Abstract
An integrative approach was used to obtain pure cultures of previously uncultivated members of the divisions Acidobacteria and Verrucomicrobia from agricultural soil and from the guts of wood-feeding termites. Some elements of the cultivation procedure included the following: the use of agar media with little or no added nutrients; relatively long periods of incubation (more than 30 days); protection of cells from exogenous peroxides; and inclusion of humic acids or a humic acid analogue (anthraquinone disulfonate) and quorum-signaling compounds (acyl homoserine lactones) in growth media. The bacteria were incubated in the presence of air and in hypoxic (1 to 2% O2 [vol/vol]) and anoxic atmospheres. Some bacteria were incubated with elevated concentrations of CO2 (5% [vol/vol]). Significantly more Acidobacteria were found on isolation plates that had been incubated with 5% CO2. A simple, high-throughput, PCR-based surveillance method (plate wash PCR) was developed. This method greatly facilitated detection and ultimate isolation of target bacteria from as many as 1,000 colonies of nontarget microbes growing on the same agar plates. Results illustrate the power of integrating culture methods with molecular techniques to isolate bacteria from phylogenetic groups underrepresented in culture.
Cultivation-independent molecular techniques have illuminated the enormous microbial diversity that exists on our planet and have served to define nearly 40 phylum-level divisions existing within the Bacteria domain alone (23). Most of these divisions, however, are poorly represented by cultured organisms, and at least 13 remain candidate divisions represented only by environmental gene sequences (23). The Acidobacteria and Verrucomicrobia divisions are among those divisions of the domain Bacteria represented by a large diversity of 16S rRNA genes, which occur in particular abundance in soils, but contain few cultured members (3, 11, 12, 17, 20, 21, 23, 36, 48). Hence, our appreciation of the physiological diversity of Acidobacteria and Verrucomicrobia is limited, as is our knowledge of their role in global biogeochemical cycles. Clearly, a better understanding of these divisions would be attained by having a greater diversity of their members available in pure culture for detailed study.
The intrinsic selectivity of any given medium and incubation condition imposes limits on the nature, number, and diversity of microbes recovered from natural samples. It follows, then, that the application of isolation procedures that better mimic conditions existing in the habitat from which the samples were obtained could increase the likelihood of retrieving previously uncultured organisms. Recent efforts to accomplish this have met with some success by using the following: (i) relatively low concentrations of nutrients (1, 13-15, 19, 43, 45, 50); (ii) nontraditional sources of nutrients, signaling molecules, or inhibitors (of undesired organisms) (9, 10, 13, 31); and (iii) relatively lengthy periods of incubation (19, 22, 24-26, 33, 39, 40), sometimes directly in the natural environment from which the inoculum was obtained (26).
For soil microbes, some of which may have become adapted to elevated concentrations of CO2 and concentrations of O2 lower than the atmospheric O2 concentrations (38), the composition of the incubation atmosphere may be an important consideration. Elevated CO2 concentrations are rarely used in incubation atmospheres for isolation of soil microbes, yet CO2 could be important for metabolic processes other than pure autotrophy. Likewise, for soil microbes, the transition to fully aerobic conditions on plating in air may be a stressful event. This would be especially true if cells were not immediately equipped to cope with reactive oxygen species such as hydrogen peroxide (H2O2), superoxide (O2−), or hydroxyl radical (OḢ) produced by their own metabolism or present in media as a result of autoclaving (reviewed in reference 29). Even with facultative anaerobes such as Escherichia coli, an abrupt transition of anaerobically grown cells to aeration can severely retard growth of certain mutants (27). It is also noteworthy that the cultivability (in air) of E. coli and Vibrio vulnificus after starvation is greatly improved if plating media are supplemented with catalase or pyruvate, two compounds known to eliminate H2O2 (5, 34). Such observations suggest that incubation atmospheres enriched with CO2 and/or limited in O2, as well as the incorporation of agents to detoxify reactive oxygen species in the plating media, should be included among treatments seeking to recover previously uncultured microbes.
Whatever cultivation approach is tried, however, one is ultimately confronted with the need to evaluate its success. This is a potentially arduous task if, as in this study, many different media and incubation conditions are being tested and little or nothing is known about the microbes sought other than their 16S rRNA gene sequences. Accordingly, some high-throughput screening method is desirable. To deal with this, we developed a simple, high-throughput, PCR-based procedure, plate wash PCR (PWPCR), that facilitated the surveillance of isolation plates for the presence of target organisms and the ultimate recognition of colonies made up of target organisms. The results of this endeavor constitute the substance of the present paper.
MATERIALS AND METHODS
Sample collection and manipulation.
Soil samples were collected between August 2001 and October 2002 from the Long Term Ecological Research (LTER) site located at the Michigan State University W. K. Kellogg Biological Station (KBS) in Hickory Corners, Mich. The KBS-LTER site includes a large-scale replicate field experiment with treatments representing different cropping systems and types of management, several successional sites, and unmanaged forested sites (http://www.lter.kbs.msu.edu). Soil core (2-cm diameter) samples (to a depth of 10 cm) were taken from each of five permanent sampling stations distributed across one of four replicate fields (replicate 1) of the never cultivated successional treatment, which is representative of “native” soil.
Collected soil cores were stored at 4°C (usually for less than 48 h) until they were homogenized under a hypoxic, CO2-enriched atmosphere (2% O2, 5% CO2, 93% N2) contained within a flexible vinyl hypoxic chamber fitted with an oxygen sensor and controller (Coy Laboratory Products, Grass Lake, Mich.). Approximately 30 g of soil was added to 100 ml of phosphate-buffered saline (pH 7.0) containing 224 mM sodium pyrophosphate as a dispersal agent and 1 mM dithiothreitol as a reducing agent (47). The suspension was stirred vigorously for 30 min and allowed to settle for 30 min. An aliquot of the supernatant was serially diluted in the same buffer and spread onto various media with at least three replicate plates per dilution.
Termites, Reticulitermes flavipes (Kollar) (Rhinotermitidae), were collected near Dansville, Mich., and either used immediately or maintained in the laboratory as described previously (8, 35). The guts from 25 to 50 worker larvae were extracted under a hypoxic atmosphere (described above) with sterile forceps and pooled in a glass tissue homogenizer containing 2 ml of a sterile basal salts solution based on the freshwater medium of Widdel and Bak (49). The basal salts solution contained the following (per liter): KH2PO4, 0.2 g; NH4Cl, 0.25 g; KCl, 0.5 g; CaCl2 · 2H2O, 0.15 g; NaCl, 1.0 g; MgCl2 · 6H2O, 0.62 g; Na2SO4, 2.84 g; and morpholinepropanesulfonic acid (MOPS) (pH 7.0), 10 mM. After homogenization, the homogenate was diluted serially in the basal salts solution and spread onto various media.
The total numbers of microbes per gram (dry weight) of soil or per termite gut were determined by direct microscopic count after staining with 5-(4,6-dichlorotriazine-2-yl) aminofluorescein (DTAF) by the method of J. Bloem (4). Soil moisture content was determined by baking three replicate samples of soil at 80°C to a constant mass. Soil moisture content was then used with total direct counts to estimate the number of cells per gram (dry weight) of soil.
Cultivation conditions and screening.
The basal medium used for cultivation of soil bacteria was a modification of the basal salts solution described above and contained the following (per liter): KH2PO4, 0.2 g; NH4Cl, 0.25 g; KCl, 0.5 g; CaCl2 · 2H2O, 0.15 g; NaCl, 1.0 g; MgCl2 · 6H2O, 0.62 g; Na2SO4, 2.84 g; HEPES (pH 6.8), 10 mM; trace element solution (discussed below), 1 ml; vitamin B12 solution (50 mg/liter), 1 ml; mixed vitamin solution (discussed below), 1 ml; and Bacto Agar (Becton Dickinson and Company, Franklin Lakes, N.J.), 15 g. The final pH of the medium was adjusted to 6.8 to 7.0. The trace element stock solution contained the following (per liter): FeCl2 · 4H2O, 1.5 g; CoCl2 · 6H2O, 190 mg; MnCl2 · 4H2O, 100 mg; ZnCl2, 70 mg; H3BO3, 6 mg; Na2MoO4 · 2H2O, 36 mg; NiCl2 · H2O, 24 mg; CaCl2 · H2O, 2 mg; and HCl (25% [vol/vol]), 10 ml (49). The mixed vitamin stock solution contained the following (per liter): 4-aminobenzoic acid, 40 mg; d-(+)-biotin, 10 mg; nicotinic acid, 100 mg; calcium d(+)-pantothenate, 50 mg; pyridoxamine dihydrochloride, 100 mg; and thiamine dihydrochloride, 100 mg (49).
All solutions were heat sterilized, except for the trace element and mixed vitamin solutions, which were passed through a 0.22-μm-pore-size filter. Variations in the medium composition above included the incorporation of some or all of the following (per liter): a mixture of organic carbon substrates (yeast extract, Bacto Protease Peptone 3, Casamino Acids, and dextrose [Becton Dickinson]), 0.05 g each; catalase (bovine liver; Sigma-Aldrich, Inc., St. Louis, Mo.), 2,000 U (spread onto individual plates containing 30 ml of solidified medium just prior to inoculation) or 130,000 U (added to 1 liter of cooled, molten agar just prior to pouring plates); soil extract (44), 100 ml; disodium anthraquinone-2,6-disulfonate (AQDS), 2 g; and an N-acyl homoserine lactone (acyl-HSL) cocktail prepared in ethyl acetate acidified with 0.1% (vol/vol) acetic acid and containing N-(butyryl, heptanoyl, hexanoyl, β-ketocaproyl, octanoyl, and tetradecanoyl)-dl-homoserine lactones (Sigma-Aldrich, Inc.), each used at a final concentration of 1 μM in the media. The pH range of the prepared media was 5.9 to 6.4, depending upon medium composition. Termite gut microbes were cultivated with the same basal medium (described above) supplemented with sodium acetate (2.46 g/liter) and/or a combination of yeast extract and peptone (0.1 g/liter each).
Incubation atmospheres used were as follows: air (unamended); CO2-enriched (5% vol/vol) air; 2% O2, 5% CO2, and 93% N2 (hypoxic); or 5% CO2, 10% H2, and 85% N2 (anoxic). Cultures incubated in atmospheres other than in unamended air were performed in glass desiccator jars, the flexible vinyl hypoxic chamber (above), or a Plexiglas anoxic chamber (Plas-Labs, Inc., Lansing, Mich.). All cultures were maintained under low light conditions at room temperature (21 to 23°C).
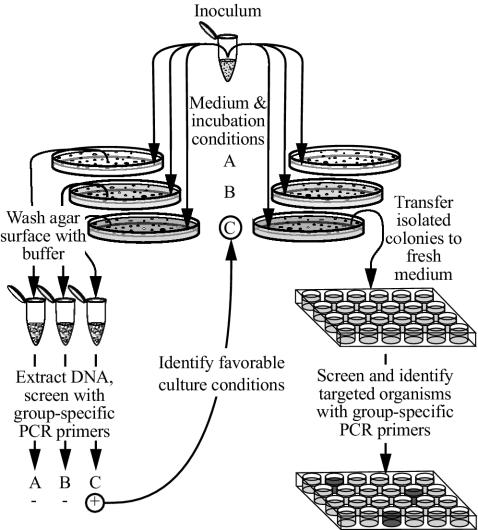
Primary screening for growth of target organisms was done after 30 days or more of incubation by sacrificing at least one replicate agar plate from selected treatments containing between 30 and 300 colonies and subjecting it to PWPCR with group-specific primers (below). Remaining plates from successful treatments were then used as a source of colonies. The organisms were picked individually or removed in groups by swabbing sectors of the plate for patching or streaking on homologous medium. For picking isolated colonies, many of which were invisible to the naked eye, the plates were held under a dissecting microscope and illuminated with cool white light from a fiber-optic illuminator positioned at an angle of about 45° from the horizontal plate surface. When subcultures were grown, individual colonies or defined pools of the colonies were again subjected to screening by the PCR with specific primers. This process was continued until individual colonies of target organisms were ultimately identified and obtained as pure cultures (Fig. 1).
FIG. 1.
PWPCR method to detect growth and monitor isolation of targeted bacteria. Of the three medium and incubation conditions shown in this diagram (conditions A, B, and C), growth of targeted bacteria (+) is represented only in condition C.
PCR and PWPCR.
PCR was performed with primers targeting regions of 16S rRNA-encoding genes common to nearly all bacteria or specific to the phyla Acidobacteria and Verrucomicrobia (Table 1). Unless otherwise stated, each 25-μl reaction mixture contained approximately 50 ng of template DNA, 1× reaction buffer (Invitrogen, Carlsbad, Calif.), 1.5 mM MgCl2, 0.25 mM concentration of each deoxynucleoside triphosphate, 0.2 μM concentration of each forward (F) and reverse (R) primer, and 0.625 U of Taq DNA polymerase (Invitrogen). PCR mixtures were incubated in a model PT-100 thermal cycler (MJ Research, Inc., Watertown, Mass.) for the following amplification schedule: (i) 3 min at 95.0°C; (ii) 30 cycles, with 1 cycle consisting of 30 s at 95.0°C, 30 s at the annealing temperature shown in Table 1, and 45 s at 72.0°C; and (iii) 10 min at 72.0°C.
TABLE 1.
PCR primers used in this study
| Primer | Positionsa | Sequence (5′→3′) | Annealing tempb | Reference or source |
|---|---|---|---|---|
| F2 | CAG TCA CGA CGT TGT AAA ACG ACG GC | 62.0 | 28 | |
| R4*c | CAG GAA ACA GCT ATG ACC ATG | 28 | ||
| Acd31F | 15-31 | GAT CCT GGC TCA GAA TC | 56.7 | 3 |
| Ver53F | 37-53 | TGG CGG CGT GGW TAA GA | 61.0 | D. Buckleyd |
| 1492R | 1492-1510 | GGT TAC CTT GTT ACG ACT T | 30 |
The positions of the target region are given using E. coli numbering system of the 16S rRNA gene. Primers F2 and R4* complement regions of the multiple cloning site of pCR2.1 or pCR4.0.
Annealing temperatures optimized for the conditions used in this study are given for the forward primer of each primer pair.
Primer R4* was modified from the primer in reference 28 by omission of five deoxynucleotides from the 5′ end.
Personal communication.
Preliminary experiments to determine the optimum PCR conditions with the Acidobacteria-targeting (Acd31F-1492R) and Verrucomicrobia-targeting (Ver53F-1492R) primer pairs were performed by using template DNA from Acidobacterium capsulatum (ATCC 51196) and Verrucomicrobium spinosum (ATCC 43997), respectively. Optimum reaction conditions were determined across a gradient of annealing temperatures (50 to 65°C) and MgCl2 concentrations (1 to 2.5 mM) by using a PTC-200 DNA Engine gradient thermocycler (MJ Research, South San Francisco, Calif.). Determining the sensitivity of target gene detection was performed by the PCRs with Acidobacteria-targeting primers and decreasing amounts of A. capsulatum DNA mixed with nontarget DNA (E. coli K-12) to yield 50 ng of total DNA per reaction mixture. Sensitivity was also determined by using the Verrucomicrobia-targeting primer set with decreasing amounts of genomic DNA from a termite-associated Verrucomicrobia division isolate TAV1 (described below) in a 1:2 mass ratio with E. coli K-12 DNA. Direct, group-specific PCR amplification of 16S rRNA genes in environmental samples was performed with group-specific primers (see above) and 50 ng of DNA from soil or from 50 termite guts. Genomic DNA was extracted using the Ultraclean soil or fecal DNA kits per the manufacturer's protocols (MoBio Laboratories, Carlsbad, Calif.).
PWPCR was simply the PCR in which template DNA was obtained from the aggregate of colonies present on an isolation plate (Fig. 1). To obtain template DNA from the aggregate of colonies present on an isolation plate, the surface of the agar medium was flooded with 2 ml of bead solution from the Ultraclean fecal DNA kit (MoBio Laboratories), and then a sterile spreader was used to suspend as much colony material as possible. The bead solution with suspended cells was transferred to a dry bead tube from the DNA kit, 50 μl of lysozyme solution (50 mg/ml; Sigma-Aldrich) was added, and the tube was incubated for 45 min in a 56°C water bath. After incubation, DNA extraction was performed according to the manufacturer's protocol, except that a Mini-BeadBeater-8 (BioSpec Products, Inc., Bartlesville, Okla.) operating at full speed for 45 s was used for physical disruption of the cells. The concentration and purity of each DNA sample were estimated from the absorbance at wavelengths from 220 to 320 nm (41).
The ability of PWPCR to detect a colony of a target organism from many nontarget organisms was examined by performing PWPCR on simulated isolation plates. A laboratory collection of 26 different bacterial isolates obtained from KBS-LTER soil (various α-, β-, and γ-Proteobacteria, Bacillus spp. [phylum Firmicutes], Arthrobacter spp. [phylum Actinobacteria], and Cytophaga- and Flavobacterium-like strains [phylum Bacteroidetes]) were each inoculated onto three or four sites (94 total) on plates of R2A agar (Difco, Detroit, Mich.), a medium commonly used for isolating environmental heterotrophs. Some plates were also inoculated with V. spinosum (ATCC 43997) at one site, and then all plates were incubated in air, at room temperature, and in the dark for 6 days. At the end of the incubation period, plates with (positive) and without (negative) V. spinosum were used for PWPCR individually, and after dilution of template DNA extracted from V. spinosum-positive plates, the plates were incubated with DNA extracted from V. spinosum-negative plates.
PCR products were analyzed by electrophoresis of 5-μl samples of reaction mixtures on 1% agarose gels at 100 V in 0.5× Tris-borate-EDTA. PCR products were visualized by UV illumination after staining with 1× Gelstar nucleic acid stain (Cambrex, East Rutherford, N.J.), and images were captured by using a Kodak electrophoresis documentation and analysis system 290 (Eastman Kodak).
Sequence determination and phylogenetic analyses.
PCR-amplified 16S rRNA genes from environmental samples, PWPCR, or bacterial isolates were cloned directly into E. coli using the plasmid vector pCR2.1 or pCR4.0 (TOPO TA cloning kit; Invitrogen). Restriction fragment length polymorphism analyses were used to identify common and unique clones. The partial sequence of each clone was determined with Applied Biosystems cycle sequencing technology (Applied Biosystems, Foster City, Calif.), the 16S rRNA gene primer 531R (5′-TAC CGC GGC TGC TGG CAC-3′), and/or vector primers. Preliminary phylogenetic affiliation of each clone was determined by sequence comparison to the GenBank nucleotide database using BLAST (2) or to the Ribosomal Database Project II database using the sequence match tool (18). Nearly full-length sequence (at least fourfold coverage) of the 16S rRNA gene from isolates and selected clones was obtained by using primers complementary to the multiple cloning site of pCR2.1 or pCR4.0 (F2 and R4* [see Table 1]); Acd31F, Ver53F, and 1492R (Table 1); and 338F (5′-CTC CTA CGG GAG GCA GCA GT-3′), 531R (above), 776F (5′-AGC AAA CAG GAT TAG ATA CCC TGG-3′), 810R (5′-GGC GTG GAC TTC CAG GGT ATC T-3′), and 1087F (5′-GGT TAA GTC CCG CAA CGA-3′) with the Applied Biosystems cycle sequencing technology and either an ABI Prism 3100 genetic analyzer or ABI Prism 3700 DNA analyzer (Applied Biosystems).
Contiguous sequences for each isolate were assembled with the Vector NTI software package (Informax). These sequences were inserted into and aligned against a 16S rRNA gene sequence database in the ARB software package (http://www.arb-home.de/) (32), along with any other available phylum-specific sequences (>500 nucleotides [nt]) from GenBank (http://www.ncbi.nlm.nih.gov/), the Ribosomal Database Project II (http://rdp.cme.msu.edu/) (18), or our own environmental clones. Aligned Acidobacteria and Verrucomicrobia sequences longer than 1,250 nt were used to generate phylogenetic trees using maximum-likelihood analysis based on 1,097 shared nucleotides for the Acidobacteria and 1,050 nucleotides for the Verrucomicrobia. The minimum evolutionary distance method in PAUP* was used for bootstrap analyses of the same data (46).
Treatment effects on cultivability.
In order to determine which, if any, treatments had a significant impact on overall cultivability or the cultivability of Acidobacteria from soil, colony counts (in CFU per gram [dry weight] of soil) and PWPCR results were compared for each treatment and used in a chi-square test for goodness of fit with Bonferroni's error rate adjustment (37, 42). Colonies used to determine colony counts had a minimum diameter of 0.2 mm and were visible using a colony counter fitted with a 1.5× magnifying lens. For overall cultivability, the average colony count was used as the expected value and that for a particular treatment was used as the observed value. For Acidobacteria cultivability, the expected value was the probability of detection using PWPCR among all treatments multiplied by the number of agar plates used for a given treatment, where as the observed value was the number of times Acidobacteria were detected for a particular treatment. A total of 63 treatments were screened for this analysis.
Nucleotide sequence accession numbers.
Partial 16S rRNA gene sequences (ca. 1,400 bases) from isolates KBS89, TAA43, TAA48, TAA166, TAV1, TAV2, TAV3, and TAV4 have been deposited in the EMBL, GenBank, and DDBJ nucleotide sequence databases under accession numbers AY587227 through AY587234.
RESULTS AND DISCUSSION
Specificities and sensitivities of PCR and PWPCR.
Only targeted 16S rRNA genes were amplified during PCR with group-specific primers in control reaction mixtures run in the presence of E. coli DNA or after PWPCR of a diverse collection of soil bacteria (Fig. 2). Amplification with the PCR mixture and as little as 16 pg of A. capsulatum DNA and 93.75 fg of V. spinosum DNA yielded a visible amplimer. The specificity of each primer set was also confirmed by sequence analysis of clones obtained after PCR amplification using soil or termite gut community DNA and after PWPCR of simulated or experimental isolation plates. Of more than 100 such clones examined, all corresponded to the 16S rRNA gene targeted by the primer pair.
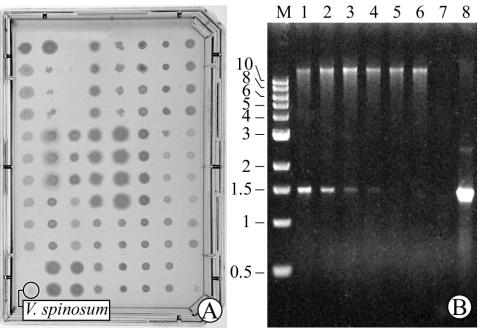
FIG. 2.
Detection of V. spinosum in a collection of diverse bacteria isolated from soil. (A) A single V. spinosum colony is shown among 94 other colonies growing on an agar plate. (B) PWPCR with Verrucomicrobia-specific primers using template in which V. spinosum colony material represented 1 part in 95 parts (i.e., plate in panel A; lane 1), 1 part in 189 parts (lane 2), 1 part in 471 parts (lane 3), 1 part in 941 parts (lane 4), and 1 part in 9,401 parts (lane 5). PWPCR of a control plate lacking V. spinosum (lane 6), negative control (no DNA; lane 7), V. spinosum DNA (50 ng; lane 8), and molecular size markers (lane M) is also shown. The positions of molecular size markers (in kilobases) are shown to the left of the gel.
By PWPCR, the equivalent of a single V. spinosum colony could be detected on plates from a background of at least 940 nontarget colonies composed of 26 different soil bacteria from six major phylogenetic groups (Fig. 2). Considering the small amount of V. spinosum colony material relative to that of the other bacteria, it should be possible to detect colonies of targeted microbes among a much larger number of nontarget colonies of similar size. Restriction fragment length polymorphism analysis revealed that only Verrucomicrobia-specific ribosomal DNA was amplified despite the diversity of nonspecific DNA in each sample (data not shown).
Treatment effects and isolation of Acidobacteria and Verrucomicrobia.
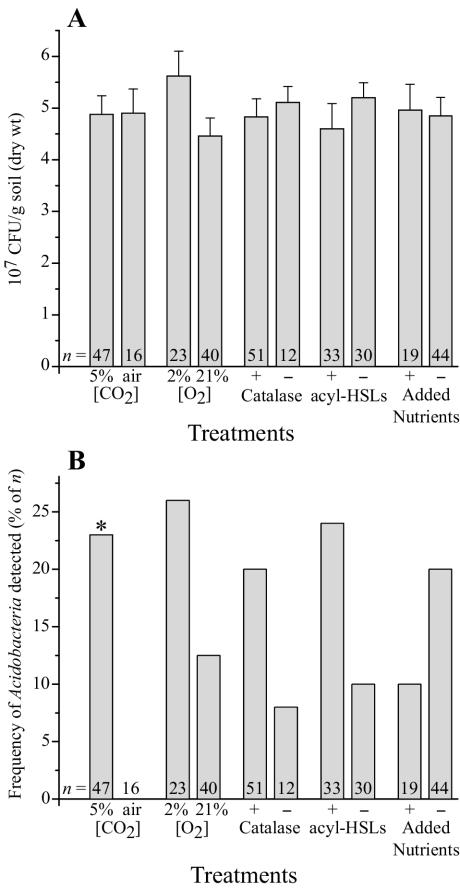
On the basis of direct microscopic counts, 1.41 × 109 ± 0.16 × 109 DTAF-stainable microbes (n = 3) were present in each gram (dry weight) of soil. In cultivation experiments, recoveries ranged from 4.0 × 107 to 9.7 × 107 CFU/g (dry weight) of soil, or roughly 4.0 to 7.0% of the total microbial community on the basis of direct counts. These recoveries were higher than the “1% or less” recoveries commonly cited but similar to those from other studies that have used low nutrient concentrations and long incubation times (19, 24). No single treatment significantly increased the overall recovery of soil bacteria (Fig. 3A), which suggests that the longer incubation times used for all experiments may be responsible for our higher recoveries of bacteria.
FIG. 3.
Influence of medium additives and incubation conditions on CFU recovered from soil (A) and on the occurrence of Acidobacteria among the isolates (B). “Air” is actually the concentration of CO2 (0.03% [vol/vol]) in normal air. Data in panel A represent the mean CFU recovered from soil among n samples. Error bars represent sample standard deviations. Data in panel B represent the frequency with which Acidobacteria were detected among the plates in panel A using PWPCR. These data were subjected to chi-square analyses with Bonferroni's error rate adjustment. Statistical significance (α = 0.10, df = 1) is indicated by an asterisk.
When PWPCR results were compiled from the same experiments, however, one treatment, used individually or in combination with other treatments, had a significant positive effect on the occurrence of soil Acidobacteria on plates: this was the presence of 5% CO2 in incubation atmospheres (Fig. 3B). Incubating the media in atmospheres with 5% CO2 resulted in a slight acidification (about half of a pH unit) and, therefore, could also be responsible for the increase in cultivation of Acidobacteria. Incubation of plates under hypoxia or supplementation of media with catalase or acyl-HSLs also tended to elicit a greater occurrence of Acidobacteria, whereas supplementation of media with an organic nutrient mixture appeared to have the opposite effect. The addition of humic acids in the form of soil extract or the humic acid analogue AQDS had no apparent effect on the occurrence of Acidobacteria (data not shown). While the increased values seen with these latter treatments were not statistically significant in this study, they may ultimately prove to be so if examined individually and systematically in a large-scale experiment.
PWPCR-based identification of primary isolation plates containing Acidobacteria enabled us to make an informed selection of companion treatment plates from which to prepare subcultures for additional PCR-based screening (see Materials and Methods). Ultimately, soil Acidobacteria strain KBS89 was isolated from soil that was plated on basal medium supplemented with catalase, acyl-HSLs, and a mixture of organic carbon substrates (described above) and incubated in CO2-enriched air.
A PWPCR-based strategy, similar to that used for the isolation of Acidobacteria from soil, was used for the isolation of previously uncultivated microbes from termite guts. The overall recovery of viable prokaryotes from termite guts (9.7% of the direct microscopic count) was marginally higher than that from soil, with an estimated 4.5 × 105 CFU per gut equivalent. As with primary isolation plates from soil inocula, PWPCR revealed the presence of Acidobacteria on some of the plates, and by using analogous procedures, Acidobacteria strains TAA43, TAA48, and TAA166 were subsequently isolated on plates containing basal medium supplemented with yeast extract and peptone (0.1% [wt/vol] each) and incubated in CO2-enriched air.
When Verrucomicrobia-specific primers were used, Verrucomicrobia were also detected by PWPCR on primary isolation plates of all compositions inoculated with termite gut homogenate and incubated in air and in a hypoxic atmosphere enriched with 5% CO2. Media used for cultivation of termite-associated microorganisms contained yeast extract and peptone with or without acetate and/or catalase. From such plates, four termite gut Verrucomicrobia (strains TAV1 through TAV4) were isolated, assisted by PWPCR surveillance of subcultures. TAV1 and TAV2 were isolated from plates containing basal medium with yeast extract, peptone, and acetate. TAV3 and TAV4 were isolated from the same medium without acetate. All TAV isolates were obtained from plates incubated in CO2-enriched air.
Properties of Acidobacteria and Verrucomicrobia isolates.
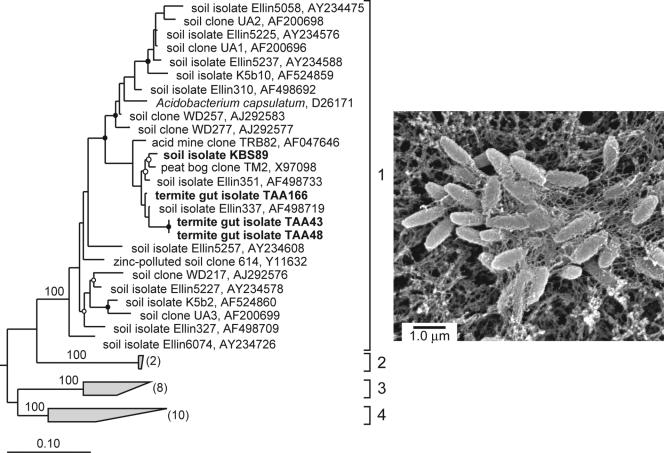
All of the Acidobacteria isolates belong to subdivision 1 of the Acidobacteria (Fig. 4) (23). On the basis of 16S rRNA gene sequence similarity, the nearest cultivated relatives to strains KBS89 and TAA166 are Ellin351 (97%) and Ellin337 (98%), respectively (40). TAA166 is the nearest cultivated relative to strains TAA43 and TAA48 (96.1%), which possess identical 16S rRNA sequences. All Acidobacteria isolates are short rods (0.5 by 1 μm) that divide via binary fission and form slightly opaque colonies after 4 or 5 days, which reach a maximum diameter of 1 mm in 14 to 16 days. Soil Acidobacteria isolate KBS89 and, to a lesser extent, the termite gut Acidobacteria isolates produce copious amounts of an extracellular (apparently capsular) material (Fig. 4), which made colonies hard to disrupt and was presumably responsible for their flocculent growth in liquid cultures.
FIG. 4.
(Left) Maximum-likelihood tree of subdivisions 1 to 4 of the phylum Acidobacteria based on 16S rRNA gene sequences from organisms in culture and PCR-generated clones from soil. Isolate designations and accession numbers are given. Isolates obtained in this study are shown in boldface type. Bootstrap values for branch points of the major subdivisions are given. Branch points conserved in all analyses with bootstrap values of >75% (closed circles) and bootstrap values of 50 to 74% (open circles) are indicated. Subdivisions 2 to 4 are labeled, bracketed, and condensed as shaded trapezia with the number of sequences represented in parentheses. 16S rRNA gene sequences of members of subdivisions 6 to 8 were used as outgroups (not shown). The scale bar represents 0.10 change per nucleotide. (Right) The scanning electron micrograph shows KBS89 cells trapped in an extracellular matrix.
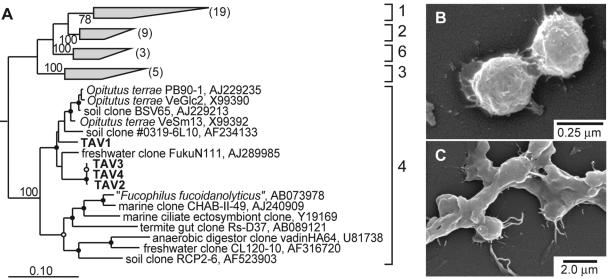
The termite-associated Verrucomicrobia isolates (TAV1 to TAV4) belong to subdivision 4 of the phylum Verrucomicrobia (Fig. 5A) (23). On the basis of 16S rRNA gene sequence similarity, the nearest cultivated relative to TAV1 is Opitutus terrae strain VeSm 13 (94.2%). Strains TAV2, TAV3, and TAV4 have 16S rRNA gene sequences virtually identical to each other, and the nearest cultivated relative to these isolates is Opitutus terrae strain PB90-1 (93%). TAV1 shares only 92.7% sequence similarity to the other TAV isolates. All of the termite-associated Verrucomicrobia isolates are facultative anaerobes, obtaining significantly higher population densities in liquid culture in CO2-enriched air and in an hypoxic atmosphere than under a CO2-enriched anoxic atmosphere. TAV cells are 0.25 to 0.50 μm in diameter and occur almost exclusively in pairs (Fig. 5B). Additionally, TAV1 produces an abundance of extracellular (apparently capsular) material (Fig. 5C). The TAV isolates were detected on primary isolation plates after 30 days and on subculture plates after 14 days. On the original isolation media, they formed very small (<0.5 mm in diameter), white, round, mucoid colonies that were visible only with a dissecting microscope. After isolation and several passages in the laboratory, however, all TAV isolates formed larger colonies (2- to 4-mm diameter) in 2 to 5 days on R2A agar in air. Preliminary results from studying the distribution and abundance of these targeted phylogenetic groups suggest that Verrucomicrobia are autochthonous to the guts of R. flavipes and not allochthonous contaminants derived from soil, whereas the opposite is true for the Acidobacteria (J. T. Wertz, B. S. Stevenson, and J. A. Breznak, Abstr. 103rd Gen. Meet. Am. Soc. Microbiol. 2003, abstr. N-223, 2003).
FIG. 5.
(A) Maximum-likelihood tree of subdivisions 1 to 4 and 6 of the phylum Verrucomicrobia based on 16S rRNA gene sequences from organisms in culture and PCR-generated clones from environmental samples. Isolate designations and accession numbers are given. Isolates obtained in this study are shown in boldface type. Bootstrap values for branch points of the major subdivisions are given. Branch points conserved in all analyses with bootstrap values of >75% (closed circles) and bootstrap values of 50 to 74% (open circles) are indicated. Subdivisions 1 to 3 and 6 are labeled, bracketed, and condensed as shaded trapezia with the number of represented sequences in parentheses to simplify presentation of the tree. 16S rRNA gene sequences of members of the phylum Planctomycetes were used as an outgroup (not shown). The scale bar represents 0.10 change per nucleotide. (B and C) The scanning electron micrograph of TAV2 (B) shows the doublet cell morphology shared by all TAV isolates, and that of TAV1 (C) shows the encapsulation of the cells in an extracellular matrix, a morphological feature not shared by TAV2, TAV3, or TAV4.
Overview of the PWPCR-based isolation procedure.
Given the variety of individual cultivation treatments and treatment combinations used in this study, as well as the various sources of inocula, the detection and isolation of Acidobacteria and Verrucomicrobia would have been extremely difficult without the PWPCR method. One of the most time-consuming aspects of any isolation procedure is the screening, selecting, and subculture of colonies from primary isolation plates, and if low nutrient conditions are used to prevent overgrowth by nondesired organisms, most colonies on such plates will be fairly small. Indeed, colonies of the Acidobacteria and Verrucomicrobia strains isolated in this study would have been easily overlooked without the aid of a dissecting microscope. However, the PWPCR procedure economizes on time by directing one to treatment plates known to contain the target organism(s). Hence, owing to its simplicity, utility, and relatively low cost, we anticipate that PWPCR will become widely used as an adjunct to creative approaches for isolation of novel, sought-after organisms. The only requirement is at least one specific and reliable primer in the pair of primers used for PCR.
As with any method, PWPCR also has some limitations. For PWPCR, the sought-after organisms must be capable of growth on plates solidified with agar (or an agar substitute) and also capable of being harvested from such plates. This would eliminate organisms that either cannot grow on solid media or that, like certain spirochetes (7) and spirilla (16), form largely subsurface colonies difficult to harvest by simple plate washing. However, the key element of PWPCR is PCR with a specific primer pair, so as long as sufficient cell material can be obtained to make a DNA template, either by harvesting cells from liquid cultures or removing subsurface colony material by coring, surveillance of cultures is possible. Thus, our results underscore the power of integrating various cultivation conditions with molecular biology to retrieve some of the “not-yet-cultured majority” of microbes on our planet (6, 39).
Acknowledgments
This work was supported in part by grants from the U.S. Department of Agriculture (2001-35107-09939) and National Science Foundation (MCB-0135880 and IBN-0114505).
We thank Les Dethlefsen for assistance with the statistical analyses.
REFERENCES
- 1.Aagot, N., O. Nybroe, P. Nielsen, and K. Johnsen. 2001. An altered Pseudomonas diversity is recovered from soil by using nutrient-poor Pseudomonas-selective soil extract media. Appl. Environ. Microbiol. 67:5233-5239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Barns, S. M., S. L. Takala, and C. R. Kuske. 1999. Wide distribution and diversity of members of the bacterial kingdom Acidobacterium in the environment. Appl. Environ. Microbiol. 65:1731-1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bloem, J. 1995. Fluorescent staining of microbes for total direct counts, section 4.1.8, p. 1-12. In A. D. L. Akkermans, J. D. van Elsas, and F. J. de Bruijn (ed.), Molecular microbial ecology manual. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 5.Bogosian, G., N. D. Aardema, E. V. Bourneuf, P. J. L. Morris, and J. P. O'Neil. 2000. Recovery of hydrogen peroxide-sensitive culturable cells of Vibrio vulnificus gives the appearance of resuscitation from a viable but nonculturable state. J. Bacteriol. 182:5070-5075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Breznak, J. A. 2002. A need to retrieve the not-yet-cultured majority. Environ. Microbiol. 4:4-5. [DOI] [PubMed] [Google Scholar]
- 7.Breznak, J. A., and E. Canale-Parola. 1975. Morphology and physiology of Spirochaeta aurantia strains isolated from aquatic habitats. Arch. Microbiol. 105:1-12. [DOI] [PubMed] [Google Scholar]
- 8.Breznak, J. A., and H. S. Pankratz. 1977. In situ morphology of gut microbiota of wood-eating termites [Reticulitermes flavipes (Kollar) and Coptotermes formosanus Shiraki]. Appl. Environ. Microbiol. 33:406-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bruns, A., H. Cypionka, and J. Overmann. 2002. Cyclic AMP and acyl homoserine lactones increase the cultivation efficiency of heterotrophic bacteria from the central Baltic Sea. Appl. Environ. Microbiol. 68:3978-3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bruns, A., U. Nubel, H. Cypionka, and J. Overmann. 2003. Effect of signal compounds and incubation conditions on the culturability of freshwater bacterioplankton. Appl. Environ. Microbiol. 69:1980-1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buckley, D. H., and T. M. Schmidt. 2003. Diversity and dynamics of microbial communities in soils from agro-ecosystems. Environ. Microbiol. 5:441-452. [DOI] [PubMed] [Google Scholar]
- 12.Buckley, D. H., and T. M. Schmidt. 2001. The structure of microbial communities in soil and the lasting impact of cultivation. Microb. Ecol. 42:11-21. [DOI] [PubMed] [Google Scholar]
- 13.Bussmann, I., B. Philipp, and B. Schink. 2001. Factors influencing the cultivability of lake water bacteria. J. Microbiol. Methods 47:41-50. [DOI] [PubMed] [Google Scholar]
- 14.Button, D. K., B. R. Robertson, and P. Quang. 2001. Isolation of oligobacteria. Methods Microbiol. 30:161-173. [Google Scholar]
- 15.Button, D. K., F. Schut, P. Quang, R. Martin, and B. R. Robertson. 1993. Viability and isolation of marine bacteria by dilution culture: theory, procedures, and initial results. Appl. Environ. Microbiol. 59:881-891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Canale-Parola, E., S. L. Rosenthal, and D. G. Kupfer. 1966. Morphological and physiological characteristics of Spirillum gracile sp. n. Antonie Leeuwenhoek 32:113-124. [DOI] [PubMed] [Google Scholar]
- 17.Chin, K. J., D. Hahn, U. Hengstmann, W. Liesack, and P. H. Janssen. 1999. Characterization and identification of numerically abundant culturable bacteria from the anoxic bulk soil of rice paddy microcosms. Appl. Environ. Microbiol. 65:5042-5049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cole, J. R., B. Chai, T. L. Marsh, R. J. Farris, Q. Wang, S. A. Kulam, S. Chandra, D. M. McGarrell, T. M. Schmidt, G. M. Garrity, and J. M. Tiedje. 2003. The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 31:442-443. [DOI] [PMC free article] [PubMed]
- 19.Connon, S. A., and S. J. Giovannoni. 2002. High-throughput methods for culturing microorganisms in very-low-nutrient media yield diverse new marine isolates. Appl. Environ. Microbiol. 68:3878-3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Felske, A., and A. D. L. Akkermans. 1998. Prominent occurrence of ribosomes from an uncultured bacterium of the Verrucomicrobiales cluster in grassland soils. Lett. Appl. Microbiol. 26:219-223. [DOI] [PubMed] [Google Scholar]
- 21.Felske, A., W. M. de Vos, and A. D. L. Akkermans. 2000. Spatial distribution of 16S rRNA levels from uncultured acidobacteria in soil. Lett. Appl. Microbiol. 31:118-122. [DOI] [PubMed] [Google Scholar]
- 22.Hattori, T. 1981. Enrichment of oligotrophic bacteria at microsites of soil. J. Gen. Appl. Microbiol. 27:43-55. [Google Scholar]
- 23.Hugenholtz, P., B. M. Goebel, and N. R. Pace. 1998. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 180:4765-4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Janssen, P. H., P. S. Yates, B. E. Grinton, P. M. Taylor, and M. Sait. 2002. Improved culturability of soil bacteria and isolation in pure culture of novel members of the divisions Acidobacteria, Actinobacteria, Proteobacteria, and Verrucomicrobia. Appl. Environ. Microbiol. 68:2391-2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Joseph, S. J., P. Hugenholtz, P. Sangwan, C. A. Osborne, and P. H. Janssen. 2003. Laboratory cultivation of widespread and previously uncultured soil bacteria. Appl. Environ. Microbiol. 69:7210-7215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaeberlein, T., K. Lewis, and S. S. Epstein. 2002. Isolating “uncultivable” microorganisms in pure culture in a simulated natural environment. Science 296:1127-1129. [DOI] [PubMed] [Google Scholar]
- 27.Kargalioglu, Y., and J. A. Imlay. 1994. Importance of anaerobic superoxide dismutase synthesis in facilitating outgrowth of Escherichia coli upon entry into an aerobic habitat. J. Bacteriol. 176:7653-7658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim, K. S., T. G. Lilburn, M. J. Renner, and J. A. Breznak. 1998. arfI and arfII, two genes encoding α-l-arabinofuranosidases in Cytophaga xylanolytica. Appl. Environ. Microbiol. 64:1919-1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krieg, N. R., and P. S. Hoffman. 1986. Microaerophily and oxygen toxicity. Annu. Rev. Microbiol. 40:107-130. [DOI] [PubMed] [Google Scholar]
- 30.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley and Sons, New York, N.Y.
- 31.Leadbetter, J. R., T. M. Schmidt, J. R. Graber, and J. A. Breznak. 1999. Acetogenesis from H2 plus CO2 by spirochetes from termite guts. Science 283:686-689. [DOI] [PubMed] [Google Scholar]
- 32.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K. H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mitsui, H., K. Gorlach, H. J. Lee, R. Hattori, and T. Hattori. 1997. Incubation time and media requirements of culturable bacteria from different phylogenetic groups. J. Microbiol. Methods 30:103-110. [Google Scholar]
- 34.Mizunoe, Y., S. N. Wai, A. Takade, and S. Yoshida. 1999. Restoration of culturability of starvation-stressed and low-temperature-stressed Escherichia coli O157 cells by using H2O2-degrading compounds. Arch. Microbiol. 172:63-67. [DOI] [PubMed] [Google Scholar]
- 35.Odelson, D. A., and J. A. Breznak. 1983. Volatile fatty acid production by the hindgut microbiota of xylophagous termites. Appl. Environ. Microbiol. 45:1602-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O'Farrell, K. A., and P. H. Janssen. 1999. Detection of verrucomicrobia in a pasture soil by PCR-mediated amplification of 16S rRNA genes. Appl. Environ. Microbiol. 65:4280-4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ott, R. L., and M. Longnecker. 2001. An introduction to statistical methods and data analysis. Duxbury, Pacific Grove, Calif.
- 38.Paul, E. A., and F. E. Clark. 1996. Soil microbiology and biochemistry, 2nd ed. Academic Press, San Diego, Calif.
- 39.Rappe, M. S., and S. J. Giovannoni. 2003. The uncultured microbial majority. Annu. Rev. Microbiol. 57:369-394. [DOI] [PubMed] [Google Scholar]
- 40.Sait, M., P. Hugenholtz, and P. H. Janssen. 2002. Cultivation of globally distributed soil bacteria from phylogenetic lineages previously only detected in cultivation-independent surveys. Environ. Microbiol. 4:654-666. [DOI] [PubMed] [Google Scholar]
- 41.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed., vol. 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 42.Samuels, M. L. 1989. Statistics for the life sciences. Dellen Publishing Company, San Francisco, Calif.
- 43.Schut, F., E. J. Devries, J. C. Gottschal, B. R. Robertson, W. Harder, R. A. Prins, and D. K. Button. 1993. Isolation of typical marine bacteria by dilution culture: growth, maintenance, and characteristics of isolates under laboratory conditions. Appl. Environ. Microbiol. 59:2150-2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stackebrandt, E., and H. Prauser. 1991. The family Cellulomodaceae, p. 1323-1345. In A. Balows, H. G. Trüper, M. Dworkin, W. Harder, and K.-H. Schleifer (ed.), The prokaryotes, vol. 4. Springer-Verlag, New York, N.Y. [Google Scholar]
- 45.Suwa, Y., and T. Hattori. 1987. Detection of proliferating bacteria in soil populations by the analysis of colony-forming curves. Soil Sci. Plant Nutr. 33:511-515. [Google Scholar]
- 46.Swofford, D. L. 2000. PAUP*: phylogenetic analysis using parsimony (and other methods), version 4.0. Sinauer Associates, Sunderland, Mass.
- 47.van Elsas, J. D., and K. Smalla. 1997. Methods for sampling soil microbes, p. 383-390. In C. J. Hurst, G. R. Knudsen, M. J. McInerney, L. D. Stetzenbach, and M. V. Walter (ed.), Manual of environmental microbiology. ASM Press, Washington, D.C.
- 48.Vandekerckhove, T. T. M., A. Willems, M. Gillis, and A. Coomans. 2000. Occurrence of novel verrucomicrobial species, endosymbiotic and associated with parthenogenesis in Xiphinema americanum-group species (Nematoda, Longidoridae). Int. J. Syst. Evol. Microbiol. 50:2197-2205. [DOI] [PubMed] [Google Scholar]
- 49.Widdel, F., and F. Bak. 1991. Gram negative mesophilic sulfate-reducing bacteria, p. 3352-3378. In A. Balows, H. G. Trüper, M. Dworkin, W. Harder, and K.-H. Schleifer (ed.), The prokaryotes, 2nd ed., vol. 4. Springer-Verlag, New York, N.Y. [Google Scholar]
- 50.Zengler, K., G. Toledo, M. Rappe, J. Elkins, E. J. Mathur, J. M. Short, and M. Keller. 2002. Cultivating the uncultured. Proc. Natl. Acad. Sci. USA 99:15681-15686. [DOI] [PMC free article] [PubMed] [Google Scholar]