Abstract.
We performed stimulated emission depletion (STED) imaging of isolated olfactory sensory neurons (OSNs) using a custom-built microscope. The STED microscope uses a single pulsed laser to excite two separate fluorophores, Atto 590 and Atto 647N. A gated timing circuit combined with temporal interleaving of the different color excitation/STED laser pulses filters the two channel detection and greatly minimizes crosstalk. We quantified the instrument resolution to be and , for the Atto 590 and Atto 647N channels. The spatial separation between the two channels was measured to be under 10 nm, well below the resolution limit. The custom-STED microscope is incorporated onto a commercial research microscope allowing brightfield, differential interference contrast, and epifluorescence imaging on the same field of view. We performed immunolabeling of OSNs in mice to image localization of ciliary membrane proteins involved in olfactory transduction. We imaged -permeable cyclic nucleotide gated (CNG) channel (Atto 594) and adenylyl cyclase type III (ACIII) (Atto 647N) in distinct cilia. STED imaging resolved well-separated subdiffraction limited clusters for each protein. We quantified the size of each cluster to have a mean value of and , for CNG and ACIII, respectively. STED imaging showed separated clusters that were not resolvable in confocal images.
Keywords: microscopy, stimulated emission depletion, super-resolution, neuroscience, olfaction
1. Introduction
For over a century of optical microscopy, the diffraction of light limited the study of cellular structures to a resolution of . Ernst Abbe predicted the resolution limit in his famous equation, showing dependence on the optical wavelength () and the numerical aperture of the collection optics.1 Recent advances in super-resolution (SR) microscopy techniques have opened up new opportunities for overcoming this limit, allowing imaging in biological samples at resolutions on the order of . There are now several widely used SR techniques including stimulated emission depletion (STED),2,3 photo-activated localization microscopy (PALM),4 stochastic optical reconstruction microscopy (STORM),5 and structured illumination microscopy (SIM).6 Other SR techniques have also been developed including microlenses7 for white light imaging or contact microspheres for fluorescence imaging of biological samples.8
STED is a powerful SR technique that allows fast, live cell, and even live mouse brain imaging at resolutions of to 60 nm, and has made it a key tool for neuroscience studies.9–11 Unlike other SR modalities, STED requires no postprocessing to the images as in PALM, STORM, or SIM where the exact resolution of the image can often be difficult to determine. One of the limitations of STED microscopy compared with other SR techniques is that it requires higher laser powers than standard fluorescence imaging to saturate the stimulated emission transition. High STED laser powers can potentially lead to sample heating, photobleaching, and photodamage that can pose a problem when imaging.
In this paper, we present details on a custom-built, two-color STED microscope for imaging biological samples. This STED microscope uses a similar design to a previously reported system12 including a single laser generating excitation and STED beams for each color, and with dedicated, multiplexed channels. Unlike the previously reported design, our STED microscope is built on an inverted research microscope with capabilities for widefield epifluorescence, brightfield, and differential interference contrast (DIC) imaging. Our STED design improves the ease of use and assists with aligning biological samples on regions of interest (ROI). Additionally, different imaging modalities can be performed on the same region. The custom-built two-color STED microscope operates in a shared user facility on a medical campus. The custom platform has the benefit of allowing future additions and modifications as SR technology continues to improve.
We quantify the instrument resolution, cross talk, and the spatial overlap for the two-color channels. We demonstrate two-color SR imaging of isolated olfactory sensory neurons (OSN) to investigate protein organization important for transduction of odorant signals to generate action potentials for transmission of information to the olfactory bulb. We performed immunofluorescence labeling to visualize the cyclic nucleotide gated (CNG) channel (Atto 594), adenylate cyclase type III (ACIII) (Atto 647N), and acetylated tubulin (Alexa 488).
The fundamental concept for STED is described in detail in several publications.13 In STED microscopy, the point spread function (PSF) of the microscope is reduced in size using an additional laser beam at a wavelength in the emission spectrum of the fluorophore. The STED laser first passes through a phase mask that imparts a vortex phase from 0 to on the beam. At the focus of the microscope, the beam is shaped into a “donut” profile with zero intensity at the center. The intensity in the STED beam drives a process of stimulated emission that suppresses fluorescence.14 As the STED laser saturates the electronic transition, the effective point spread function of the microscope decreases according to15
represents the saturation intensity whereby half the excited molecules are depleted to the ground state and is the maximum STED intensity.
According to the above equation, resolution improves as one increases the STED laser power; however, in practice resolution is limited by photobleaching and sample damage. Recently, methods have been developed to reduce photobleaching and sample photo-damage, including using photo-switchable fluorophores to lower the required laser power.16,17 STED imaging at extremely fast frame rates () also helps with photobleaching as it allows the fluorophore to decay to the ground state before re-excitation.18
2. Methods
2.1. Custom Two-Color Stimulated Emission Depletion Microscope
The two-color custom STED microscope was constructed based on previous designs, with modifications to couple it with a commercial Olympus IX-71 microscope platform, simplifying the process of aligning the sample to the exact region of interest to be imaged. Additionally, we are able to perform widefield epi-fluorescence imaging of the two color channels or an additional GFP channel prior to STED imaging. This feature is particularly useful for sparse samples such as the olfactory cilia in dissociated olfactory sensory neurons. Because only two laser beam paths are used for alignment instead of four, the daily alignment is no more complex than for a traditional one color STED microscope. With a custom STED system we have the option in the future to make modifications, such as adding axial resolution enhancement for imaging into samples with z-structure,19 use of a spatial light modulator to correct for aberrations,20 or implementing STED-fluorescence correlation spectroscopy.21 Another modification option is to use two-photon excitation so that we can image deeper into tissue.22 Additionally, we can incorporate future advancements in STED microscopy as they become available.
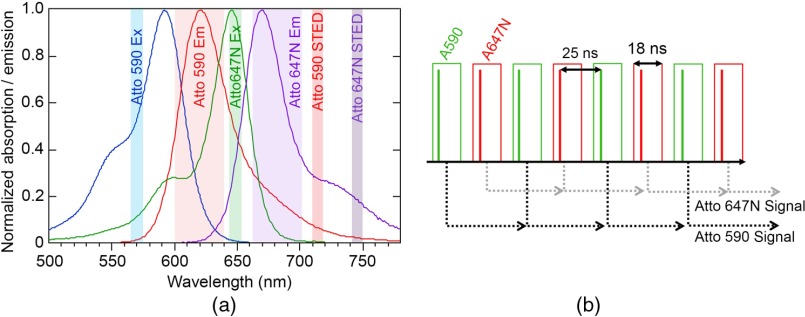
The laser source is a fiber-based laser (ALP-710-745-SC, Fianium Ltd.) operating at a repetition rate of 20 MHz as the master oscillator. The laser has two pulsed narrow-spectrum (FWHM of ) outputs centered at and (500 and 480 mW maximum average power, respectively) for the STED beams, and a pulsed supercontinuum output ranging from 400 to 2000 nm, with pulses (1.64 W total average power) that generates the excitation sources. All outputs originate from the same master oscillator and are therefore time-locked. The excitation/STED wavelengths can be used with the fluorophores Atto 590/STAR 580/Atto 594/Dylight 594 and Atto 647N/STAR 635/STAR 635P (ATTO-TEC GmbH, Abberior GmbH, Thermo Fisher Scientific Inc.). Figure 1(a) shows the absorption and emission spectra of Atto 590 and Atto 647N and the excitation and STED laser wavelengths used in our system. Minimizing the crosstalk between the two-color channels requires careful selection of filters and dichroics combined with temporal gating for simultaneous two-channel imaging, to be discussed in further detail in Sec. 2.2.
Fig. 1.
(a) Absorption and emission spectra of Atto 590 and Atto 647N, the two colors used in the STED, are shown, along with the excitation and STED laser wavelengths used. Emission detection bands shown are 600 to 640 nm for Atto 590 and 660 to 700 nm for Atto 647N. (b) Interleaved excitation/STED pulses for each color in the time domain and an illustration of the temporal gated collection are shown. Green boxes indicate the gates for the Atto 590 signal and the red boxes indicate the gates for the Atto 647N signal. The output signal from each gating circuit is sent to the computer.
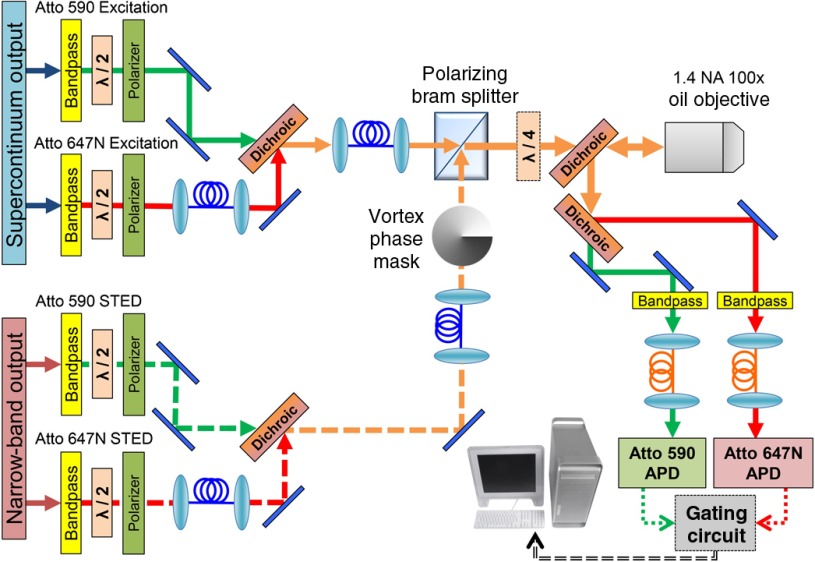
Figure 2 shows the schematic for our STED microscope. The STED beams originate from separate output ports of the laser and are first optically filtered with a bandpass filter (Z710/10x, Chroma Technology) for Atto 590 and two filters (H745/45M and LP02-785RE-25, Chroma Technology), for Atto 647N. The supercontinuum output is split into two linearly polarized beams using a polarizing beamsplitter, after which a bandpass filter generates the excitation wavelengths, Z570/10x (Chroma Technology) and HQ647/10x (Chroma Technology). The maximum power in each excitation beam is 1.78 and 2.36 mW for Atto 590/Atto 647N. Each beam in the system passes through a half-wave plate (Thorlabs Inc.) in a computer-controlled rotation mount (ACONEX-AG-PR100P, Newport Corporation) and a polarizer for control of the power and to ensure clean polarization before coupling into the polarization maintaining (PM) fibers. Temporal interleaving of the excitation/STED pulses for the two channels allows simultaneous imaging with minimal crosstalk, shown in Fig. 1(b). Here, the STED and excitation pulses for Atto 590 arrive at a 25 ns delay from the STED and excitation pulses for Atto 647N. Extra fiber is used to achieve the correct time separation for the different beams. A single PM fiber (P5-630PM-FC-2, Thorlabs Inc.) couples both STED beams while another PM fiber (P5-488PM-FC-2, Thorlabs Inc.) couples both excitation beams from the laser to the microscope. The inputs of the fibers are polished at a small angle to avoid back reflection into the laser. At the exit of the fibers achromatic lenses (49-664, Edmund Optics Inc.) collimate the output light. In addition, both STED beams pass through the same vortex phase plate (VPP-1a, RPC Photonics Inc.). With both excitation beams aligned together in a single fiber, they are automatically overlapped spatially, similarly with both STED beams, greatly simplifying the alignment process to overlap all four beams at the focus. All mirrors and dichroics used in the beam paths from the PM fibers to the microscope objective are on 6 mm thickness substrates to ensure the preservation of the spatial phase of the STED beams.
Fig. 2.
Optical schematic for the two-color STED microscope. Each beam is bandpass filtered and then sent through a half-wave plate and polarizer in succession, allowing clean polarization and individual power control. Excitation beams are combined into one polarization maintaining (PM) fiber and the STED beams are combined into another PM fiber using dichroic mirrors. STED beams pass through a vortex phase plate to generate a donut shape at the focus of the objective. A polarizing beamsplitter combines the beams and a quarter-wave plate makes the polarization circular. The beams are sent through a custom dichroic, which passes the four STED/excitation laser beams and reflects the fluorescence emission. The two emission bands are then separated using another dichroic beamsplitter. Fluorescence is focused into a multimode fiber that is coupled to an APD. The signal from the APDs goes through the custom gating circuit to counter inputs on the data acquisition (DAQ) board in the computer. The same DAQ board also controls the scanning of the piezo stage to acquire an image.
The beams pass through a custom dichroic beamsplitter designed for 15 deg angle of incidence (Chroma Technology), that has high transmission for STED/excitation wavelengths from 520 to 580 nm, 637 to 657 nm, and 700 to 770 nm and high reflection for emission wavelengths from 580 to 637 and 657 to 700 nm. After the dichroic, all beams pass through a quarter-wave plate (RAC 4.4.15, Bernhard Halle) to impart circular polarization and are then directed into the bottom port of an inverted research microscope (IX-71, Olympus, Inc.). A high NA objective (100X, 1.4 NA UPlanSApo Oil, Olympus Inc.) focuses all four beams onto the sample. The fluorescence emission is collected by the objective and separated by the custom dichroic.
The two-color channels are spectrally separated using a dichroic (T650lpxr, Chroma Technology) and emission filters remove any additional background (Atto 647N: HQ660LP, HQ675/50M and HQ620/40M, Atto 590: HQ680/40M-HC, Chroma Technology) before coupling into two core multimode fibers (M31L01, Thorlabs Inc.). Each multimode fiber serves as both a confocal pinhole and couples the light to an avalanche photodiode (APD) detector (SPCM-AQRH-13-FC, Perkin Elmer, Inc.). The image is constructed using a high-resolution three-dimensional (3-D) piezoelectric stage that scans the sample with a maximum travel and travel (P-733.3DD, Physik Instrumente) with an analog/digital controller (E-725.3CDA, Physik Instrumente). A image with 10(20) nm pixels is acquired in 79(38) s. The stage is controlled from the computer’s data acquisition board (PCIe-6353, National Instruments, Inc.). The piezo stage is mounted to a motorized stage (MD5422LOU, Physik Instrumente) connected to a controller (C-867.260, Physik Instrumente) that scans the sample at lower resolutions with a joystick, for locating the region of the sample of interest for imaging. The user is able to align the sample using widefield epifluorescence, brightfield, and/or DIC imaging. For epifluorescence imaging, we use a light source (Lumen 200, Prior Scientific Inc.) that has higher emission at red wavelengths to visualize Atto 647N. A CCD camera (Axiocam MRm, Carl Zeiss Microscopy GmbH) on the right side port is used to visualize the fluorescence that is mostly outside visible range for far-red fluorophores.
The system requires several precision alignment steps for reliable STED imaging:
-
•
Ensure clean linear polarization out of the PM fibers by optimizing the alignment of the polarizer with the axis of the PM fiber for each laser beam.
-
•
Remove the objective and use a collimated alignment laser propagating backwards from the objective lens position to the PM fibers to align the mirrors in the optical path so light is hitting the center of the fiber. The above steps need to be done only during the initial setup.
-
•
Ensure the two STED beams are circularly polarized after the quarter-wave plate to the highest possible degree. In reality, the quarter-wave plate is not exactly optimized for both wavelengths, but is rotated to an angle in between the optimal for each. It may be necessary to rotate the wave plate around an axis perpendicular to the table to obtain optimal circular polarization.
-
•
Precisely set the timing of the excitation and STED pulses for efficient STED. We accomplish this with a fast photodiode and oscilloscope (Newport AD-70xr, 6 GHz detector with Agilent Infiniium 54831B DSO, 600 MHz, oscilloscope), adjusting the delay using translation stages and delay fibers. The pulses should be interleaved with proper timing and the excitation and STED pulses for each color must overlap in time.
-
•
Optimize the STED donut mode at the focus. A 45% reflective pellicle beam splitter (BP145B1, Thorlabs Inc.) is placed before the quarter-wave plate to direct back scattered light from a sample of 80 nm gold beads onto a photomultiplier (PMM02, Thorlabs Inc.). Visualization of the shape of the STED beam at the focus is performed by repeated scans in , , or across a single gold bead while looking at the intensity traces from the photomultiplier tube. Typically both the position of the vortex phase plate and the steering of the beam into the microscope are adjusted to make the STED donut mode as cylindrically symmetric in and linearly symmetric in as possible. It is essential that the null in the center of the donut mode is as close as possible to zero intensity. If it is far from zero after optimization, try flipping the phase mask around.
-
•
Align the excitation beams so they overlap well with the null of the donut STED beams at the focus and check that the and scans are symmetric in . The axial focus of the beams must also be aligned in . Since the separation of the excitation wavelengths is , chromatic aberration can cause a shift in the foci. Use an appropriate collimation lens that allows both excitation beams to be focused at a similar -position when used in combination with the objective lens.
-
•
Align the detection beam path by measuring the fluorescence signal of a dilute dye sample. Maximize the signal on each APD independently by adjusting the mirrors that direct the emission light onto the multimode fibers. If no fluorescence is detected, light can be sent through the multimode fiber and then checked for overlap at the focus by backscattering from the gold beads sample as a course method to adjust the beam path to the detectors.
2.2. Resolution and Cross-Talk of the Two-Color Stimulated Emission Depletion Microscope
A custom time-gated detection circuit and pulse interleaving between the two excitation/STED beams for the two colors is required to minimize crosstalk between channels. It is important to image in both colors simultaneously rather than sequentially because a small spatial drift error between scans can be significant when compared to the high spatial resolution of the system. Without time gating, the 711-nm STED laser (for the Atto 590 channel) causes excitation of Atto 647N dye. The excitation wavelengths for both channels can also excite fluorescence for either dye due to overlap in the absorption spectra. Therefore, simultaneous two-color SR imaging requires electronically time-gated detection.
The laser emits pulses every 50 ns (20-MHz repetition rate), while the fluorescence lifetime of Atto 590/Atto 647N is . Thus, most of the fluorescence emission photons arrive in the 25 ns between the interleaved excitation pulses. The gating circuit selectively passes photon counts that arrive within after the excitation pulse to the corresponding counter on the DAQ board, while suppressing counts outside that time window. Electronic gating of counts is accomplished using two high-speed logical AND gates, which gate the signals from each APD with an adjustable duration and delay. The gating pulses are triggered by a common 20-MHz laser-clock signal but are otherwise independently adjustable. Each circuit has an 8-bit variable-pulse generator and a 120-ns variable delay line (3D7608 and DDU37F, Data Delay Devices Inc.). The temporal duration and delay of each gating signal are tuned while monitoring the counts from diluted dye samples to maximize the expected fluorescence signal from each channel while minimizing crosstalk.
2.3. Computational Methods for Characterizing Resolution
Custom Matlab (Mathworks, Inc.) routines were used to analyze images. Images of bead samples were first thresholded and a peak-finding routine was used to identify the locations for each bead. ROIs were defined around each bead. A least squares fit to a two-dimensional (2-D) Gaussian function was performed and the center-of-mass position and FWHM in and for each ROI was determined. Fits with large residual values and by inspection were eliminated. For the dual-color labeled bead samples, we calculated the differences in center-of-mass positions for a given bead in the two-colors channels.
Cluster analysis of fluorescently labeled proteins on the cilia was done on user selected ROI centered on the peaks. A least squares fit using a 2-D rotated Gaussian distribution was performed. We calculated the statistical average size of the clusters using the average of the and FWHM, removing those fits with high residual values.
2.4. Sample Preparation
Samples were prepared of polystyrene beads (red and dark red, Invitrogen, at 590 nm) to match the Atto 590 and Atto 647N wavelengths along with samples of custom dual-color labeled beads emitting fluorescence in both channels ( diameter measured by the manufacturer with transmission electron microscopy). The beads were first attached to a coverslip coated with poly-L-lysine (Sigma Aldrich Co.). Immersion oil index matched to the objective lens was used as the medium between the slide and coverslip (Olympus immersion oil type F, , coverslip #1.5, 0.17-mm thickness).
For the imaging of OSNs, olfactory epithelial tissue was dissected from adult mice (2 to 4 months). All experiments followed protocols approved by the University of Colorado Anschutz Medical Campus Institutional Animal Care and Use Committee. OSN cells were dissociated by mechanical tweezing and gentle pipetting in mammalian Ringer’s solution and allowed to settle on precoated slides as previously described in Ref. 23. Cells were fixed with 4% paraformaldehyde for 10 min, followed by washing with phosphate-buffered saline, and incubation in a blocking solution containing bovine serum albumin and 3% normal donkey serum for one hour. We then incubated the slides overnight at 4°C with two primary antibodies against CNGA2, the subunit of the CNG channel, ACIII, and acetylated-tubulin. We then incubated the slides for 2 h at room temperature in custom secondary antibodies conjugated with Atto 647N and Atto 594 fluorescent tags (Rockland Immunochemicals Inc.) to label ACIII and CNGA2, respectively, along with Alexa Fluor 488 to label acetylated-tubulin. OSNs, identified under epifluorescence by their morphology, were imaged at resolution under both confocal and STED. Excitation power was 1 to and STED power was 10 to 20 mW at the sample during imaging.
3. Results and Discussion
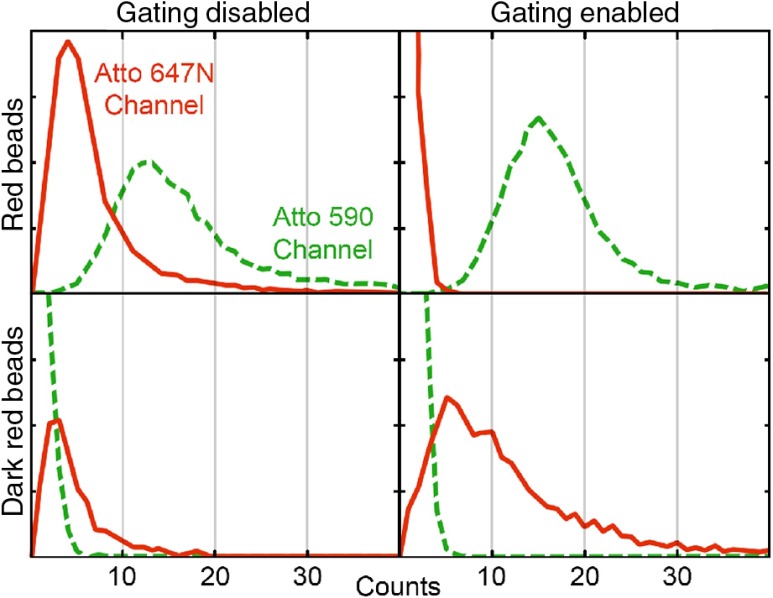
The resolution and spatial overlap between the two channels on the STED microscope were measured using samples of fluorescent beads. We first imaged Red and Dark Red beads separately in one color, shown in Figs. 3(a) and 3(b). Bead samples were then imaged using two-color simultaneous excitation with pulse interleaving, with and without time gated detection to quantify crosstalk between the channels. Figure 4 shows that the first order crosstalk signal is nearly eliminated with gated detection compared with when no gating is used. In addition, the amount of actual signal is not reduced when using gated detection, though we did note that the resolution achieved when imaging the individual channels was better than imaging both simultaneously.
Fig. 3.
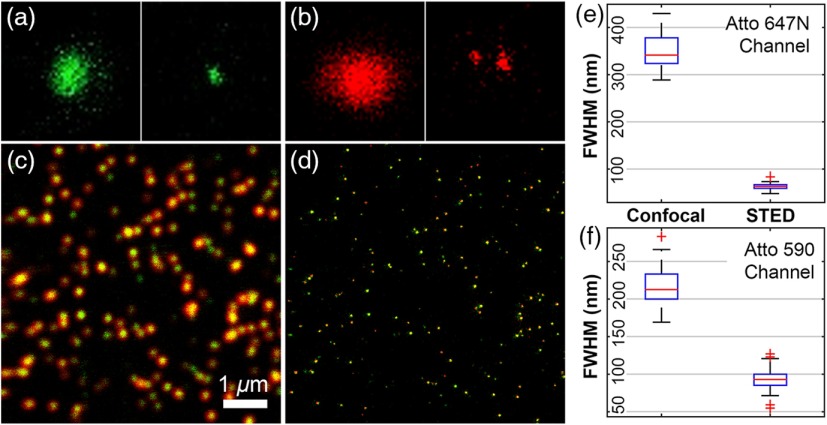
Confocal and STED images of fluorescent beads to determine resolution and spatial overlap of the two channels. (a) Red beads imaged in the Atto 590 channel, (b) dark red beads imaged in the Atto 647N channel, (frame size: ). (c) Confocal and (d) STED images of custom double-labeled beads taken simultaneously in the Atto 590 channel (green) and Atto 647N channel (red), (). Right: Boxcar plots for the fitted FWHM of the beads in the (e) Atto 647N and (f) Atto590 channels indicate increased resolution obtained with STED over confocal imaging. A least squares fit to a 2-D-Gaussian function was performed to calculate the FWHM for individual beads shows: (confocal) and (STED) for Atto 590 and (confocal) and (STED) for the Atto 647N channel, beads (STED), beads (confocal). The average separation between the fitted centers of the Gaussian in the two channels was and . Two-color image pixel size: 19.5 nm, pixel dwell time: 50 microseconds and total imaging time: 0:38.
Fig. 4.
Effects of cross-talk rejection using gating circuit for two-color STED. Samples of either red or dark red 40 nm fluorescent beads were imaged in both color channels with or without gating. Image histograms are reported by summing the intensity over ROI drawn around each individual bead. Green (dashed): Atto 590 channel. Red: Atto 647N channel. The gating circuit greatly reduces the background signal without any reduction in counts in the correct channel.
We then imaged dual-color labeled nanobeads to measure colocalization on both channels [shown in Figs. 3(c) and 3(d)]; and determined the distribution in size for the dual-color beads for each color channel. The calculations show a FWHM of (confocal) and (STED) for Atto 590, and (confocal) and (STED) for the Atto 647N channel. Using the actual size of the nanobeads (), we calculate the PSF of our instrument to be for Atto 590 and for Atto 647N. We additionally measured an average separation between the center positions for each bead in the two channels of and , indicating the spatial overlap is well below our resolution.
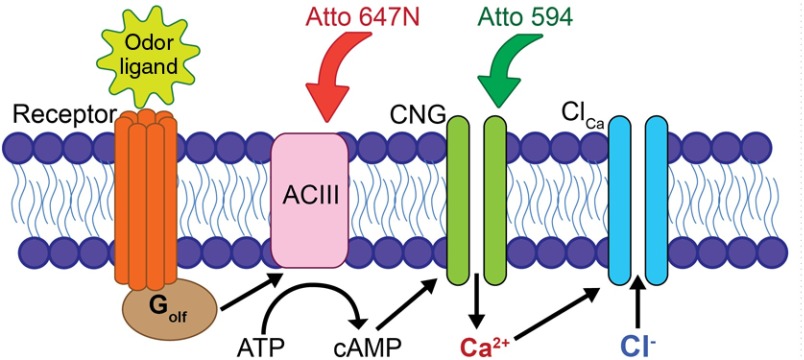
We demonstrate use of the two-color STED microscope by imaging transduction proteins on the cilia of OSNs to reveal sub-diffraction-limited structures involved in olfactory transduction. In the canonical olfactory transduction pathway (shown in Fig. 5), an odorant ligand binds to a G-protein coupled odorant receptor, resulting in activation of ACIII and the initiation of the olfactory signal amplification cascade. ACIII increases intraciliary cyclic adenosine monophosphate (cAMP) concentration, which then opens the CNG channel, allowing an influx of and . High local concentration in the cilia activates downstream -sensitive channels, such as the -activated () channels, which further amplify the signal.24 Despite over two decades of study, the molecular-scale dynamics of olfactory transduction are still not clearly understood. Localized signaling domains have been shown to be crucial for efficient retinal phototransduction25 and are suspected to be important in olfactory transduction as well.26,27 But the existence and importance of protein colocalization in olfactory transduction has been controversial.23 Recently, STED microscopy was used to show that the CNG channel and several -activated chloride channels of the anoctamin family localize to discrete structures on the olfactory cilia, but did not show the relative spatial arrangement of interacting proteins.28 Also, the experiments were performed on whole olfactory epithelial tissue in which there could be an overlap of proteins in the axial dimension. Here we demonstrate simultaneous two-color STED microscopy on isolated OSNs to investigate the colocalization and clustering of the CNG and ACIII proteins. Because these proteins are known to interact via intraciliary cAMP and , it has been previously hypothesized that they would be spatially colocalized along the OSN cilia.27 By isolating OSNs from the epithelium and imaging the thin (100 to 200 nm diameter) cilia individually, we are able to minimize the chance that there will be axial overlap of protein clusters that could lead to false spatial correlations.
Fig. 5.
Canonical transduction pathway in the cilia of OSNs. An odor ligand binding to a specific receptor activates the G-protein , which then activates ACIII to convert intracellular ATP to cAMP. The CNG channel is opened by binding nearby cAMP and allows depolarizing cations, including , to enter the cell. High concentration may also activate downstream channels, which further depolarizes the cell. Also illustrated are representations of the antibodies conjugated with Atto 594 and Atto 647N that label the CNG and ACIII proteins.
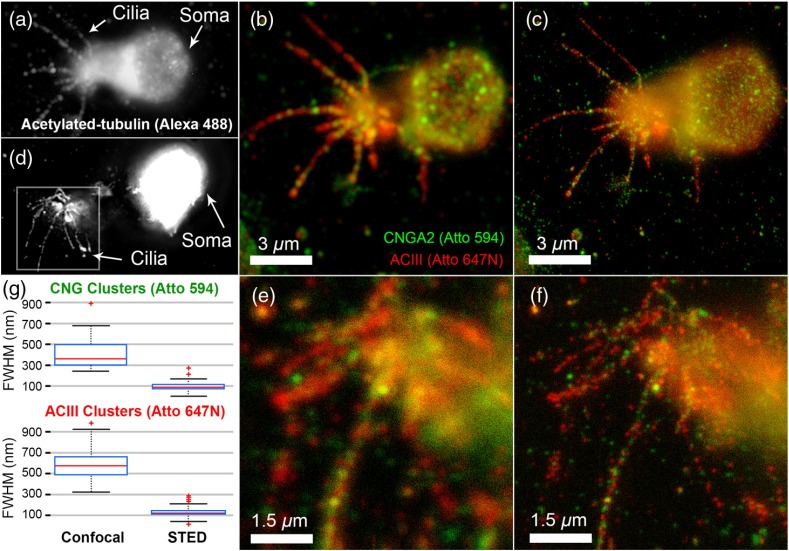
Figures 6(a) and 6(d) shows epifluorescence imaging of acetylated-tubulin, which was used to identify the ciliary morphology of the OSNs. Under normal confocal imaging, shown in Figs. 6(b) and 6(e), we see that CNG and ACIII localize to, and are nonuniformly distributed along, the cilia. STED imaging, shown in Figs. 6(c) and 6(f) of the same ROI revealed an underlying spatial complexity of CNG and ACIII proteins not previously observed. These clusters were also seen under STED imaging of OSNs singly-labeled for the CNG protein, indicating that the clustering is likely not due to steric hindrance or competitive binding of these antibodies. We characterized the size of these clusters calculating FWHM values of , (Atto 594/CNGA2) and , (Atto 647N/ACIII) in confocal and , (Atto 594/CNGA2) and , (Atto 647N/ACIII) in STED [shown in Fig. 6(g)].
Fig. 6.
Two-color STED images of OSNs labeled for ACIII, CNGA2, and acetylated tubulin. Panel (a) and (d) show the widefield epifluorescence images of the OSN labeled with Alexa Fluor 488 against acetylated-tubulin, while panel (b),(e) and (c),(f) show the two-color images in confocal and STED, respectively, with green indicating Atto 594-labeled CNGA2 and red indicating Atto 647N-labeled ACIII. (g) Boxcar plots for clusters on OSN cilia, combining clusters on both OSNs. Top: Atto 594 channel STED enhancement, showing the decrease in imaged cluster FWHM in going from confocal (, ) to STED (, ). Bottom: Atto 647N channel STED enhancement, confocal (, ) and STED (, ). The FWHM was calculated from a rotated 2-D Gaussian fit to () ROIs that were centered on the peaks of clusters. All images taken with pixel size: 9.8 nm, pixel dwell time: 30 microseconds and total imaging time: 3:56.
STED imaging of transduction proteins on OSN cilia can help researchers answer the question of functional spatial interactions in olfactory transduction. STED can resolve the spatial clustering of the transduction proteins compared with confocal imaging. Since the CNG and ACIII proteins are functionally coupled and membrane-bound, we may expect them to colocalize within the cilia. Interestingly, these preliminary results challenge the hypothesis that these proteins would cluster together in the cilia. As with all super-resolution techniques, it is important to continue with controls to ensure that nanoscale labeling artifacts such as postfixation diffusion, competitive binding, and steric hindrance are not affecting localization of these clusters. We plan to follow up these results with more extensive controls for labeling and also confirm our results by imaging cilia in intact olfactory epithelium compared with isolated cells. Multicolor STED imaging of other membrane proteins involved in olfaction, particularly the -activated anoctamin channels whose role in transduction is not settled, can improve our understanding of how these proteins interact. Additionally, with our custom system we have the option to add more than two-color channels. Three- or four- channel imaging could be done by using two dyes with overlapping excitation/emission spectra but different fluorescence lifetimes12 or by spectral detection and linear unmixing of the emission from two fluorophores using a single excitation and STED wavelength.22
4. Conclusions
We describe construction of a custom-built two-color STED microscope and demonstrate capabilities for imaging protein–protein localization in biological samples. We characterized the resolution in both channels and spatial overlap. Images were obtained of membrane proteins, CNG and ACIII, which are involved in olfactory transduction in the cilia of olfactory sensory neurons. These proteins were found to cluster in sizes averaging , below the optical diffraction limit. Additional spatial features showing separation between clusters were also seen in the STED images. Future work will involve two-color labeling of combinations of transduction-related proteins, especially downstream -activated channels. We also plan to combine two-color three-dimensional STED imaging, with fast video-rate beam scanning that will allow imaging cilia in epithelial tissue slices and in live cells. Live cell imaging is possible using tag-probe methods combined with near-infrared fluorophores for membrane or intracellular protein labeling.29,30 Live cell STED imaging of ciliary proteins in OSNs would allow dynamical changes to be observed during odor transduction and is something we wish to pursue in future work.
Acknowledgments
Stefan Hell and members of his lab at the Max Planck Institute in Göttingen, Germany, were indispensable in the implementation of STED at University of Colorado Denver Anschutz Medical Campus in not only the basic design but also details of alignment, software for image acquisition, and hands-on training. The staff and faculty involved in the Advanced Light Microscopy Core at UC Denver were instrumental in supporting this work. The authors thank Bill Betz for pioneering the effort from the beginning to bring STED microscopy to UC Denver and Dr. Silvio Rizzoli for scientific advice. The study was funded by NSF DBI-1337573 (EAG, DR), NIH S10 RR023381 (DR, EAG), and NIH DC014253 (DR).
Biographies
Stephanie A. Meyer is a research associate at the University of Colorado Denver Anschutz Medical Campus in the Department of Bioengineering. She currently works for the Advanced Light Microscopy Core on STED microscopy. She received her PhD from the Department of Physics at the University of Colorado Boulder in 2011 with a thesis titled “A Frequency-stabilized, Diode-pumped Yb:KYW Optical Frequency Comb for Metrology,” under the advisement of Dr. Scott A. Diddams at the National Institute of Standards and Technology in Boulder. Her current interest is in the development of advanced optical tools that allow biologists to open up new realms of research questions for exploration.
Baris N. Ozbay is a doctoral candidate at the University of Colorado Denver Anschutz Medical Campus in the Department of Bioengineering. He received his Bachelor of Science degree in electrical engineering from the University of Colorado Denver in 2010. His doctoral work involves translating innovative optical microscopy techniques for use by neuroscience researchers.
Mariana Potcoava is a physicist at the Intelligent Imaging Innovation in Denver, Colorado. Prior to that she worked as a research associate at the University of Colorado Denver Anschutz Medical Campus in the Department of Bioengineering. Her interests include the development of new optical systems for super-resolution microscopy.
Ernesto Salcedo is a senior instructor in the Department of Cell and Developmental Biology at the University of Colorado Anschutz Medical Campus. He received his PhD from the Molecular Medicine program at the University of Texas Health Science Center San Antonio. His thesis was titled “Investigating the Molecular Mechanism of Spectral Tuning in Invertebrate Visual Pigments.” His current interests include the use of digital image processing and modeling of 3-D image datasets to explore how anatomy can organize function in the brain.
Diego Restrepo received his MSc and PhD degrees from the Department of Biophysics at the University of Rochester and is a professor of cell and developmental biology at the University of Colorado Anschutz Medical Campus. He is an expert in systems neurobiology of sensory systems with a focus on the chemical senses. His interests include development of new approaches to measure and modulate neural activity and computational tools for analysis of neural circuit function in awake behaving animals with an emphasis on understanding the role of neural networks in sensory decision making.
Emily A. Gibson received her degree in physics from JILA, University of Colorado Boulder, specializing in nonlinear optics. She is an assistant professor in the Bioengineering Department at the University of Colorado Denver Anschutz Medical Campus. Her research interests include the development and application of advanced microscopy for neuroscience and clinical diagnostics.
References
- 1.Abbe E., “Beiträge zur theorie des mikroskops und der mikroskopischen wahrnehmung,” Arch. Mikrosk. Anat. 9(1), 413–418 (1873). 10.1007/BF02956173 [DOI] [Google Scholar]
- 2.Donnert G., et al. , “Two-color far-field fluorescence nanoscopy,” Biophys. J. 92(8), L67–L69 (2007). 10.1529/biophysj.107.104497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klar T., et al. , “Fluorescence microscopy with diffraction resolution barrier broken by stimulated emission,” Proc. Natl. Acad. Sci. U. S. A. 97(15), 8206–8210 (2000). 10.1073/pnas.97.15.8206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Betzig E., et al. , “Imaging intracellular fluorescent proteins at nanometer resolution,” Science 313(5793), 1642–1645 (2006). 10.1126/science.1127344 [DOI] [PubMed] [Google Scholar]
- 5.Rust M. J., Bates M., Zhuang X., “Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM),” Nat. Methods 3(10), 793–796 (2006). 10.1038/nmeth929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gustafsson M., “Nonlinear structured-illumination microscopy: Wide-field fluorescence imaging with theoretically unlimited resolution,” Proc. Natl. Acad. Sci. U. S. A. 102(37), 13081–13086 (2005). 10.1073/pnas.0406877102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Z., et al. , “Optical virtual imaging at 50 nm lateral resolution with a white-light nanoscope,” Nat. Commun. 2, 218 (2011). 10.1038/ncomms1211 [DOI] [PubMed] [Google Scholar]
- 8.Darafsheh A., et al. , “Optical super-resolution imaging by high-index microspheres embedded in elastomers,” Opt. Lett. 40(1), 5–8 (2015). 10.1364/OL.40.000005 [DOI] [PubMed] [Google Scholar]
- 9.Pellett P. A., et al. , “Two-color STED microscopy in living cells,” Biomed. Opt. Express 2(8), 2364–2371 (2011). 10.1364/BOE.2.002364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berning S., et al. , “Nanoscopy in a living mouse brain,” Science 335(6068), 551 (2012). 10.1126/science.1215369 [DOI] [PubMed] [Google Scholar]
- 11.Westphal V., et al. , “Video-rate far-field optical nanoscopy dissects synaptic vesicle movement,” Science 320(5873), 246–249 (2008). 10.1126/science.1154228 [DOI] [PubMed] [Google Scholar]
- 12.Bueckers J., et al. , “Simultaneous multi-lifetime multi-color STED imaging for colocalization analyses,” Opt. Express 19(4), 3130–3143 (2011). 10.1364/OE.19.003130 [DOI] [PubMed] [Google Scholar]
- 13.Hell S. W., “Far-field optical nanoscopy,” Science 316(5828), 1153–1158 (2007). 10.1126/science.1137395 [DOI] [PubMed] [Google Scholar]
- 14.Einstein A., “Zur quantentheorie der strahlung,” Physik. Z. 18, 121–128 (1917). [Google Scholar]
- 15.Westphal V., Hell S. W., “Nanoscale resolution in the focal plane of an optical microscope,” Phys. Rev. Lett. 94(14), 143903 (2005). 10.1103/PhysRevLett.94.143903 [DOI] [PubMed] [Google Scholar]
- 16.Hofmann M., et al. , “Breaking the diffraction barrier in fluorescence microscopy at low light intensities by using reversibly photoswitchable proteins,” Proc. Natl. Acad. Sci. U. S. A. 102(49), 17565–17569 (2005). 10.1073/pnas.0506010102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Testa I., et al. , “Dual channel RESOLFT nanoscopy by using fluorescent state kinetics,” Nano Lett. 15(1), 103–106 (2015). 10.1021/nl503058k [DOI] [PubMed] [Google Scholar]
- 18.Schneider J., et al. , “Ultrafast, temporally stochastic STED nanoscopy of millisecond dynamics,” Nat. Methods 12(9), 827–830 (2015). 10.1038/nmeth.3481 [DOI] [PubMed] [Google Scholar]
- 19.Osseforth C., et al. , “Simultaneous dual-color 3D STED microscopy,” Opt. Express 22(6), 7028–7012 (2014). 10.1364/OE.22.007028 [DOI] [PubMed] [Google Scholar]
- 20.Lenz M. O., et al. , “3-D stimulated emission depletion microscopy with programmable aberration correction,” J. Biophoton. 7(1-2), 29–36 (2014). 10.1002/jbio.201300041 [DOI] [PubMed] [Google Scholar]
- 21.Honigmann A., et al. , “Scanning STED-FCS reveals spatiotemporal heterogeneity of lipid interaction in the plasma membrane of living cells,” Nat. Commun. 5, 5412 (2014). 10.1038/ncomms6412 [DOI] [PubMed] [Google Scholar]
- 22.Tønnesen J., et al. , “Two-color STED microscopy of living synapses using a single laser-beam pair,” Biophys. J. 101(10), 2545–2552 (2011). 10.1016/j.bpj.2011.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reisert J., et al. , “The Ca-activated Cl channel and its control in rat olfactory receptor neurons,” J. Gen. Physiol. 122(3), 349–364 (2003). 10.1085/jgp.200308888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kleene S. J., “The electrochemical basis of odor transduction in vertebrate olfactory cilia,” Chem. Senses 33(9), 839–859 (2008). 10.1093/chemse/bjn048 [DOI] [PubMed] [Google Scholar]
- 25.Yau K.-W., Hardie R. C., “Phototransduction motifs and variations,” Cell 139(2), 246–264 (2009). 10.1016/j.cell.2009.09.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Castillo K., Restrepo D., Bacigalupo J., “Cellular and molecular Ca2+ microdomains in olfactory cilia support low signaling amplification of odor transduction,” Eur. J. Neurosci. 32(6), 932–938 (2010). 10.1111/j.1460-9568.2010.07393.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen C., Nakamura T., Koutalos Y., “Cyclic AMP diffusion coefficient in frog olfactory cilia,” Biophys. J. 76(5), 2861–2867 (1999). 10.1016/S0006-3495(99)77440-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Henkel B., et al. , “Co-expression of anoctamins in cilia of olfactory sensory neurons,” Chem. Senses 40(2), 73–87 (2015). 10.1093/chemse/bju061 [DOI] [PubMed] [Google Scholar]
- 29.Yano Y., Matsuzaki K., “Tag-probe labeling methods for live-cell imaging of membrane proteins,” Biochim. Biophys. Acta 1788(10), 2124–2131 (2009). 10.1016/j.bbamem.2009.07.017 [DOI] [PubMed] [Google Scholar]
- 30.Lukinavičius G., et al. , “A near-infrared fluorophore for live-cell super-resolution microscopy of cellular proteins,” Nat. Chem. 5(2), 132–139 (2013). 10.1038/nchem.1546 [DOI] [PubMed] [Google Scholar]