Abstract
Bacillus thuringiensis vegetative cells are known to be highly pathogenic when injected into the hemocoel of susceptible insect larvae. This pathogenicity is due to the capacity of B. thuringiensis to cause septicemia in the host. We screened a B. thuringiensis mini-Tn10 insertion library for loss of virulence against Bombyx mori larvae on injection into the hemocoel. Three clones with attenuated virulence were isolated, corresponding to two different mini-Tn10 insertions mapping to the yqgB/yqfZ locus. Single disruptions of the yqgB and yqfZ genes did not affect virulence against B. mori. In contrast, the inactivation of both genes simultaneously reproduced the effect of the mini-Tn10 insertion and resulted in a significant delay to infection. The double ΔyqgB ΔyqfZ mutant was also nonmotile, and its growth was affected at 25°C. We analyzed lacZ transcriptional fusions and detected promoter activity upstream from yqgB at 25 and 37°C. Overall, our findings suggest that the yqgB and yqfZ genes encode adaptive factors that may act in synergy, enabling the bacteria to cope with the physical environment in vivo, facilitating colonization of the host.
The entomopathogenic properties of Bacillus thuringiensis, a gram-positive spore-forming bacterium of the Bacillus cereus group, result partly from the production of insecticidal crystalline inclusions. The crystal inclusion consists of toxins known as δ-endotoxins or Cry proteins, which are synthesized during the stationary phase. Upon ingestion by susceptible insects, crystal toxins bind to midgut epithelial cell-specific receptors, disrupting the integrity of the gut and providing a means of entry into the host (27). This specific toxemia may itself be fatal to insect larvae. However, B. thuringiensis can also kill insects by means of a mechanism not mediated by Cry toxins. Indeed, B. thuringiensis strains lacking crystals are fully virulent when injected into insect larvae or pupae. For some B. thuringiensis strains, inocula containing small numbers of cells have been shown to multiply to high titers and to invade the insect hemocoel, causing septicemia (5, 13, 33). The involvement of the bacteria itself in the killing of insects provides evidence for the existence of additional virulence factors allowing B. thuringiensis to survive and to multiply in the hostile in vivo environment and to withstand the immune defenses of the host.
B. thuringiensis is known to produce several putative virulence factors, including phospholipases C, enterotoxins, hemolysins, cell surface proteins, and metalloproteases, all of which may be involved in establishing infection. Genes encoding these factors are controlled by a transcriptional pleiotropic activator, PlcR (1, 18). However, none of these factors has been shown to play an essential role in the pathogenic mechanisms by which B. thuringiensis causes systemic septicemia. Indeed, inactivation of the PlcR regulon has no effect on B. thuringiensis virulence in intrahemocoelic infection models (26). A zinc-metalloprotease, InhA, which had been reported to hydrolyze Hyalophora cecropia antibacterial peptides specifically (3, 5) and to be highly toxic when injected into the hemolymph of insects (20, 28), was found to make no major contribution to the pathogenic properties of B. thuringiensis (7). To date, only clpP1, the gene encoding the proteolytic subunit of Clp ATP-dependent proteases, has been demonstrated to be involved in B. thuringiensis virulence in the Bombyx mori infection model. However, ClpP1 seems to be required for efficient cell division at low temperatures rather than for the pathogenic properties of B. thuringiensis cells (6).
We carried out transposon mutagenesis in the pathogenic acristalliferous B. thuringiensis strain 407 Cry−, with the aim of identifying virulence determinants. The transposon library was screened for loss of virulence in a model of bacteremia, the silkworm B. mori, which is known to be particularly susceptible to low-titer inocula of B. thuringiensis strain 407 Cry−. We report here the identification and characterization of two genes—yqgB and yqfZ—of unknown function, mutations in which decreased the pathogenicity of B. thuringiensis against B. mori larvae.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The acrystalliferous B. thuringiensis strain 407 Cry−, belonging to serotype 1 (19), was used throughout this study. The nonmotile flhA null mutant (B. thuringiensis strain 407 Cry− [plcA′-lacZ; flhA::mini-Tn10]) carrying a disrupted flhA gene, designated MP02 by Ghelardi and colleagues (9), was used as a negative control in motility assays. Escherichia coli K-12 strain TG1 [Δ(lac-proAB) supE thi hsdΔ5 (F′ traD36 proA+ proB+ lacIq lacZΔM15)] (10) was used as a host for cloning experiments. ET12567 (F− dam-13::Tn9 dcm-6 hsdM hsdR recF143 zjj-202::Tn10 galK2 galT22 ara-14 pacY1 xyl-5 leuB6 thi-1) was used to generate unmethylated plasmid DNA for the electrotransformation of B. thuringiensis. B. thuringiensis and E. coli strains were transformed by electroporation as previously described (4, 19).
E. coli and B. thuringiensis cells were cultured in Luria-Bertani (LB) broth, with vigorous shaking, at 37 and 30°C, respectively. The antibiotic concentrations used for bacterial selection were as follows: 100 μg of ampicillin ml−1 for E. coli; 100 μg of spectinomycin ml−1 for E. coli and 300 μg of spectinomycin ml−1 for B. thuringiensis; 20 μg of kanamycin ml−1 for E. coli and 200 μg of kanamycin ml−1 for B. thuringiensis; and 10 μg of erythromycin ml−1 for B. thuringiensis. If used concomitantly, the doses of antibiotics were adjusted to 250 μg of spectinomycin ml−1 and 3 μg of erythromycin ml−1.
For phenotypic studies, various stress conditions were established as follows. Frozen glycerol stocks (exponentially growing cells; optical density at 600 nm [OD600] of 1) of the various strains were diluted 1:100 in LB medium devoid of antibiotics and cultured with vigorous shaking at 37°C. At an OD600 of 0.1 to 0.2, the culture was divided in two. One of the resulting half-cultures was incubated at 37°C (control), whereas the other half was mixed with sodium chloride at a final concentration of 6% (wt/vol) or with 1.7% (wt/vol) lithium chloride.
Motility assays were performed on LB soft agar swarm plates (final concentration, 0.3% agar) by spotting 2 μl of a culture with an OD600 of 1 at the center of the plate and then incubating the plate for 24 h at 37°C. During this period, we measured colony diameters at 8-h intervals and scored motility.
We used Columbia medium agar plates (Biomérieux) containing 5% sheep blood to assay the hemolytic activity of B. thuringiensis strains.
Sporulation assays were performed as follows. Frozen glycerol stocks of exponentially growing cells were diluted 1:100 in sporulation-specific medium (hydrolysate of casein tryptone) (16) devoid of antibiotics and incubated for 24 h at 37°C. Sporulating cells were subjected to heat treatment (80°C, 12 min), with serial dilutions plated before and after heat treatment. Sporulation frequencies were established on the basis of viable-cell and heat-resistant spore counts.
DNA manipulations.
Plasmid DNA was extracted from E. coli by standard alkaline lysis by using QIAprep spin columns (QIAGEN). Chromosomal DNA was extracted from B. thuringiensis cells harvested in the mid-exponential growth phase, as previously described (22). Restriction enzymes and T4 DNA ligase were used as recommended by the manufacturer (New England Biolabs). Oligonucleotide primers (Table 1) were synthesized by Proligo (Paris, France). PCR was performed in a GeneAmp PCR system 2400 thermal cycler (Perkin-Elmer). Amplified DNA fragments were purified with the QIAquick PCR purification kit (QIAGEN). Digested DNA fragments were separated by electrophoresis and eluted from agarose gels with a centrifugal filter device (Montage Genomics; Millipore, Billerica, Mass.).
TABLE 1.
Primer sequences used in this study
| Primer | Nucleotide sequencea | Restriction site |
|---|---|---|
| YqgB1 | 5′-ACCAAGCCCAACCAGAACC-3′ | |
| YqgB2 | 5′-CATGCCATGGTTGTCCTCCGCCGTTTGG-3′ | NcoI |
| YqgB3 | 5′-CGGTAAAACCATTTTGTATATTA-3′ | |
| YqgB4 | 5′-CCATGCCAACACCATCCTGGAT-3′ | |
| YqgB5 | 5′-CGCGGATCCCTAGGAATATATGCTTGTCC-3′ | BamHI |
| YqgB6 | 5′-CGCGGATCCGGAACGGCAACGCGGACGAC-3′ | BamHI |
| YqgB7 | 5′-AACTGCAGATTCCGTAAAAAAAGCTTCCTTC-3′ | PstI |
| YqgB8 | 5′-CGCGGATCCATGCCTACACCATCTTGG-3′ | BamHI |
| YqgB9 | 5′-AAACTGCAGGTTCTGCTATGAGCGGTC-3′ | PstI |
| YqgB10 | 5′-AAACTGCAGAAAACCTTTTGTATATTA-3′ | PstI |
| YqgB11 | 5′-GGAATTCCGGTTCTGCTATGAGCGGTC-3′ | EcoRI |
| YqgB12 | 5′-TCCCCCGGGTTCTTGGAATAGTAG-3′ | SmaI |
| YqfZ1 | 5′-TCTCGTTCAGCAGCCATTTCAGCA-3′ | |
| YqfZ2 | 5′-CATGCCATGGCCTATCGGATATGCTCTTTAAG-3′ | NcoI |
| YqfZ3 | 5′-CCGGAATTCGGAAAGTCTTATAAATTCCCG-3′ | EcoRI |
| YqfZ4 | 5′-CGCGGATCCGTTTGGGCAGCAGTTG-3′ | BamHI |
| YqfZ5 | 5′-TCCCCCGGGCCGAGAACAAGCACGAATAG-3′ | SmaI |
| YqfZ6 | 5′-CGCGGATCCACGTGCACCGGCAATACC-3′ | BamHI |
Restriction sites are underlined.
Generation and screening of a B. thuringiensis transposon bank.
An insertion library was constructed in B. thuringiensis strain 407 Cry− by using the mini-Tn10 as described by Gominet et al. (11). The mini-Tn10 is a derivative of the Tn10 transposon of Salmonella enterica serovar Typhimurium, delivered by the thermosensitive plasmid pIC333 and used for random insertion mutagenesis (24, 29). The plasmid pIC333 includes a thermosensitive origin of replication and a gene conferring resistance to erythromycin. This plasmid is eliminated by a transposition event triggered by moving the culture to a nonpermissive temperature, 40°C. Insertion mutants were screened for loss of virulence in B. mori larvae, following their inoculation into the hemocoel. The chromosomal DNA regions flanking the insertion locus were cloned, and their nucleotide sequences were determined as previously described (11).
Insects and experimental infections in vivo.
Eggs of B. mori strain Nistari provided by the Institut National de la Recherche Agronomique (Unité Nationale Séricicole, Lyon, France) were incubated at 25°C. The hatched larvae were reared on a commercially available artificial diet (Fukui and Co., Ltd., Yokohama, Japan). Pathogenicity assays were carried out with B. thuringiensis vegetative cells.
For in vivo screening, B. thuringiensis clones isolated from the insertion library were plated on LB agar medium devoid of antibiotics and incubated for 18 h at 30°C. Colonies were picked with an entomologic pin, which was then used to prick the hemocoel of three successive B. mori larvae such that the inoculum was most concentrated for the first larva, less concentrated for the second larva, and least concentrated for the third larva. The B. mori larvae were inoculated on the first day of the third instar. Infection experiments were repeated three times, and infected larvae were incubated in plastic containers at 25°C. Mortality was recorded 24 h after infection.
For the determination of 50% lethal doses (LD50s), cells of wild-type and mutant strains of B. thuringiensis were cultured in LB medium devoid of antibiotics at 30°C, with shaking. Bacterial densities were monitored by determining the OD600 and checked by plating dilutions on LB agar plates. Various dilutions of exponentially growing B. thuringiensis cells were used to inoculate groups of 30 B. mori larvae (10 μl of cell suspension larva−1). The control group was injected with sterile water. B. mori larvae were used on the first day of the fourth instar and weighed about 150 to 200 mg. The injections were delivered into the intersegmental membrane between the fourth and fifth abdominal leg of the larva by using a 1-ml Terumo syringe and a microapplicator (Buckard type LV. 65). Inoculated larvae were incubated individually in plastic containers at 25°C. Mortality was recorded daily over a 3-day period.
We counted B. thuringiensis cells in the living and dead insects as follows. Five infected larvae in each infection experiment were individually crushed and homogenized in 1 ml of sterile water, and dilutions were plated on LB agar plates containing the appropriate antibiotics.
Database comparisons and sequence analysis.
Sequences were compared and aligned with sequences from the GenBank database by using the BLAST program of the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov/genomes/MICROBES/Complete.html) network server. Bacillus anthracis genome sequences were provided by the NCBI network server. The Bacillus subtilis sequence was obtained from SubtiList (http://genolist.pasteur.fr/SubtiList), and Listeria monocytogenes and Listeria innocua sequences were obtained from ListiList (http://genolist.pasteur.fr/ListiList). The B. cereus genome sequence was obtained from http://integratedgenomics.com.
PCR amplification and sequencing of the B. thuringiensis yqgB and yqfZ genes.
Two synthetic oligonucleotides, YqgB1 and YqfZ1 (Table 1), were designed from the B. cereus strain ATCC 14579 (http://www.integratedgenomics.com) genomic region encompassing the yqgB and yqfZ genes, from positions −940 to +1240 with respect to the ATG start codon and the TAA terminal codon of yqgB and yqfZ, respectively. YqgB1 and YqfZ1 were used to amplify a 3,210-bp fragment, with 407 Cry− chromosomal DNA used as the template. PCR was carried out in a reaction volume of 100 μl with Pfx DNA polymerase, as recommended by the manufacturer (Invitrogen, Life Technologies). Purified PCR products were sequenced on both strands by Genome Express (Paris, France) by using oligonucleotides YqgB2, YqgB3, YqgB4, YqgB5, YqgB6, YqfZ2, and YqfZ3 (Table 1) based on the B. cereus strain ATCC 14579 yqgB/yqfZ genomic region.
Site-directed mutagenesis.
A deletion-replacement mutant of yqgB was constructed as follows. A 963-bp NcoI/SmaI DNA fragment and a 988-bp EcoRI/BamHI DNA fragment, corresponding to the DNA chromosomal regions located immediately upstream and downstream from the yqgB gene, respectively, were generated by PCR using B. thuringiensis strain 407 Cry− chromosomal DNA as a template and oligonucleotide pairs YqgB2-YqgB12 and YqgB11-YqgB6, respectively (Table 1). A Kmr cassette, conferring resistance to kanamycin, was purified from pDG783 as a 1.5-kb SmaI/EcoRI fragment carrying the aphA3 gene from Enterococcus faecalis (30). The amplified DNA fragments and the Kmr cassette were digested with the appropriate enzymes and inserted between the NcoI and BamHI sites of the thermosensitive plasmid pMAD (M. Arnaud, A. Chastanet, and M. Debarbouillé, unpublished data), a derivative of the thermosensitive plasmid pE194 (31) conferring resistance to erythromycin in gram-positive hosts and to ampicillin in E. coli and harboring a constitutively expressed transcriptional fusion with the Bacillus stearothermophilus bgaB gene encoding the thermostable β-galactosidase (14). The resulting plasmid was checked by restriction mapping and used to transform the wild-type strain 407 Cry−. Integrants resistant to kanamycin, sensitive to erythromycin, and appearing white on LB agar medium supplemented with X-Gal arose through a double-crossover event in which the chromosomal wild-type copy of yqgB was deleted and replaced with the Kmr cassette, as previously described (17). The chromosomal allele exchange was checked by PCR with the appropriate oligonucleotide primers. The corresponding mutant strain was named 407 Cry− ΔyqgB.
A deletion-replacement mutant of yqfZ and a double deletion-replacement mutant of both yqfZ and yqgB were constructed as reported for the 407 Cry− ΔyqgB mutant using the following primer pairs: YqfZ2-YqfZ5 and YqfZ3-YqfZ6 for the 407 Cry− ΔyqfZ mutant and YqgB2-YqgB6 and YqfZ3-YqfZ6 for the 407 Cry− ΔyqgB ΔyqfZ double mutant.
Construction of lacZ fusions and determination of β-galactosidase activity.
We constructed lacZ reporter gene fusions to yqgB and yqfZ by inserting, between the BamHI and PstI sites of pHT304-18′Z (2), PstI-BamHI-digested PCR fragments amplified with Taq polymerase, with B. thuringiensis strain 407 Cry− chromosomal DNA used as a template. The synthetic oligonucleotide pairs YqgB7-YqgB8 and YqgB9-YqfZ4 were used to generate two fragments of 555 bp and 392 bp, respectively, corresponding to the yqgB and yqfZ upstream regions. The recombinant plasmids pHT-yqgB′Z and pHT-yqfZ′Z were introduced by electroporation into the B. thuringiensis wild-type strain (407 Cry−).
Colonies expressing lacZ fusions were detected on media containing 5-bromo-4-chloro-3-indolyl-d-galactopyranoside (X-Gal) (40 μg ml−1) and erythromycin (10 μg ml−1). Cells were cultured in LB medium devoid of antibiotics at 25 and 37°C, with vigorous shaking, and the specific activity of β-galactosidase was determined as previously described (12).
Statistical analysis.
Mortality data were analyzed by calculating 50% lethal doses (LD50s) with the Log-Probit program (8, 25).
Nucleotide sequence accession number.
The nucleotide sequence of the B. thuringiensis strain 407 Cry− genomic region encompassing yqgB and yqfZ has been deposited in the GenBank database under the accession number AY455944.
RESULTS AND DISCUSSION
Screening of mutant bank and isolation of a clone (Bt11) with reduced virulence against B. mori larvae.
We tried to identify the B. thuringiensis determinants required for bacterial virulence by individually screening 1,200 B. thuringiensis mini-Tn10 insertion mutants for loss of virulence on injection into the hemocoel of B. mori larvae. The screening procedure involved pricking the hemocoels of three consecutive larvae with a pin contaminated with a single B. thuringiensis colony. Only mutants failing to kill the second and third larvae were considered to display attenuated virulence. We isolated 18 attenuated mutants in total. We investigated the virulence of these mutants further by injecting a dose of exponentially growing cells corresponding to the 90% lethal dose of the parental strain. Three mutants—clones Bt3, Bt11, and Bt19—gave results confirming those obtained in the primary screening. Analysis of the nucleotide sequences of the ends adjacent to the insertion sites revealed that the mini-Tn10 transposon mapped to the same locus in these three clones. In the Bt3 and Bt11 mutants, the mini-Tn10 had inserted at the same position in the genome. In the Bt19 mutant, the transposon had inserted at a position 120 bp upstream from that in the other two mutants. As these three clones were isolated in independent mutagenesis and screening experiments but nonetheless contained insertions at the same locus, it seems likely that this locus plays an important role in virulence. We then characterized clone Bt11 further. We first checked that the virulence of Bt11 was indeed attenuated by establishing its LD50 in intrahemocoelically infected B. mori larvae. The LD50 of Bt11 was found to be seven times higher than that of the wild-type parental strain (33.2 CFU larva−1 for Bt11 versus 4.78 CFU larva−1 for B. thuringiensis 407 Cry−). Thus, the virulence of the insertion mutant was strongly attenuated.
Molecular characterization of clone Bt11.
Southern hybridization, using the internal part of mini-Tn10 (spectinomycin resistance gene) as a probe and the chromosomal DNA of clone Bt11 digested with either EcoRI or HindIII, showed that the clone contained a single transposon insertion (data not shown). BLAST analysis of the DNA sequences surrounding the transposon insertion sites and of homologous sequences deposited in the NCBI database revealed that mini-Tn10 was inserted 14 bp upstream from the 3′ end of a putative 756-bp open reading frame (ORF), encoding a protein with a deduced amino acid sequence 98% identical to that of the putative YqgB protein of B. cereus strain ATCC 14579. An ORF was identified 65 bp downstream from the yqgB TAA codon, encoding a 121-amino-acid protein that is 98.4% identical to the B. cereus strain ATCC 14579 YqfZ protein (Fig. 1). By analogy with B. cereus, we named these B. thuringiensis genes yqgB and yqfZ. The ATG codons of both the yqgB and yqfZ genes of B. thuringiensis were preceded by a potential ribosome-binding sequence at an appropriate distance. The TAA stop codons of both ORFs were followed by a possible rho-independent transcription terminator. The stem-loop sequences corresponding to these putative terminators were AAAAAAATGCCTAAATAGGCATTTTTTT for yqgB and GAAACCGAGCACTTCGGTTTC for yqfZ. These genes were found to be highly conserved and similarly organized in other organisms such as B. subtilis, L. monocytogenes, L. innocua, B. anthracis, and Bacillus halodurans, except that in B. subtilis, yqfZ and yqgB are separated by the yqgA gene transcribed in the opposite orientation. However, no function has yet been assigned to these genes in any of these other organisms. Analysis of the predicted amino acid sequences of YqgB and YqfZ by the Signal P and TMHMM predictor servers (15, 23) suggested that YqgB is probably a transmembrane protein, whereas YqfZ is probably secreted.
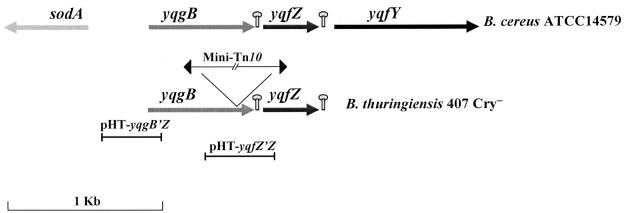
FIG. 1.
Schematic representation of the yqgB and yqfZ genes in B. thuringiensis strain 407 Cry− and B. cereus strain ATCC 14579. The mini-Tn10 insertion in the B. thuringiensis yqgB gene is indicated. Potential stem-loop structures are represented. The segments indicate the DNA fragments used to construct the lacZ plasmid transcriptional fusions.
Disruption of the yqgB and yqfZ genes and effects on virulence.
Based on the molecular characteristics of clone Bt11, we thought that the observed attenuation of the insertion mutant was probably linked either to the abolition of YqgB function, resulting from disruption of the carboxy-terminal end of this protein, or to a polar effect on the expression of the downstream yqfZ gene. We tested these hypotheses by constructing single and double mutants of yqfZ and yqgB by deletion and replacement and assessing the pathogenicity of the resulting exponentially growing cells during the intrahemocoelic infection of B. mori larvae. The virulence of the ΔyqgB and ΔyqfZ mutants did not differ significantly from that of the 407 Cry− parental strain, as the confidence intervals for the LD50s overlapped (Table 2). In contrast, the ΔyqgB ΔyqfZ double mutant was markedly less pathogenic (about one-tenth as pathogenic as the wild-type parental strain) (Table 2). However, the avirulent phenotype of the double mutant strain was limited to the first 24 h following infection. Larvae injected with the cells of the double mutant strain became sick after 24 h but did not die immediately. Instead, they started to die 1 day later. Indeed, the LD50 of the double mutant reached values similar to the LD50 of the parental strain, which remained unchanged, 2 days after infection (Table 2). The time lag before the first observed deaths indicates an effect on the progression of the infection for the double mutant strain.
TABLE 2.
Pathogenicity of vegetative cells from B. thuringiensis wild-type and mutant strains injected into the hemocoel of B. mori larvae
| Strain | LD50 (95% CL)a
|
|
|---|---|---|
| At 24 h (CFU/injected larva) | At 48 h (CFU/injected larva) | |
| 407 Cry− | 12.55 (4.82-22.51) | Unchanged |
| 407 Cry− ΔyqgB | 19 (8.58-30.82) | Unchanged |
| 407 Cry− ΔyqfZ | 14 (4.97-28.44) | Unchanged |
| 407 Cry− ΔyqgB ΔyqfZ | 178 (70.4-407.33) | 12.71 (0.24-33.55) |
LD50 was calculated by log-probit analysis. CL, confidence limit (8).
This time lag may result from an effect on the growth rate during the infection process in the double mutant strain. We investigated this possibility by following the fate of the bacteria in monoinfection experiments. Larvae were injected with an equivalent dose (about 100 CFU larva−1) of the parental strain or of the ΔyqgB ΔyqfZ double mutant. After 24 h, 3.8 × 107 CFU larva−1 were counted in living larvae infected with the mutant strain versus 5.37 × 108 CFU larva−1 for the B. thuringiensis parental strain. Thus, mutant cell in vivo counts were one-tenth of the parental strain number, which suggests that the delayed death of the larvae was due to a defect in bacterial growth.
Phenotypic analysis of the mutant strains.
We investigated the possible functions of YqgB and YqfZ by looking for changes in the phenotypes of the mutants in terms of chemical stresses and stationary phase adaptive responses (see Materials and Methods). All mutants displayed growth rates similar to the rate of the parental strain in the presence of 6% NaCl or 1.7% LiCl, the highest concentrations compatible with the growth and survival of B. thuringiensis 407 Cry− (data not shown). We also determined hemolytic activity on sheep blood agar. All mutant strains were fully hemolytic on sheep erythrocytes, indicating that yqgB and yqfZ are not involved in hemolysis in B. thuringiensis strain 407 Cry− (data not shown). Sporulation assays, in which sporulation frequencies were established after 24 h of incubation in hydrolysate of casein tryptone medium, showed that the mutations did not significantly affect the ability of B. thuringiensis cells to sporulate (data not shown). To investigate whether the partially anaerobic conditions encountered in the insect hemolymph could prevent mutant strain growth, we assessed bacterial survival in LB medium incubated at 37°C without shaking. We found that mutations in yqgB and yqfZ did not affect B. thuringiensis growth in partial anaerobiosis (data not shown). We also assessed bacterial motility by recording, at 8-h intervals, the colony size of the strains spotted on 0.3% LB agar plates. The single mutant strains displayed wild-type motility (Table 3). However, the yqgByqfZ mutant was markedly affected in its motility because colony size after 24 h was only one-third that of the wild type. A similar defect was observed for the MP02 variant lacking the flhA gene, a mutation that has been shown to inhibit swarming motility (9). Overall, these results provided no evidence for a change in phenotype in these tests, with an effect on motility the only apparent phenotype obtained. Examination of the predicted amino acid sequences of YqgB and YqfZ showed them to be probable transmembrane and secreted proteins, respectively. The probable association of these proteins with the cell membrane is consistent with a role for these proteins in motility.
TABLE 3.
Results of motility assaysa
| B. thuringiensis strain | Colony size (cm)
|
||
|---|---|---|---|
| After 8 h | After 16 h | After 24 h | |
| 407 Cry− | 1.3 | 3.5 | 6 |
| 407 Cry− ΔyqgB | 1.4 | 3.2 | 6.4 |
| 407 Cry− ΔyqfZ | 1.5 | 3.2 | 5 |
| 407 Cry− ΔyqgB ΔyqfZ | 0.5 | 1.5 | 2.5 |
| MP02b | 0.5 | 0.9 | 1.1 |
Assays were conducted on soft 3% LB agar plates incubated at 37°C as described in Materials and Methods.
MP02 is a nonmotile B. thuringiensis variant lacking the flhA gene (9).
Mutations in yqgB and yqfZ altered bacterial growth at low temperatures.
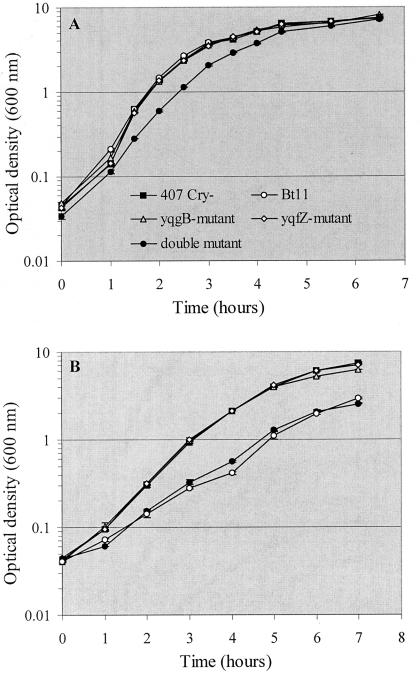
As the insect larvae were reared at 25°C, we assessed the effects of the mutations on the bacterial growth rate at this temperature (Fig. 2). B. thuringiensis mutant and wild-type strains were grown at 37 and 25°C. No major difference in growth rates was observed between single mutants and the parental strain during in vitro exponential growth at 37 and 25°C (Fig. 2). However, the growth of the double mutant was slightly affected at 37°C (Fig. 2A), with a marked reduction at 25°C (Fig. 2B). In addition, the transposon insertion in clone Bt11 was also found to have a significant effect on the ability of the mutant to grow in the same temperature conditions (Fig. 2B). This and other results demonstrate that the double mutant phenotype is correlated with the transposon insertion mutation. Presumably, the mini-Tn10 insertion had a polar effect on yqfZ expression. In contrast, yqgB deletion did not prevent yqfZ transcription, because the single mutant phenotypes were similar to the phenotype of the wild-type strain.
FIG. 2.
Growth curves of B. thuringiensis wild-type and mutant strains in LB medium at 37°C (A) and at 25°C (B).
The phenotypic features of the double mutant resemble those of the ClpP1− mutant in terms of virulence in this infection model and growth at 25°C (6). The clpP1 gene is known to encode the proteolytic subunit of the Clp ATP-dependent protease (21). The precise role of this gene in regulating growth in response to temperature is unknown. It has been reported that the avirulence of B. thuringiensis ΔclpP1 against B. mori larvae infected via the hemocoel results essentially from a growth defect at 25°C. Indeed, B. mori, like all invertebrates, is a poikilotherm host, the internal temperature of which is not regulated. At the temperature at which the ΔclpP1 infection assays were conducted (25°C)—assuming that this temperature is similar to that encountered in the B. mori hemolymph—the clpP1 mutant strain cannot grow and therefore displays an avirulent phenotype (6). The same may be true for the ΔyqgB ΔyqfZ mutant in B. mori. Consistent with this, in vivo bacterial counts demonstrated that the double mutant did not multiply to give high titers as rapidly as the wild-type strain.
Analysis of gene expression.
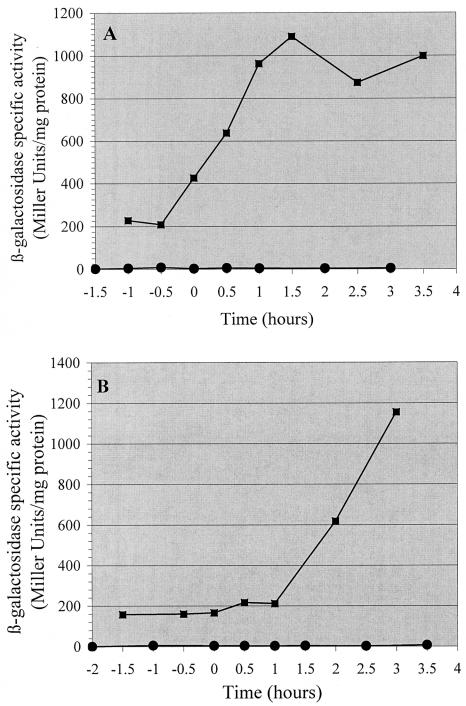
The phenotypic changes resulting from disruption of the B. thuringiensis yqgB and yqfZ genes led us to investigate the expression of these two genes at two different growth temperatures. Expression of the yqgB and yqfZ genes was analyzed by using plasmid transcriptional fusions with the lacZ reporter gene. Two fusions, pHT-yqgB′Z and pHT-yqfZ′Z, harboring the regions immediately upstream from the yqgB and yqfZ genes, respectively, were constructed and introduced into B. thuringiensis strain 407 Cry− (Fig. 1). Transformants were cultured in LB medium at 25 and 37°C. β-Galactosidase activity was determined at various stages of growth between 1 h before the onset of the stationary phase and 4 h after the onset of the stationary phase. The yqgB′-Z fusion was expressed during the exponential and stationary growth phases at both temperatures (Fig. 3). In contrast, no promoter activity was detected upstream from yqfZ because cells harboring the yqfZ′-Z fusion gave no β-galactosidase activity at either of the temperatures tested (Fig. 3). This suggests that the transcription of yqgB and yqfZ is under control of the promoter located upstream from yqgB.
FIG. 3.
Expression of pHT-yqgB′Z (▪) and pHT-yqfZ′Z (•) transcriptional fusions in B. thuringiensis strain 407 Cry−. Cells were grown in LB medium at 37°C (A) and at 25°C (B).
Conclusions.
This investigation is the first example of in vivo screening of B. thuringiensis insertion mutants and led to the identification of previously uncharacterized virulence genes. Wassenaar and Gaastra (32) proposed a refined definition of virulence genes in which the function of the gene in the complex process of virulence is taken into account. In this way, three subclasses of virulence genes have been proposed. According to this classification, yqgB and yqfZ genes should not be considered true virulence factors but instead lifestyle determinants, the products of which act in concert, enabling the bacteria to cope with its suboptimal physical environment and thus facilitating host colonization. This agrees with the presence of both genes in the genome of bacteria with distinct pathogenic properties, some of which, like B. subtilis, are not even pathogens. The swarming motility of the ΔyqgB ΔyqfZ mutant strain on semisolid agar was almost completely abolished. This lack of motility, together with the replication defect of this strain, may contribute to the inability of cells of the mutant strain to cause septicemia in vivo. Motility may enable the bacteria to adhere to and to colonize diverse insect tissues, which may act as alternative nutrient sources required by B. thuringiensis cells for a particular stage in the infection process.
Acknowledgments
We thank Michel Gohar for the analysis of predicted amino acid sequences. We thank Angela Jackson for her participation in the screening experiments.
This work was supported by a grant from Santé des Plantes et Environnement department-INRA (project no. 0071-2001-02: colonisation du biotope insecte par des bactéries pathogènes).
REFERENCES
- 1.Agaisse, H., M. Gominet, O. A. Økstad, A. B. Kolstø, and D. Lereclus. 1999. PlcR is a pleiotropic regulator of extracellular virulence factor gene expression in Bacillus thuringiensis. Mol. Microbiol. 32:1043-1053. [DOI] [PubMed] [Google Scholar]
- 2.Agaisse, H., and D. Lereclus. 1994. Expression in Bacillus subtilis of the Bacillus thuringiensis cryIIIA toxin gene is not dependent on a sporulation-specific sigma factor and is increased in a spo0A mutant. J. Bacteriol. 176:4734-4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dalhammar, G., and H. Steiner. 1984. Characterization of inhibitor A, a protease from Bacillus thuringiensis which degrades attacins and cecropins, two classes of antibacterial proteins in insects. Eur. J. Biochem. 139:247-252. [DOI] [PubMed] [Google Scholar]
- 4.Dower, W. J., J. F. Miller, and C. W. Ragsdale. 1988. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 16:6127-6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Edlund, T., I. Siden, and H. G. Boman. 1976. Evidence for two immune inhibitors from Bacillus thuringiensis interfering with the humoral defense system of saturniid pupae. Infect. Immun. 14:934-941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fedhila, S., T. Msadek, P. Nel, and D. Lereclus. 2002. Distinct clpP genes control specific adaptive responses in Bacillus thuringiensis. J. Bacteriol. 184:5554-5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fedhila, S., P. Nel, and D. Lereclus. 2002. The InhA2 metalloprotease of Bacillus thuringiensis strain 407 is required for pathogenicity in insects infected via the oral route. J. Bacteriol. 184:3296-3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Finney, D. J. 1971. Probit analysis. Cambridge University Press, Cambridge, United Kingdom.
- 9.Ghelardi, E., F. Celandroni, S. Salvetti, D. J. Beecher, M. Gominet, D. Lereclus, A. C. Wong, and S. Senesi. 2002. Requirement of flhA for swarming differentiation, flagellin export, and secretion of virulence-associated proteins in Bacillus thuringiensis. J. Bacteriol 184:6424-6433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gibson, T. J. 1984. Studies on the Epstein-Barr virus genome. Ph.D. thesis. University of Cambridge, Cambridge, United Kingdom.
- 11.Gominet, M., L. Slamti, N. Gilois, M. Rose, and D. Lereclus. 2001. Oligopeptide permease is required for expression of the Bacillus thuringiensis plcR regulon and for virulence. Mol. Microbiol. 40:963-975. [DOI] [PubMed] [Google Scholar]
- 12.Grandvalet, C., M. Gominet, and D. Lereclus. 2001. Identification of genes involved in the activation of the Bacillus thuringiensis inhA metalloprotease gene at the onset of sporulation. Microbiology 147:1805-1813. [DOI] [PubMed] [Google Scholar]
- 13.Heierson, A., I. Sidén, A. Kivaisi, and H. G. Boman. 1986. Bacteriophage-resistant mutants of Bacillus thuringiensis with decreased virulence in pupae of Hyalophora cecropia. J. Bacteriol. 167:18-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hirata, H., T. Fukazawa, S. Negoro, and H. Okada. 1986. Structure of a beta-galactosidase gene of Bacillus stearothermophilus. J. Bacteriol. 166:722-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krogh, A., B. Larsson, G. von Heijne, and E. L. Sonnhammer. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567-580. [DOI] [PubMed] [Google Scholar]
- 16.Lecadet, M. M., M. O. Blondel, and J. Ribier. 1980. Generalized transduction in Bacillus thuringiensis var. berliner 1715, using bacteriophage CP54 Ber. J. Gen. Microbiol. 121:203-212. [DOI] [PubMed] [Google Scholar]
- 17.Lereclus, D., H. Agaisse, M. Gominet, and J. Chaufaux. 1995. Overproduction of encapsulated insecticidal crystal proteins in a Bacillus thuringiensis spo0A mutant. Bio/Technology 13:67-71. [DOI] [PubMed] [Google Scholar]
- 18.Lereclus, D., H. Agaisse, M. Gominet, S. Salamitou, and V. Sanchis. 1996. Identification of a Bacillus thuringiensis gene that positively regulates transcription of the phosphatidylinositol-specific phospholipase C gene at the onset of the stationary phase. J. Bacteriol. 178:2749-2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lereclus, D., O. Arantes, J. Chaufaux, and M.-M. Lecadet. 1989. Transformation and expression of a cloned delta-endotoxin gene in Bacillus thuringiensis. FEMS Microbiol. Lett. 60:211-218. [DOI] [PubMed] [Google Scholar]
- 20.Lövgren, A., M. Zhang, A. Engström, G. Dalhammar, and R. Landén. 1990. Molecular characterization of immune inhibitor A, a secreted virulence protease from Bacillus thuringiensis. Mol. Microbiol. 4:2137-2146. [DOI] [PubMed] [Google Scholar]
- 21.Maurizi, M. R., W. P. Clark, S. H. Kim, and S. Gottesman. 1990. ClpP represents a unique family of serine proteases. J. Biol. Chem. 265:12546-12552. [PubMed] [Google Scholar]
- 22.Msadek, T., F. Kunst, D. Henner, A. Klier, G. Rapoport, and R. Dedonder. 1990. Signal transduction pathway controlling synthesis of a class of degradative enzymes in Bacillus subtilis: expression of the regulatory genes and analysis of mutations in degS and degU. J. Bacteriol. 172:824-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 24.Petit, M. A., C. Bruand, L. Janniere, and S. D. Ehrlich. 1990. Tn10-derived transposons active in Bacillus subtilis. J. Bacteriol. 172:6736-6740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raymond, M., G. Prato, and D. Ratsira. 1993. PROBIT analysis of mortality assays displaying quantal response. License L93019. Avenix, St. Georges d'Orques, France.
- 26.Salamitou, S., F. Ramisse, M. Brehelin, D. Bourguet, N. Gilois, M. Gominet, E. Hernandez, and D. Lereclus. 2000. The PlcR regulon is involved in the opportunistic properties of Bacillus thuringiensis and Bacillus cereus in mice and insects. Microbiology 146:2825-2832. [DOI] [PubMed] [Google Scholar]
- 27.Schnepf, E., N. Crickmore, J. Van Rie, D. Lereclus, J. Baum, J. Feitelson, D. R. Zeigler, and D. H. Dean. 1998. Bacillus thuringiensis and its pesticidal crystal proteins. Microbiol. Mol. Biol. Rev. 62:775-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Siden, I., G. Dalhammar, B. Telander, H. G. Boman, and H. Somerville. 1979. Virulence factors in Bacillus thuringiensis: purification and properties of a protein inhibitor of immunity in insects. J. Gen. Microbiol. 114:45-52. [DOI] [PubMed] [Google Scholar]
- 29.Steinmetz, M., and R. Richter. 1994. Easy cloning of mini-Tn10 insertions from the Bacillus subtilis chromosome. J. Bacteriol. 176:1761-1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trieu-Cuot, P., and P. Courvalin. 1983. Nucleotide sequence of the Streptococcus faecalis plasmid gene encoding the 3′5"-aminoglycoside phosphotransferase type III. Gene 23:331-341. [DOI] [PubMed] [Google Scholar]
- 31.Villafane, R., D. H. Bechhofer, C. S. Narayanan, and D. Dubnau. 1987. Replication control genes of plasmid pE194. J. Bacteriol. 169:4822-4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wassenaar, T. M., and W. Gaastra. 2001. Bacterial virulence: can we draw the line? FEMS Microbiol. Lett. 201:1-7. [DOI] [PubMed] [Google Scholar]
- 33.Zhang, M.-Y., A. Lövgren, M. G. Low, and R. Landén. 1993. Characterization of an avirulent pleiotropic mutant of the insect pathogen Bacillus thuringiensis: reduced expression of flagellin and phospholipases. Infect. Immun. 61:4947-4954. [DOI] [PMC free article] [PubMed] [Google Scholar]