Abstract
Reductive dehalogenation of vinyl chloride (VC) to ethene is the key step in complete anaerobic degradation of chlorinated ethenes. VC-reductive dehalogenase was partially purified from a highly enriched culture of the VC-respiring Dehalococcoides sp. strain VS. The enzyme reduced VC and all dichloroethene (DCE) isomers, but not tetrachloroethene (PCE) or trichloroethene (TCE), at high rates. By using reversed genetics, the corresponding gene (vcrA) was isolated and characterized. Based on the predicted amino acid sequence, VC reductase is a novel member of the family of corrinoid/iron-sulfur cluster containing reductive dehalogenases. The vcrA gene was found to be cotranscribed with vcrB, encoding a small hydrophobic protein presumably acting as membrane anchor for VC reductase, and vcrC, encoding a protein with similarity to transcriptional regulators of the NosR/NirI family. The vcrAB genes were subsequently found to be present and expressed in other cultures containing VC-respiring Dehalococcoides organisms and could be detected in water samples from a field site contaminated with chlorinated ethenes. Therefore, the vcrA gene identified here may be a useful molecular target for evaluating, predicting, and monitoring in situ reductive VC dehalogenation.
Contamination of groundwater with the chlorinated solvents tetrachloroethene (PCE) and trichloroethene (TCE) threatens numerous drinking water supplies (6, 36). Conventional approaches for groundwater remediation have placed a multi-billion-dollar burden on society and have consequently stimulated research in alternative clean-up strategies (22). One such strategy, the removal of these contaminants by naturally occurring, chloroethene-degrading microorganisms (bioremediation), appears to be a viable and cost-effective alternative. The microbial degradation of PCE and TCE has been observed most frequently under anaerobic conditions where the chlorinated ethenes can be reductively dehalogenated via the less chlorinated ethenes cis-1,2-dichloroethene (cDCE) and vinyl chloride (VC) to harmless ethene. However, at many chloroethene-contaminated sites, reductive dehalogenation ceases or is significantly slowed down at the level of VC, resulting in its accumulation. Because VC is a known human carcinogen and is the most toxic compound of all chloroethenes, reduction of VC to ethene is the key step in the complete anaerobic degradation of these compounds.
Reductive dehalogenation of VC has been linked to the genus Dehalococcoides (3, 7-9, 18). Dehalococcoides ethenogenes strain 195, the first microorganism isolated in pure culture that dehalogenates VC to ethene (18), catalyzes this reduction only in a slow, cometabolic reaction (14, 15, 19). Recently, enrichment cultures containing Dehalococcoides-like organisms which couple VC reduction with energy conservation have been reported (3, 7). The isolation of an axenic culture of one of those organisms, strain BAV1, was subsequently described (8).
While reductive dehalogenation of higher chlorinated ethenes and of some chlorinated aromatic compounds has been studied on a biochemical, chemical, and genetic level (14-16, 20, 23-25, 26, 31-35), little is known about molecular features of reductive dehalogenation of VC. In D. ethenogenes 195, VC reduction is mediated by the TCE-reductive dehalogenase (TceA) (14, 15). The VC reduction rate of TceA is, however, less than 1% of its activity of TCE and cDCE reduction. In a previous study with the VC-degrading enrichment culture maintained in our laboratory, the presence of a membrane-bound activity that reduced VC and cDCE, but not TCE or PCE, with high rates was shown (29).
By using the highly enriched Dehalococcoides sp. strain VS, we provide here, for the first time, insights into the molecular composition of a VC reductase as well as the encoding gene. This gene was subsequently found by molecular probing to be present both in VC-dehalogenating mixed cultures used in bioremediation approaches and in groundwater samples from a contaminated field site undergoing bioremediation. These observations suggest that the here-described enzyme is an environmentally relevant VC reductase and that molecular probing methods, based on the vcrA gene sequence, can be useful tools to assess the capacity for VC reduction at contaminated sites.
MATERIALS AND METHODS
Titanium(III)-nitrilotriacetic acid (NTA) stock solutions contained 100 mM Ti3+ chelated by 150 mM NTA and were prepared as described elsewhere (21). FeS was prepared according to Ehrenreich and Widdel (4). All other chemicals and gases used were of reagent grade or better and were from standard commercial sources. Fast-protein liquid chromatography columns were obtained from Pharmacia.
Bacterial culture and growth conditions.
The VC-dehalogenating culture (Victoria culture) was cultivated under strictly anoxic conditions in a morpholinepropanesulfonic acid (MOPS)-buffered (20 mM, pH 7.2) mineral salt water medium essentially as described previously (29), with alteration of the buffering system. The culture was grown under an N2/H2 (90:10 [vol/vol]) atmosphere in 5-liter carboy bottles containing 4 liters of medium with FeS (∼0.5 mmol/liter) and 0.2 mM titanium(III)-NTA as reducing agents. Addition of 10% sterile-filtered supernatant of a mixed culture containing Dehalococcoides sp. strain VS grown with bicarbonate buffer was needed to obtain sufficient growth. The electron acceptors VC or 1,1-DCE were added discontinuously over time in 200-μmol/liter increments up to a total of 5 to 10 mmol/liter; the electron donor H2 was replenished when needed. The cultures were stirred in the dark at 30°C for several weeks with frequent exchange of the headspace of the bottles in an anoxic chamber (Coy Laboratory Products, Ann Arbor, Mich.). The mixed cultures KB-1, Pinellas, and WS were grown in reduced mineral medium with VC as electron acceptor essentially as described previously (37).
Enzyme assays.
Cells were harvested by centrifugation (20,000 × g, 20 min, 4°C) under strictly anoxic conditions using the anoxic chamber and were washed and resuspended in degassed MOPS buffer (20 mM, pH 7.2) supplemented with 2 mM 1,4-dithioerythritol (DTE) and 0.2 mM titanium(III)-NTA (buffer A). The buffer was incubated overnight in the anoxic chamber before addition of reducing agents. Cell extracts were obtained by anoxic disruption of cells by French press treatment (138 MPa; two passages) followed by two centrifugation steps (30,000 × g, 15 min, 4°C).
Assays of VC reduction were conducted as described previously in 2-ml glass vials under an N2/H2 atmosphere with Ti(III)-reduced methyl viologen as artificial electron donor (29). The protein concentration in the assay varied between 10 and 100 μg/ml. The test was started by addition of gaseous VC to the assay mix (total aqueous volume, 0.3 ml), and change in VC and ethene concentrations were followed with time by gas chromatography. Experiments testing for reduction of liquid chlorinated ethenes (PCE, TCE, and DCE isomers) were carried out essentially as described above with the appropriate substitution of the chlorinated ethene. The chlorinated compounds were added from aqueous stock solutions. Oxygen sensitivity of VC-reducing activity in cell extracts was investigated in buffer amended with resazurin as redox indicator. Prior to addition of VC, the assay was exposed to air until the resazurin underwent a color change from colorless to pink. Subsequently the assay was rendered anoxic again by exchanging the gas phase inside the anoxic chamber followed by addition of Ti(III)-NTA (5 mM final concentration). All enzyme assays were carried out at ambient temperature, and given activities are means of at least three independent measurements.
Partial purification of VC reductase.
Protein purification was performed at 4°C in the anoxic chamber. The membrane fraction was obtained by centrifugation of cell extract at 100,000 × g for 90 min at 4°C. The pellet was resuspended in 1 ml of buffer A supplemented with 2 mM 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS) and incubated for 1 h on ice. After subsequent centrifugation (100,000 × g, 90 min, 4°C) the solubilized membrane fraction was diluted with an equal volume of buffer B, which contained 50 mM 1,3-bis[Tris(hydroxymethyl)methylamino]propane (pH 9.6), 2 mM dithiothreitol, 0.2 mM titanium(III)-NTA, 20 mM CHAPS, and was loaded on a 1-ml HighTrap Q column (Amersham Pharmacia) equilibrated with buffer C (same as buffer B but with 2 mM CHAPS). VC-reductive dehalogenase activity was eluted as a single peak (at 550 to 580 mM NaCl) with 10 ml of buffer C followed by a 40-ml linear gradient from 0 to 700 mM NaCl in buffer C at a flow of 1 ml/min.
Fractions containing the highest activity were pooled and mixed with an equal volume of buffer A containing 20 mM CHAPS and were applied to a Superose 6 column equilibrated with the same buffer. Enzyme activity was eluted with buffer A at a flow rate of 0.2 ml/min. The protein concentration of the samples was determined according to Bradford (1a) with bovine serum albumin as a standard.
Amino acid sequencing and tryptic digest.
Peptides present in active enzyme fractions were separated by sodium dodecyl sulfate (SDS)-12% polyacrylamide gel electrophoresis and were adsorbed onto polyvinylidene difluoride membrane by electroblotting. The transferred peptides were excised, and their N termini were sequenced at the Stanford Protein and Nucleic Acid (PAN) Facility. To obtain internal peptides, active enzyme fractions were separated by SDS gel electrophoresis and bands were excised, digested with trypsin, separated by high-performance liquid chromatography, and sequenced.
PCR amplification, cloning, and sequencing.
Standard protocols were used for DNA cloning and transformation (30). Chromosomal DNA was purified according to Owen and Borman (27). Purification of PCR products and plasmids was performed with Qiaprep spin columns (QIAGEN). Clones were generated in pETBlue-1 (Novagen) or in pDrive (QIAGEN). Sequencing was carried out on an ABI Prism 373 sequencer with ABI Big-Dye sequencing chemistry (PE Applied Biosystems) at the Stanford Protein and Nucleic Acid (PAN) Facility. Southern blot analysis was performed with digoxigenin-labeled probes following the Genius kit protocol (Boehringer Mannheim).
Based on the N-terminal amino acid sequence (EANSTKDQPWYVKHREHFDP) and on one internal amino acid sequence (DALFYAVTQPF) of the 62-kDa peptide (see below), the following degenerate oligonucleotides were designed: Nterm13F (5′-ACVAARGAYCARCCDTGGTA-3′) and Intern4R (5′-TTYTAYGCMGTIACVCARCC-3′). PCR conditions were 100 ng of genomic DNA, 200 nM each primer, 200 μM deoxynucleoside triphosphates, and 1 U of Taq polymerase in PCR buffer with 1.5 mM MgCl2 (Roche). PCR parameters were 3 min at 92°C, 30 cycles of 1 min at 94°C, 1 min at 47°C, and 1 min at 72°C, followed by 5 min at 72°C. A single amplification product of 292 bp was cloned into pET-Blue and was used to generate a probe for Southern blot analysis.
Southern blot analysis with genomic DNA from the VC-degrading mixed culture identified a 3.0-kb HindIII fragment, a 3.0-kb AccI fragment, and a 1.6-kb SspI fragment that hybridized to the probe. Subsequently, inverse PCR with genomic DNA from the VC-degrading mixed culture was carried out as described below. Genomic DNA was digested with SspI, the reaction mixture was purified with Qiaprep spin columns, and the digested DNA was ligated (0.5 ng/μl) with T4 DNA ligase in the presence of ATP overnight at 12°C. The ligated DNA was purified with QIAGEN plasmid preparation and was used in inverse PCR amplification. Based on the previously determined 292-bp DNA sequence, primers vcrSspf (5′-CTATTTTACGCCGTCACCCAACCT-3′) and vcrSspr (5′-TGTAATCGTAGGGTCAAAATGCTC-3′) were designed. The reaction mixture contained 25 ng of circularized DNA/μl, 150 nM each primer, 200 μM deoxynucleoside triphosphates, 1 U of Taq polymerase in PCR buffer with 1.5 mM MgCl2, and Q solution (QIAGEN) in a total volume of 20 μl. PCR parameters were 3 min at 92°C, 35 cycles of 1 min at 94°C, 1 min at 56°C, and 2 min at 72°C, followed by 7 min at 72°C. A single 1.4-kb fragment was amplified, cloned, and sequenced. Based on this sequence, the primer pair vcrAcc/Hinf (5′-ACGCGAGATGGGGTTTGTA-3′) and vcrAcc/Hinr (5′-AATTCGCTTCTTTTGCTCTTCAC-3′) was designed for a second round of inverse PCR with genomic DNA digested with AccI or HindIII, respectively. The MgCl2 concentration in the PCR mixtures was raised to 2.0 mM. PCR parameters were 3 min at 92°C, 35 cycles of 1 min at 94°C, 1 min at 51°C, and 3 min at 72°C, followed by 7 min at 72°C. A single 2.8-kb fragment from each PCR was amplified, cloned, and sequenced. To obtain further downstream sequence, a new probe for Southern blotting was generated from the 3′ end of the HindIII fragment. This probe hybridized to a 1.5-kb NcoI fragment. Inverse PCR with NcoI-digested genomic DNA yielded a 1.1-kb fragment. Primers were vcrNcof (5′-GCAAAACGGCAGACAGGTATTATC-3′) and vcrNcor (5′-GCCACGCCCAACTGAATAGG-3′), and PCR parameters were 3 min at 92°C, 35 cycles of 1 min at 94°C, 1 min at 56°C, and 3 min at 72°C, followed by 7 min at 72°C. The sequences of vcrA and vcrB were verified by PCR amplification with Pfu polymerase (Stratagene) and by sequencing products from three independent PCR assays.
For PCR amplification of vcrAB operons from other VC-degrading mixed cultures (KB-1, Pinellas-culture, WS) the following degenerate primers were designed: 5′-ACVAARGAYCARCCDTGGTA-3′ and 5′-TYGGTCCYTCYTCYTTCC-3′. A single 1,392-bp product was obtained from each culture, cloned, and sequenced. The flanking regions of those products were PCR amplified with primers designed from the vcrAB genomic locus of strain VS. The possibility of a contamination of the three mixed cultures with strain VS was excluded by amplification of the vcrAB operon from genomic DNA isolated externally and by testing for the presence of the 16S rRNA gene of strain VS in those cultures.
For PCR amplification of vcrAB operon from groundwater samples, total DNA from sediment material of 2 liters of groundwater samples was isolated with the UltraClean Soil DNA Isolation kit (Mo Bio Laboratories, Inc.). The following primer pair was used: 5′-CTATGAAGGCCCTCCAGATGC-3′ and 5′-GTAACAGCCCCAATATGCAAGTA-3′.
Reverse-transcriptase PCR (RT-PCR).
Total RNA was prepared from cells in mid-log phase by a combination of TRIzol extraction (Invitrogen) and RNA clean-up with the RNeasy Mini Kit (QIAGEN). DNA was removed from the RNA by three treatments with RNase-free DNase I (QIAGEN). cDNA was synthesized from 0.2 to 0.8 μg of RNA and 2 pmol of specific primer with SuperScript II RNase H− Reverse Transcriptase (Invitrogen) as described by the supplier. The PCR amplification mixtures contained 6 μl of cDNA, 200 nM each primer, 200 μM dNTP, 1 U of Taq polymerase in PCR buffer with 1.5 mM MgCl2.
For amplification of parts of vcrA, vcrB, and vcrC as well as intergenic regions of vcrA and vcrB and also vcrB and vcrC, five sets of primers were chosen for RT-PCR: vcrAf (5′-TGCTGGTGGCGTTGGTGCTCT-3′) and vcrAr (5′-TGCCCGTCAAAAGTGGTAAAG-3′); vcrBf (5′-CTTGGCATATTGGGGCTGTTAC-3′) and vcrBr (5′-ATTTGTCTACCCTGCGTCTTACTG-3′); vcrCf (5′-GTGGCCCTCTTACGGTTGTT-3′) and vcrCr (5′-CTAAGTGGCGAGAAAGAATAATG-3′); vcrABf (5′-AAAATAGTAAAAGGTGTTGTTGC-3′) and vcrABr (5′-TATTTGTCTACCCTGCGTCTTA-3′); and vcrBCf (5′-TGCGGCAAGATCAGTAAGACG-3′) and vcrBCr (5′-GTAAGAGGGCCACCATAACCATAG-3′). PCR parameters were 3 min at 92°C, 30 cycles of 0.45 min at 94°C, 1 min at 55°C for the primer pairs vcrAf-vcrAr, vcrBf-vcrBr, vcrCf-vcrCr, and vcrBCf-vcrBCr and at 50°C for primer pair vcrABf-vcrABr, 1 min at 72°C, followed by 7 min at 72°C.
To determine the region of the transcriptional start site of vcrABC, the following primer pairs were used: vcrT1f (5′-TATCTTTGCGTATTTTGTGC-3′) and vcrT1r (5′-GCCCGCTGATCCCCTCTC-3′); vcrT2f (5′-TTGTACTGAGGAAACGCTTATGG-3′) and vcrT2r (5′-GCCCGCTGATCCCCTCTCC-3′); vcrT3f (5′-CTTATGGATATTTGGCGTTCAGGA-3′) and vcrT3r (5′-AATTCGCTTCTTTTGCTCTTCACC-3′). PCR parameters were essentially as described above, with an annealing temperature of 51°C.
Computer analysis.
The nucleotide and amino acid sequences were analyzed by using the DNAStar software package (DNASTAR, Madison, Wis.) as well as the Simple Modular Architecture Research Tool (SMART; http://smart.embl-heidelberg.de). Preliminary sequence data was obtained from The Institute for Genomic Research (Bethesda, Md.) website at http://www.tigr.org and from the Joint Genome Institute (http://www.jgi.doe.gov).
Nucleotide accession number.
All sequence data have been deposited in the GenBank database under accession number AY322364.
RESULTS
Cultivation of Dehalococcoides sp. strain VS.
Anaerobic dehalogenation of VC was studied with a mixed bacterial culture (Victoria culture) comprised of greater than 99% small cocci. The culture was grown in reduced mineral medium with VC or 1,1-DCE as electron acceptor, H2 as electron donor, and acetate as a carbon source. An axenic culture of those small cocci has been obtained since then (J. A. Müller, G. Meshulam Simon, and A. M. Spormann, unpublished data), and its 16S rRNA gene sequence was found to be identical to that of the previously described Dehalococcoides-like bacterium VS (3). The mixed culture contained small numbers of three morphologically different types of organisms. These microorganisms were rapidly enriched for with either pyruvate or vanillate instead of VC as catabolic substrate. After four consecutive transfers (1%, vol/vol) in the presence of pyruvate or vanillate, these cultures had lost the ability to reduce VC and the small cocci could not be detected microscopically.
Partial purification and characterization of VC reductase.
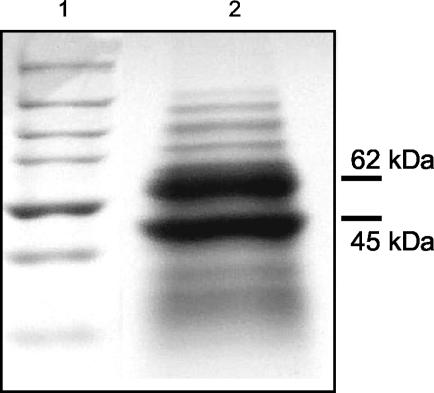
Previous work had shown that the VC-reductive dehalogenating activity of the mixed culture was associated with the bacterial membrane fraction (29). For purification of VC reductase, the highly enriched mixed culture was grown for 2 months in 20 liters of medium containing either VC or 1,1-DCE as electron acceptor and H2 as an electron donor. Cell extract was prepared anoxically, and VC-reductive dehalogenating activity was solubilized from membranes in the presence of 2 mM CHAPS. The VC reductase was partially purified about fourfold to an apparent electrophoretic homogeneity of 50% by means of anion exchange chromatography and gel filtration (Table 1 and Fig. 1). A single peak of VC reductase activity was obtained at both chromatographic steps, suggesting that only one such enzyme was present in the culture. However, the activity eluted during gel filtration as a relatively broad peak, which was likely due to aggregation and/or micell formation of the detergent. During the anion exchange chromatography step, a high percentage (up to 90%) of the enzyme activity was irreversibly lost. Slightly better recovery of the activity was achieved with the pH of the elution buffer adjusted to pH 9.6 from 7.4 or 8.5. Addition of glycerol (20%) to the buffer or varying the detergent concentration (0.5 to 10 mM) did not increase the recovery of activity. Attempts to further purify VC reductase with additional chromatographic steps (sequential anion exchange chromatography at different pH values, hydrophobic interaction chromatography, hydroxyapatite, native and blue native gel electrophoresis) were unsuccessful. In those latter cases, recovery of activity was essentially zero. Therefore, low yields after chromatographic steps in combination with low available biomass at the onset impeded purification of the protein to homogeneity. After gel filtration, protein fractions with VC reductase activity displayed two major bands on SDS-polyacrylamide gel electrophoresis gels, corresponding to an apparent molecular mass of 62 and 45 kDa (Fig. 1). These two peptides were estimated to comprise 95% of the total protein in active fractions. Minor peptide bands of 30, 34, 76, 107, 140, and 175 kDa were also present. However, the bands at 30, 34, 76, 140, and 175 kDa were not detectable in all fast-protein liquid chromatography fractions where VC reductase activity was found (data not shown).
TABLE 1.
Purification scheme for VC reductive dehalogenase of Dehaloccoides strain VS
| Step | Amt of protein (mg) | Activitya (mU) | Sp act (mU/mg of protein) | Yield (%) | Purification factor |
|---|---|---|---|---|---|
| Cell extract | 40.5 | 8,910 | 220 | 100 | |
| Membrane fraction | 16.2 | 5,612 | 346 | 63 | 1.5 |
| Solubilized membrane fraction | 9.9 | 3,047 | 308 | 34 | 1.4 |
| Anionic exchange chromotagraphy | 0.9 | 607 | 638 | 7 | 2.9 |
| Gel filtration | 0.5 | 449 | 990 | 5 | 4.5 |
Activity was monitored as described in Materials and Methods by gas chromatography with reduced methyl viologen as electron donor (29), and given activities are means of at least three independent measurements. One milliunit (mU) of activity is defined as the rate of reduction of 1 nmol of VC per min.
FIG. 1.
SDS-polyacrylamide gel of partially purified VC-reductive dehalogenase (7 μg) of Dehalococcoides sp. strain VS. Lane 1, molecular mass standard (Bio-Rad) with sizes of 250, 150, 100, 75, 50, 37, and 25 kDa; lane 2, active fraction after gel filtration with sizes of major peptides indicated. The gel was stained with Coomassie brilliant blue R-250.
Active protein fractions obtained after gel filtration catalyzed the reduction of VC (350 nmol min−1 mg of protein−1) and all three DCE isomers at high rates (350 to 390 nmol min−1 mg of protein−1) with reduced methyl viologen as electron donor. The DCE isomers were first dehalogenated to VC, which was then further reduced to ethene. Reduction of TCE to cDCE was very slow and occurred at only 5% of the reduction rate for cDCE to VC. PCE was not transformed during the course of the enzyme assay (2 h). The reduction rate of VC was not affected in the presence of saturating concentrations of PCE or TCE. Thus, the enriched enzyme has preferred substrate specificity to VC and DCEs over TCE and PCE and was therefore designated VC reductase. The VC reductase activity was sensitive to exposure to air, with an activity half-life of 5 ± 3 min.
The N-terminal amino acid sequences of the two major peptide bands of 62 and 45 kDa as well as of the minor peptide band of 107 kDa were obtained. Furthermore, internal peptides of the 62-kDa band were obtained after a tryptic digest of the excised band. The amino acid sequence of the N terminus of the 62-kDa peptide (EANSTKDQPWYVKHREHFDP) was found to be similar to that of a 20-amino-acid region of the N terminus of TceA from D. ethenogenes (14). The sequence run showed no indication of background protein contamination. Three internal peptide fractions obtained after tryptic digestion of the 62-kDa peptide band were sequenced. Two of those fractions appeared to be homogenous [peptide1, VYEGPPDA(P)FT(S/T); peptide2, VGTLVQMF(L); ambiguous amino acid residues are in parentheses]. The third fraction contained two different peptides (peptide3, DALFYAVTQPFPG; and peptide4, ESIXTFTLP) of roughly equal abundance. All four sequences were found to have moderate identity (25 to 33%) with internal sequences of TceA.
The N terminus of the 45-kDa peptide (AVREQVYGFFIPSVTLIGIG) was nearly identical to N termini of some alcohol dehydrogenases. A BLASTP search in the deduced proteome of D. ethenogenes using the complete sequence of the alcohol dehydrogenase with the most similar N terminus (accession number ZP_00128696) revealed a putative alcohol dehydrogenase gene located about 30 kb from tceAB. Interestingly, this 30-kb region contains almost exclusively phage-related genes and genes involved in DNA recombination.
The N terminus of the 107-kDa peptide [ANQD(W)SKISLPGSGATG(G/A)YV] was highly similar (90% identity) to a amino acid sequence in the N-terminal region of a deduced protein of D. ethenogenes. This protein, with a predicted molecular mass of 106 kDa, showed no significant similarities to any other protein in the databases.
Based on the abundance of the 62-kDa peptide in active protein fractions, the molecular similarity of this protein to other known reductive dehalogenases, and the absence of any other obvious candidate protein, we concluded that the 62-kDa peptide constitutes the catalytically active subunit of the VC reductase.
Cloning of the genes encoding VC reductase.
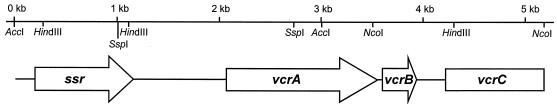
Degenerate oligonucleotide primers were designed from the N-terminal amino acid sequence and one internal sequence of the 62-kDa peptide (see Material and Methods). PCR amplification with those primers yielded a single 292-bp product which, after sequencing, was found to be comprised of a single open reading frame (ORF). The predicted amino acid sequence of this ORF included the amino acid sequences of the N terminus as well as those of peptide1 and peptide3 of the 62-kDa peptide. A probe generated from the 292-bp fragment was found in Southern blot analysis to hybridize to a 1.6-kb SspI fragment, a 2.8-kb AccI fragment, and a 2.8-kb HindIII fragment of restriction enzyme-digested genomic DNA of the mixed culture. The flanking DNA regions of the 292-bp product were subsequently amplified by several inverse PCRs using circularized SspI-, AccI-, HindIII-, and NcoI-digested genomic DNA as templates. The respective PCR products were sequenced, and the four sequences were assembled into a 5.1-kb contig revealing four ORFs (Fig. 2). The N terminus and all four internal peptide sequences of the 62-kDa peptide were present in one ORF, designated vcrA (for vinyl chloride reductase). The deduced VcrA sequence, however, contained an isoleucine at position 330 instead of the (ambiguous) leucine as indicated by the peptide2 sequence. A second ORF, designated vcrB, was found 41 bp downstream of vcrA (Fig. 2). An ORF that started 228 bp downstream of vcrB was found and designated vcrC. One additional ORF upstream of vcrA was identified and designated ssr (for site-specific recombinase; see below).
FIG. 2.
Physical map of the vcr gene locus in Dehalococcoides sp. strain VS. The location and direction of ORFs are indicated by arrows. Relevant restriction enzyme recognition sites are shown.
Computational analysis of vcrA and VcrA.
The vcrA gene is 1,560 bp in length and is predicted to encode a polypeptide, VcrA, of 519 amino acids with a calculated molecular mass of 57,506 Da. The G+C content of vcrA (44.6%) is similar to the G+C content of the D. ethenogenes genome (48.9%) and the average G+C content of the putative dehalogenase genes (48.3%) in this organism. A putative ribosome binding site and a σ70-type promoter sequence (see below) were found upstream of the predicted start codon of vcrA. The N-terminal amino acid sequence of the partially purified VC reductase matches amino acid residues at positions 44 to 63, respectively, of VcrA, consistent with the predicted polypeptide containing a leader sequence, which is cleaved off, leaving a mature polypeptide of 476 amino acids with a calculated molecular mass of 53,115 Da. The proposed leader sequence containing a twin-arginine motif (Tat motif) is predicted by motif search using SMART and is similar to those found in other reductive dehalogenases (14, 16, 25, 33, 35). Two motifs for iron-sulfur clusters were identified at positions 400 to 411 and 444 to 456. Both motifs are similar to the ferredoxin-type 4Fe4S cluster (CX2CX2CX3CP), with the variations that the first motif contains a valine after the fourth cysteine instead of the canonical proline and the second motif displays three amino acids between the first two cysteines instead of two, as in the consensus sequence. As for all other reductive dehalogenases, the described binding motif for corrinoids (DXHX2G) found in various corrinoid-containing enzymes (12) is absent in VcrA.
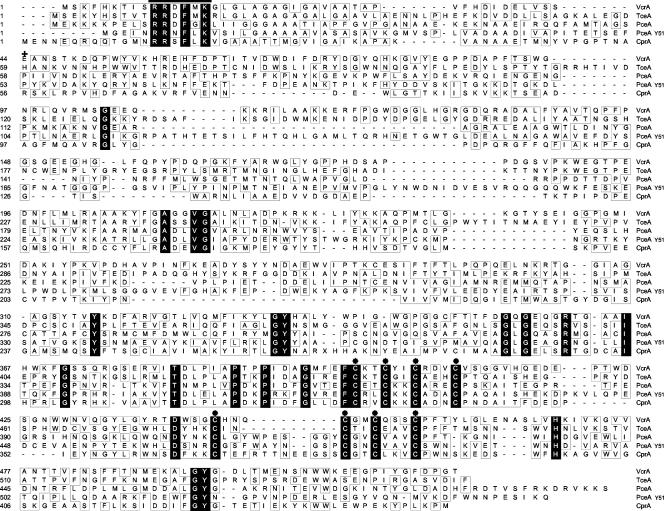
The sequence of VcrA was aligned with those of described reductive dehalogenases (Fig. 3) as well as with putative dehalogenases identified in the genomes of D. ethenogenes and Desulfitobacterium frappieri. Closest similarity (36% identity) was with TceA from D. ethenogenes. Identities to PCE dehalogenases from Sulfurospirillum (formerly Dehalospirillum [13]) multivorans (PceA-Sm) (25), Dehalobacter restrictus (PceA-Dr) (17), and Desulfitobacterium sp. strain Y51 (PceA-Y51) (33) as well as from ortho-chlorophenol-reductive dehalogenase from Desulfitobacterium dehalogenans (CprA) (35) were between 14 and 19%. VcrA was more identical to the putative dehalogenases from D. ethenogenes (up to 34% identity) than to that of Desulfitobacterium hafniense (up to 23% identity). In addition to the leader sequence and the C-terminal two iron-sulfur cluster motifs, several highly conserved amino acid residues, including a conserved histidine, H469, are present in VcrA and all other reductive dehalogenases. Furthermore, the sequence in VcrA ranging from amino acid 198 to 215 displays strong similarity to a region in TceA and all putative dehalogenases from D. ethenogenes.
FIG. 3.
Amino acid sequence alignment of VcrA from Dehalococcoides sp. strain VS with TceA from D. ethenogenes (accession number AF228507), PceA from S. multivorans (accession number AF022812), PceA from Desulfitobacterium sp. strain Y51 (accession number AB070709), and CprA from D. dehalogenans (accession number AF204275). Amino acid residues identical in all five sequences are highlighted in black. Functionally similar amino acid residues (2 distance units) and amino acid residues that are conserved in only some of the sequences are boxed. Horizontal bar, twin-arginine motif; plus sign, first amino acid residue, E44, of the mature VcrA; asterisks, conserved cysteines.
Computational analysis of vcrB and VcrB.
vcrB, a gene of 282 bp, is located 41 bp downstream of vcrA and is preceded by a putative ribosome binding site. The 94 predominantly hydrophobic amino acids account for a calculated molecular mass of 10,641 Da (VcrB). Sequence analysis of VcrB with Protean (DNAStar) and SMART predicted the presence of three transmembrane-spanning regions (data not shown). The predicted VcrB sequence shows some identity to proposed membrane anchors (B proteins) for described as well as putative reductive dehalogenases. The most similar sequence to VcrB was TceB, with 43% identity. Canonical binding motifs for redox-active cofactors, e.g., iron-sulfur cluster, heme, NAD/FAD, or flavin, were not detected in VcrB.
Computational analysis of vcrC and VcrC.
The presumed start codon of vcrC is 228 bp downstream of vcrB. No transcriptional stop codon for vcrC was found in the DNA fragment analyzed. The partial vcrC gene translates into a protein, VcrC, of at least 305 amino acids with a calculated molecular mass of at least 33,363 Da. Four potential membrane-spanning helices can be predicted to occur in VcrC (data not shown). The amino acid sequence of VcrC displays some similarity (25% identity) to CprC from D. dehalogenans. CprC was postulated to function as a NosR/NirI-type transcriptional regulator in ortho-chlorophenol respiration (31). Four ORFs similar to VcrC were also found in the deduced proteome of D. ethenogenes (38 to 47% identity). Two of those are located at loci next to genes encoding putative dehalogenases. The other two ORFs are located directly downstream of homologs of tatB and tatC, components of the translocation apparatus for proteins carrying the Tat motif.
Transcriptional organization of vcrA, vcrB, and vcrC.
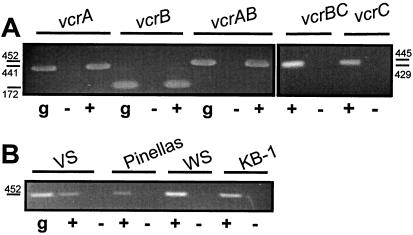
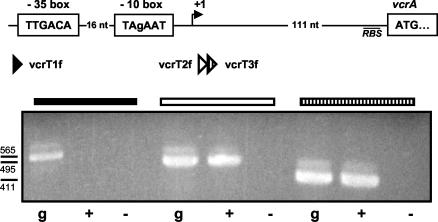
Transcription of vcrA, vcrB, and vcrC was investigated in a series of RT experiments. RT-PCR was performed on RNA isolated from VC-grown cells. RT-PCR assays with primer pairs designed to amplify internal fragments of vcrA, vcrB, and vcrC, respectively, yielded products of the expected sizes (Fig. 4A). PCR products were also obtained for the intergenic regions of vcrA and vcrB as well as of vcrB and vcrC. The RNA-specific recovery of these products demonstrates that vcrA, vcrB, and vcrC are expressed as a polycistronic unit during growth on VC. Two DNA sequences upstream of vcrA were nearly identical to the −10 and −35 regions of an E. coli σ70 promoter (Fig. 5). RT-PCR assays were conducted to determine whether the transcriptional start site of the vcrABC operon is located downstream of that σ70 promoter consensus sequence. PCR products were obtained only with forward primers matching regions downstream but not upstream of the putative transcriptional start site. These results map the transcriptional start of the vcr operon to a region containing a σ70-type promoter sequence.
FIG. 4.
RT-PCR analysis of vcrA, vcrB, and vcrC in Dehalococcoides sp. strain VS. (A) Transcriptional organization of vcrA, vcrB, and vcrC as an operon in strain VS. Agarose gel electrophoresis of RT-PCR assays with primers targeting vcrA, vcrB, and vcrC and intergenic regions of vcrA and vcrB as well as vcrB and vcrC, respectively. (B) Transcription of vcrA in strain VS and in the mixed cultures Pinellas, WS, and KB-1. g, genomic DNA as template; +, assays with RNA as template and conducted with RT; −, assays with RNA as template conducted without RT. Sizes of products are indicated and were as predicted (for vcrA, 441 bp; for vcrB, 172 bp; for vcrAB, 452 bp; for vcrBC, 429 bp; for vcrC, 445 bp). Absolute positions of used primers are the following: vcrAf, 2691 to 2711; vcrAr, 3111 to 3131; vcrBf, 3773 to 3793; vcrBr, 3921 to 3944; vcrCf, 4346 to 4365; vcrCr, 4767 to 4790; vcrABf, 3472 to 3494; vcrABr, 3924 to 3945; vcrBCf, 3910 to 3930; vcrBCr, 4335 to 4358.
FIG. 5.
Transcriptional start and promoter region of vcrABC. Schematic diagram of the nucleotide region upstream of vcrA in Dehalococcoides sp. strain VS (A) and RT-PCR analysis of the region of the transcriptional start site (B). The putative −10 and −35 regions and the putative translational start site of vcrA are boxed, the putative transcriptional start site is indicated by a bend arrow, and the position of a putative ribosome binding site (RBS) is underlined. The positions of forward primers used in RT-PCR are indicated by shaded triangles (position of reverse primers not shown), and corresponding PCR products are indicated by shaded horizontal bars (see the text for details). Absolute nucleotide positions are the following: vcrT1f, 1904 to 1923; vcrT2f, 1974 to 1996; vcrT3f, 1990 to 2013; −35 region, 1926 to 1931; −10 region, 1948 to 1953; predicted transcription initiation site, 1960; translational start site, 2065. g, genomic DNA as template; +, assays with RNA as template and conducted with RT; −, assays with RNA as template conducted without RT. Sizes of products are indicated and were as expected (for vcrT1, 565 bp; for vcrT2, 495 bp; for vcrT3, 411 bp).
DNA region upstream of the vcr operon.
Upstream (850 bp) of the start codon of vcrA, an ORF, ssr, of 972 bp was identified (Fig. 2). It translates into a predicted protein of 324 amino acid residues with a calculated mass of 36,553 Da. A BLASTP search revealed low similarity (25 to 28% identity) to site-specific recombinases of the resolvase family containing a Pin domain (28). A putative ribosome binding site was not identified immediately upstream of the start codon.
Presence of vcrAB homologs in other cultures containing Dehalococcoides spp.
Degenerate PCR primers were designed and used to probe for the presence of the vcrA gene in three mixed cultures, KB-1 (17), Pinellas (5), and WS (W. Wu, unpublished data). These cultures contain Dehalococcoides-like organisms that can grow by reductive dehalogenation of VC to ethene (A. M. Cupples, A. M. Spormann, and P. L. McCarty, submitted for publication). PCR products 1,392 bp in size were obtained from genomic DNA of all three cultures. The products were cloned and sequenced and were found to be highly similar to the corresponding sequence of vcrA previously identified in Dehalococcoides sp. strain VS. Additional primers were designed to amplify the complete vcrA and vcrB genes from those cultures. Nucleotide sequences thereby obtained showed high identities (>98%) to vcrAB of Dehalococcoides sp. strain VS, and the deduced amino acid sequences contained only a few conservative changes, suggesting that they are true homologs. The expression of vcrA during growth of KB-1, Pinellas, and WS with a VC electron acceptor was demonstrated by RT-PCR analysis (Fig. 4B). Dehalococcoides sp. strains 195 and CBDB-1 (1) and a culture containing strain FL-2 (11) were examined for the presence of a vcrA homolog by PCR. None of these cultures have been reported to grow by reductive dehalogenation of VC. With the primer set used, no PCR product was obtained with genomic DNA from those cultures as template (data not shown).
Detection of a vcrAB homolog in contaminated groundwater samples.
The strong correlation of the presence of a vcrA homolog with reductive VC dehalogenation, as observed in the above-described dehalogenating culture, suggested that the vcrAB sequence could be useful as a molecular probe for testing for in situ VC reduction potential at contaminated field sites. Such molecular probes could prove valuable when evaluating, monitoring, or predicting complete removal of chlorinated ethenes from groundwater in field-scale bioremediation projects. Therefore, we queried for the presence of the vcrAB genes in groundwater samples from a chlorinated ethene-contaminated aquifer at Moffett Field, Calif., where complete reductive dehalogenation to ethene was achieved through biostimulation of indigenous microorganisms (2). DNA extracted from groundwater wells exhibiting VC reduction as well as from areas where VC dehalogenation did not occur was used as template in PCR experiments with the primers described above. PCR products were obtained only from groundwater samples undergoing VC reduction to ethene. One of the PCR products was sequenced, and its vcrAB genes were found to be nearly identical (>98%) to those obtained from Dehalococcoides sp. strain VS.
DISCUSSION
Reductive dehalogenation of VC has long been considered the most critical step for complete anaerobic removal of PCE and TCE from groundwater and contaminated soils (22), but the molecular features of reductive VC dehalogenation have been largely unknown. Here, for the first time, a VC-reductive dehalogenase was partially purified, and its encoding genes, vcrAB, were identified and characterized. Furthermore, a strong correlation between the presence of vcrAB in other cultures and catabolic reductive VC dehalogenation was found. A PCR assay testing for the in situ presence of vcrAB was successfully developed and used on groundwater samples from a site contaminated with chlorinated ethenes. The data suggest that the vcrAB genes identified here may be widely distributed, may be of relevance for in situ VC-reductive dehalogenation, and may be a useful target for molecular probing of samples from chlorinated ethene-contaminated sites.
The VC-reductive dehalogenase was partially purified from a highly enriched culture of Dehalococcoides sp. strain VS. The partially purified enzyme reduced VC and the three DCE isomers with similar high rates. TCE was reduced with significantly lower activity, and PCE was not converted. This is in agreement with earlier results on chloroethene reduction rates in cell extracts of the parent mixed culture (29). As with most other chloroethene-reductive dehalogenases, VC-reductive dehalogenase was associated with the membrane fraction, which is consistent with this enzyme being involved in energy conservation during VC reduction to ethene. Analysis of the identified genes encoding VC reductase revealed that this enzyme is a novel member of the family of corrinoid/iron-sulfur cluster-containing reductive dehalogenases. The hydrophobic VcrB protein presumably acts as a membrane anchor for the catalytic subunit of VC reductase. The VcrC protein likely plays a role in regulation of transcription of the vcr operon. The precise function of VcrC, however, remains to be elucidated.
Chlorinated ethenes have been assumed to be introduced into the environment essentially due to human activities within the last few decades, although this has been questioned recently for VC (10). Interestingly, the vcr operon is downstream of a putative site-specific recombinase gene. It is noteworthy that also tceAB in D. ethenogenes 195 (The Institute for Genomic Research) and pceAB in Desulfitobacterium sp. strain Y51 (33) are located in close proximity to putative transposase genes. While the evolutionary origin of those reductive dehalogenases remains unknown, it is tempting to speculate that the parent genes might have encoded enzymes catalyzing the reductive dehalogenation of naturally occurring chlorinated compounds, and that through recent gene duplication and transposition, mutation, and selection the substrate specificity towards the anthropogenic chlorinated ethenes was acquired.
So far, only microorganisms affiliated with the Dehalococcoides group are known to be able to grow on VC. Because not all Dehalococcoides strains use VC as catabolic electron acceptors (8), simply demonstrating the presence of members of the Dehalococcoides group (based on phylogenetic analysis) at chlorinated ethene-contaminated sites is insufficient for predicting growth-linked, and hence rapid, VC degradation. The occurrence and expression of homologs of vcrAB in cultures containing Dehalococcoides-like organisms growing on VC, but not in members of the Dehalococcoides group which are unable to grow by reductive dehalogenation of VC, strongly suggest that the vcrAB gene sequence can be used to determine the presence of VC-respiring bacteria at contaminated field sites. Indeed, during this study the presence of vcrAB in a contaminated aquifer undergoing complete reductive dehalogenation of chlorinated ethene to ethene was demonstrated. This methodology could allow for better assessing the potential for biological degradation of chlorinated ethenes at numerous contaminated field sites and, in combination with bioaugmentation, may become a powerful tool for the clean-up of chlorinated solvents.
Acknowledgments
This work was supported by the Strategic Environmental Research Development Program (SERDP) (grant DACA72-00-0010/CU1169) of the Department of Energy, by E. I. DuPont de Nemours, Inc., and by the Western Region Hazardous Substance Research Center, sponsored through the U.S. Environmental Protection Agency.
We thank T. P. Hoelen and M. Reinhard for providing access to Moffett Field; E. A. Edwards, W. Wu, F. E. Löffler, and M. Harkness for providing culture material; Y. Yang for helpful discussions; and M. J. Ward for comments on the manuscript.
REFERENCES
- 1.Adrian, L., U. Szewzyk, J. Wecke, and H. Görisch. 2000. Bacterial dehalorespiration with chlorinated benzenes. Nature 408:580-583. [DOI] [PubMed] [Google Scholar]
- 1a.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principal of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 2.Cunningham, J. A., T. P. Hoelen, G. D. Hopkins, C. A. LeBron, and M. Reinhard. 2003. Enhanced natural attenuation of commingled plumes, final report. Stanford University, Stanford, Calif.
- 3.Cupples, A. M., A. M. Spormann, and P. L. McCarty. 2003. Growth of a Dehalococcoides-like microorganism on vinyl chloride and cis-dichloroethene as electron acceptors as determined by competitive PCR. Appl. Environ. Microbiol. 69:953-959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ehrenreich, A., and F. Widdel. 1994. Anaerobic oxidation of ferrous iron by purple bacteria, a new type of phototrophic metabolism. Appl. Environ. Microbiol. 60:4517-4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ellis, D. E., E. J. Lutz, J. M. Odom, R. J. Buchanan, C. L. Bartlett, M. D. Lee, M. R. Harkness, and K. A. DeWeerd. 2000. Bioaugmentation for accelerated in situ anaerobic bioremediation. Environ. Sci. Technol. 34:2254-2260. [Google Scholar]
- 6.Ensley, B. D. 1991. Biochemical diversity of trichloroethylene metabolism. Annu. Rev. Microbiol. 45:283-299. [DOI] [PubMed] [Google Scholar]
- 7.He, J., K. M. Ritalahti, M. R. Aiello, and F. E. Löffler. 2003. Complete detoxification of vinyl chloride by an anaerobic enrichment culture and identification of the reductively dechlorination population as a Dehalococcoides species. Appl. Environ. Microbiol. 69:996-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He, J., K. M. Ritalahti, K. L. Yang, S. S. Koenigsberg, and F. E. Löffler. 2003. Detoxification of vinyl chloride to ethene coupled to growth of an anaerobic bacterium. Nature 424:62-65. [DOI] [PubMed] [Google Scholar]
- 9.Hendrickson, E. R., J. A. Payne, R. M. Young, M. G. Starr, M. P. Perry, S. Fahnestock, D. E. Ellis, and R. C. Ebersole. 2002. Molecular analysis of Dehalococcoides 16S ribosomal DNA from chloroethene-contaminated sites throughout North America and Europe. Appl. Environ. Microbiol. 68:485-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keppler, F., R. Borchers, J. Pracht, S. Rheinberger, and H. Schöler. 2002. Natural formation of vinyl chloride in the terrestrial environment. Environ. Sci. Technol. 36:2479-2483. [DOI] [PubMed] [Google Scholar]
- 11.Löffler, F. E., Q. Sun, J. Li, and J. M. Tiedje. 2000. 16S rRNA gene-based detection of tetrachloroethene (PCE)-dechlorinating Desulfuromonas and Dehalococcoides species. Appl. Environ. Microbiol. 66:1369-1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ludwig, M. L., and R. G. Matthews. 1997. Structure-based perspectives on B12-dependent enzymes. Annu. Rev. Biochem. 66:269-313. [DOI] [PubMed] [Google Scholar]
- 13.Luijten, M. L., J. de Weert, H. Smidt, H. T. Boschker, W. M. de Vos, G. Schraa, and A. J. Stams. 2003. Description of Sulfurospirillum halorespirans sp. nov., an anaerobic, tetrachloroethene-respiring bacterium, and transfer of Dehalospirillum multivorans to the genus Sulfurospirillum as Sulfurospirillum multivorans comb. nov. Int. J. Syst. Evol. Microbiol. 53:787-793. [DOI] [PubMed] [Google Scholar]
- 14.Magnuson, J. K., M. F. Romine, D. R. Burris, and M. T. Kingsley. 2000. Trichloroethene reductive dehalogenase from Dehalococcoides ethenogenes: sequence of tceA and substrate range characterization. Appl. Environ. Microbiol. 66:5141-5147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Magnuson, J. K., R. V. Stern, J. M. Gossett, S. H. Zinder, and D. R. Burris. 1998. Reductive dechlorination of tetrachloroethene to ethene by a two-component enzyme pathway. Appl. Environ. Microbiol. 64:1270-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maillard, J., W. Schumacher, F. Vazquez, C. Regeard, W. R. Hagen, and C. Holliger. 2003. Characterization of the corrinoid iron-sulfur protein tetrachloroethene reductive dehalogenase of Dehalobacter restrictus. Appl. Environ. Microbiol. 69:4628-4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Major, D. W., M. L. McMaster, E. E. Cox, E. A. Edwards, S. M. Dworatzek, E. R. Hendrickson, M. G. Starr, J. A. Payne, and L. W. Buonamici. 2002. Field demonstration of successful bioaugmentation to achieve dechlorination of tetrachloroethene to ethene. Environ. Sci. Technol. 36:5106-5116. [DOI] [PubMed] [Google Scholar]
- 18.Maymó-Gatell, X., Y.-T. Chien, J. M. Gossett, and S. H. Zinder. 1997. Isolation of a bacterium that reductively dechlorinates tetrachloroethene to ethene. Science 276:1568-1571. [DOI] [PubMed] [Google Scholar]
- 19.Maymó-Gatell, X., I. Nijenhuis, and S. H. Zinder. 2001. Reductive dechlorination of cis-1,2-dichloroethene and vinyl chloride by “Dehalococcoides ethenogenes.” Environ. Sci. Technol. 35:516-521. [DOI] [PubMed] [Google Scholar]
- 20.Miller, E., G. Wohlfarth, and G. Diekert. 1998. Purification and characterization of the tetrachloroethene reductive dehalogenase of strain PCE-S. Arch. Microbiol. 169:497-502. [DOI] [PubMed] [Google Scholar]
- 21.Moench, T. T., and J. G. Zeikus. 1983. An improved preparation method for a titanium (III) media reductant. J. Microbiol. Methods 1:199-202. [Google Scholar]
- 22.National Research Council. 1994. Alternatives for ground water cleanup. National Academy Press, Washington, D.C.
- 23.Neuman, A., A. Siebert, T. Trescher, S. Reinhardt, G. Wohlfarth, and G. Diekert. 2002. Tetrachloroethene reductive dehalogenase of Dehalospirillum multivorans: substrate specificity of the native enzyme and its corrinoid cofactor. Arch. Microbiol. 163:276-281. [DOI] [PubMed] [Google Scholar]
- 24.Neuman, A., G. Wolfarth, and G. Diekert. 1996. Purification and characterization of tetrachloroethene reductive dehalogenase of Dehalospirillum multivorans. J. Biol. Chem. 271:16515-16519. [DOI] [PubMed] [Google Scholar]
- 25.Neuman, A., G. Wolfarth, and G. Diekert. 1998. Tetrachloroethene dehalogenase from Dehalospirillum multivorans: cloning, sequencing of the encoding genes, and expression of the pceA gene in Escherichia coli. J. Bacteriol. 180:4140-4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Okeke, B. C., Y. C. Chang, M. Hatsu, T. Suzuki, and K. Takamizawa. 2001. Purification, cloning, and sequencing of an enzyme mediating the reductive dechlorination of tetrachloroethylene (PCE) from Clostridium bifermentans DPH-1. Can. J. Microbiol. 47:448-456. [PubMed] [Google Scholar]
- 27.Owen, R. J., and P. Borman. 1987. A rapid biochemical method for purifying high molecular weight bacterial chromosomal DNA for restriction enzyme analysis. Nucleic Acids Res. 15:3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Plasterk, R. H., A. Brinkman, and P. van de Putte. 1983. DNA inversions in the chromosome of Escherichia coli and in bacteriophage Mu: relationship to other site-specific recombination systems. Proc. Natl. Acad. Sci. USA 80:5355-5358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rosner, B. M., P. L. McCarty, and A. M. Spormann. 1997. In vitro studies on reductive vinyl chloride dehalogenation by an anaerobic mixed culture. Appl. Environ. Microbiol. 63:4139-4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd. ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 31.Schumacher, W., C. Holliger, A. J. B. Zehnder, and W. R. Hagen. 1997. Redox chemistry of cobalamin and iron-sulfur cofactors in the tetrachloroethene reductase of Dehalobacter restrictus. FEBS Lett. 409:421-425. [DOI] [PubMed] [Google Scholar]
- 32.Smidt, H., M. van Leest, J. van der Oost, and W. M. de Vos. 2000. Transcriptional regulation of the cpr gene cluster in ortho-chlorophenol-respiring Desulfitobacterium dehalogenans. J. Bacteriol. 182:5683-5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Suyama, A., M. Yamashita, S. Yoshino, and K. Furukawa. 2002. Molecular characterization of the PceA reductive dehalogenase of Desulfitobacterium sp. strain Y51. J. Bacteriol. 184:3419-3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van de Pas, B. A., J. Gerritse, W. M. de Vos, G. Schraa, and A. J. M. Stams. 2001. Two distinct enzyme systems are responsible for tetrachloroethene and chlorophenol reductive dehalogenation in Desulfitobacterium strain PCE1. Arch. Microbiol. 176:165-169. [DOI] [PubMed] [Google Scholar]
- 35.van de Pas, B. A., H. Smidt, W. R. Hagen, J. van der Oost, G. Schraa, A. J. M. Stams, and W. M. de Vos. 1999. Purification and molecular characterization of ortho-chlorophenol reductive dehalogenase, a key enzyme of halorespiration in Desulfitobacterium dehalogenans. J. Biol. Chem. 274:20287-20292. [DOI] [PubMed] [Google Scholar]
- 36.Westrick, J. J., J. W. Mello, and R. F. Thomas. 1984. The groundwater supply survey. J. Am. Water Works Assoc. 76:52-59. [Google Scholar]
- 37.Yang, Y., and P. L. McCarty. 1998. Competition for hydrogen within a chlorinated solvent dehalogenating anaerobic mixed culture. Environ. Sci. Technol. 32:3591-3597. [Google Scholar]