection cycle. As was the case for the cro-like repressor gene from Sfi21, the distinct cro-like repressor gene from phage Sfi19 is transcribed as a middle gene and needs protein synthesis for transcription, excluding a crucial early regulatory role for this gene in Sfi19-infected cells. A major difference between phage Sfi21 and Sfi19 transcription was found for the genome segment between the origin of replication and the cos site, a region tentatively described as a transcriptional regulation module in Sfi21 (15). Instead of a series of 2.6-, 3.7-, and 4.8-kb early transcripts in Sfi21 initiating at the ORF 143 promoter, Sfi19 showed a single early transcript of about 1.5 kb initiating at the ORF 143 promoter and a single 2.5-kb middle transcript probably originating at ORF 67. Interestingly, ORF 67 marks the start of the second region of major genomic differences between Sfi21 and Sfi19 that contains a transcriptional regulator with a distinct DNA binding domain. A further major difference between Sfi21 and Sfi19 transcription concerns regulation. Three of the four kinetically defined middle Sfi19 transcripts do not depend on protein synthesis for their transcription, as assessed by CA inhibition experiments. This is in contrast to middle transcripts in Sfi21, which need a positive regulator for transcription. In fact, in the presence of a protein synthesis inhibitor, the Sfi19 middle transcripts became early transcripts, as if they had been relieved from negative control by an early phage protein. The genetic switch region in S. thermophilus phages has been investigated (7), but its role in transcriptional regulation during the lytic mode of infection seems to be limited. The region between the DNA replication module and the cos site seems to fulfill the regulatory role in transcription during lytic phage growth, but data on activators and repressors in this region in dairy phages are still limited (1, 12, 17, 18), and this topic warrants further investigation. Some differences in transcriptional patterns observed between Sfi19 and a similar temperate phage (Sfi21) are attributed to different regulators being produced by these phages. This hypothesis concurred with the observation deduced by comparative genomics (10, 11). Phages are certainly useful tools for the molecular dissection of the transcriptional regulator mechanisms. Transcription and promoter mapping analysis provides valuable information for generating hypotheses and for directing genetic and/or biochemical approaches, which are needed to achieve a significant increase in our understanding of such mechanisms and provide a measure of their diversity. Future studies will deal with the experimental testing of the interaction of the putative regulators with putative promoter regions identified in this study by performing DNA binding assay experiments.
Acknowledgments
We thank the Swiss National Science Foundation for the financial support of M.V. (research grant 5002-057832).
We also thank Douwe van Sinderen and Jim O'Mahony (National University of Ireland, Cork) for constructive and critical reading of the manuscript.
REFERENCES
- 1.Brøondsted, L., M. Pedersen, and K. Hammer. 2001. An activator of transcription regulates phage TP901-1 late gene expression. Appl. Environ. Microbiol. 67:5626-5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brüssow, H., M. Fremont, A. Bruttin, J. Sidoti, A. Constable, and V. Fryder. 1994. Detection and classification of Streptococcus thermophilus bacteriophages isolated from industrial milk fermentation. Appl. Environ. Microbiol. 60:4537-4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brüssow, H. 2001. Phages of dairy bacteria. Annu. Rev. Microbiol. 55:283-303. [DOI] [PubMed] [Google Scholar]
- 4.Brüssow, H., and F. Desiere. 2001. Comparative phage genomics and the evolution of Siphoviridae: insights from dairy phages. Mol. Microbiol. 39:213-222. [DOI] [PubMed] [Google Scholar]
- 5.Brüssow, H., and R. W. Hendrix. 2002. Phage genomics: small is beautiful. Cell 108:13-16. [DOI] [PubMed] [Google Scholar]
- 6.Bruttin, A., and H. Brüssow. 1996. Site-specific spontaneous deletions in three genome regions of a temperate Streptococcus thermophilus phage. Virology 219:96-104. [DOI] [PubMed] [Google Scholar]
- 7.Bruttin, A., S. Foley, and H. Brüssow. 2002. DNA-binding activity of Streptococcus thermophilus phage Sfi21 repressor. Virology 303:100-109. [DOI] [PubMed] [Google Scholar]
- 8.Canchaya, C., C. Proux, G. Fournous, A. Bruttin, and H. Brüssow. 2003. Prophage genomics. Microbiol. Mol. Biol. Rev. 67:238-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lopez, P. J., I. Marchand, O. Yarchuk, and M. Dreyfus. 1998. Translation inhibitors stabilize Escherichia coli mRNAs independently of ribosome protection. Proc. Natl. Acad. Sci. USA 95:6067-6072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lucchini, S., F. Desiere, and H. Brüssow. 1999. Comparative genomics of Streptococcus thermophilus phage species support a modular evolution model. J. Virol. 73:8647-8656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lucchini, S., F. Desiere, and H. Brüssow. 1999. The genetic relationship between virulent and temperate Streptococcus thermophilus bacteriophages: whole genome comparison of cos-site phages Sfi19 and Sfi21. Virology 260:232-243. [DOI] [PubMed] [Google Scholar]
- 12.Parreira, R., R. Valyasevi, A. L. S. Lerayer, S. D. Ehrilch, and M. C. Chopin. 1996. Gene organization and transcription of a late expressed region of a Lactococcus lactis phage. J. Bacteriol. 178:6158-6165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 14.Stanley, E., G. F. Fitzgerald, C. Le Marrec, B. Fayard, and D. van Sinderen. 1997. Sequence analysis and characterization of O1205, a temperate bacteriophage infecting Streptococcus thermophilus CNRZ 1205. Microbiology 143:3417-3429. [DOI] [PubMed] [Google Scholar]
- 15.Ventura, M., S. Foley, A. Bruttin, S. Chibani Chennoufi, C. Canchaya, and H. Brüssow. 2002. Transcription mapping as a tool in phage genomics: the case of the temperate Streptococcus thermophilus phage Sfi21. Virology 296:62-76. [DOI] [PubMed] [Google Scholar]
- 16.Ventura, M., A. Bruttin, C. Canchaya, and H. Brüssow. 2002. Transcription analysis of Streptococcus thermophilus phages in the lysogenic state. Virology 302:21-32. [DOI] [PubMed] [Google Scholar]
- 17.Walker, S. A., C. S. Dombroski, and T. R. Klaenhammer. 1998. Common elements regulating gene expression in temperate and lytic bacteriophages of Lactococcus species. Appl. Environ. Microbiol. 64:1147-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walker, S. A., and T. R. Klaenhammer. 1998. Molecular characterization of a phage-inducible middle promoter and its transcriptional activator from the lactococcal bacteriophage φ31. Appl. Environ. Microbiol. 180:921-931. [DOI] [PMC free article] [PubMed] [Google Scholar]