Abstract
A new principle for expression of heat-sensitive recombinant proteins in Escherichia coli at temperatures close to 4°C was experimentally evaluated. This principle was based on simultaneous expression of the target protein with chaperones (Cpn60 and Cpn10) from a psychrophilic bacterium, Oleispira antarctica RB8T, that allow E. coli to grow at high rates at 4°C (maximum growth rate, 0.28 h−1) (M. Ferrer, T. N. Chernikova, M. Yakimov, P. N. Golyshin, and K. N. Timmis, Nat. Biotechnol. 21:1266-1267, 2003). The expression of a temperature-sensitive esterase in this host at 4 to 10°C yielded enzyme specific activity that was 180-fold higher than the activity purified from the non-chaperonin-producing E. coli strain grown at 37°C (32,380 versus 190 μmol min−1 g−1). We present evidence that the increased specific activity was not due to the low growth temperature per se but was due to the fact that low temperature was beneficial to folding, with or without chaperones. This is the first report of successful use of a chaperone-based E. coli strain to express heat-labile recombinant proteins at temperatures below the theoretical minimum growth temperature of a common E. coli strain (7.5°C).
The number of available enzymes that have great potential for industrial chemical reactions has increased considerably since the 1980s, mainly as a result of enormous achievements in the cloning and expression of enzymes from diverse culturable microorganisms or, recently, as a result of the environmental DNA pool of nonculturable organisms, the metagenome (12); this increase was stimulated by an increasing demand for biocatalysts (18). Among these enzymes, cold-adapted enzymes from psychrophiles, which are organisms with optimal temperatures for growth between 4 to 15°C, offer novel opportunities for biotechnological applications (4, 5, 9). However, when expression of thermolabile proteins from psychrophiles is induced on a large scale in mesophilic hosts (e.g., Escherichia coli) in order to study their biochemical properties in detail, quite often the production of inclusion bodies, which are enzymatically inactive, is observed. The reason for the inactivity is misfolding of the polypeptide chains, which cannot attain their natural active conformation when they are expressed in a common E. coli strain at 37°C. In support of this hypothesis, cultivation at low temperatures, which is often beneficial to folding, may significantly increase the activity of thermolabile enzymes. Following this observation, genes encoding cold-active enzymes from bacteria were cloned and expressed in E. coli grown at low temperatures. For example, Feller et al. (6) obtained expression of the psychrophilic α-amylase from the Antarctic psychrophile Alteromonas haloplanktis in E. coli by lowering the cultivation temperature of the transformed expression host to 18°C. These workers demonstrated that expression of the active recombinant enzyme could be as high as expression of the enzyme in wild-type A. haloplanktis and, moreover, that the recombinant enzyme had the same kinetic parameters as the wild-type enzyme produced at 4°C. Feller et al. therefore concluded that the psychrophilic enzyme was folded correctly when it was expressed in E. coli at 18°C. However, a drawback of low cultivation temperatures in mesophilic host organisms (e.g., E. coli) is the dramatic reduction in the growth rate and, consequently, the reduced rate of synthesis of the heterologous protein. For this reason, Antarctic bacteria have also been screened for characteristics that may enable development of low-temperature recombinant gene expression systems for heat-labile proteins. Bacterial strains TA1 and TAD1 and the psychrophile Pseudoalteromonas haloplanktis TAC 125 exhibited high gene transfer efficiencies, and levels of induction of cold-adapted enzymes of up to 250-fold were achieved at 4 and 15°C (15, 19). However, the expression was nearly 2 orders of magnitude lower than that obtained in E. coli grown at 18°C. A cspA promoter system has also been demonstrated to be a valuable tool for production of unstable gene products in E. coli (14); however, the cspA-based expression system was effective only at intermediate temperatures (i.e., 15 to 23°C).
The objective of the present study was to test a previously engineered E. coli strain bearing the chaperonin 60 gene (cpn60) and the cochaperonin 10 gene (cpn10) from the psychrophilic bacterium Oleispira antarctica RB8T, which is able to grow at a high rate (0.28 to 0.45 h−1) at temperatures ranging from 4 to 15°C (7, 8), as a host for protein expression at low temperatures. Also, an esterase, EstRB8, cloned from the same organism, O. antarctica RB8T, was used as a model thermolabile protein in the present study.
MATERIALS AND METHODS
Plasmid and strains.
O. antarctica RB8T (DSMZ 14852T) and E. coli strains XL1-Blue MRF′ (Stratagene), XLOLR (Stratagene), and TOP10 (Invitrogen) were maintained and cultivated by using the recommendations of the suppliers and standard protocols described elsewhere (16, 21). The plasmids and strains used in this work are listed in Table 1.
TABLE 1.
Strains and plasmids used in this work
| Strain or plasmid | Description | Source and/or reference |
|---|---|---|
| O. antarctica RB8T (DSMZ 14852T) | Marine psychrophilic hydrocarbon-degrading bacterium | Yakimov et al. (21) |
| E. coli XL1-Blue MRF′ | Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 endA1 supE44 thi-1 recA1 gyrA96 relA1 lac [F′ proAB lacqZΔM15 Tn10 (Tetr)]; used to propagate lambda phages and to screen for esterase | Stratagene |
| E. coli XLOLR | Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 endA1 thi-1 recA1 gyrA96 relA1 lac [F′ proAB lacqZΔM15 Tn10 (Tetr)] Su-(nonsuppressing) λr (lambda resistant); used to obtain pBK1Est from esterase-positive lambda clone after excision procedure | Stratagene |
| E. coli TOP10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 deoR recA1 araD139 Δ(ara-leu)7697 galU galK rpsL (Strr)endA1 nupG; used as expression host for pBK1CpnEst | Invitrogen |
| pBK1Est | pBK-CMV (Stratagene) derivative; contains 999-bp fragment coding for EstRB8 | This study EMBL accession no. AJ606098 |
| pCRWT | pCR2.1 plasmid derivative (Invitrogen); harbors cpn10::cpn60 gene cassette PCR amplified with oligonucleotides CpnF and CpnR from chromosome of O. antarctica RB8T | This study |
| pBK1CpnEst | Derived from pBK1Est and pCRWT; SacI-SaII cpn10::cpn60-containing DNA fragment from pCRWT was ligated upstream of estRB8 gene in pBK1Est treated with the same endonucleases | This study EMBL accession no. AJ606099 |
Materials and buffers.
p-Nitrophenyl esters, Fast Blue RR, and α-naphthyl acetate were purchased from Sigma Chemical Co. (St. Louis, Mo.). Molecular mass markers for sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) were provided by Novagen. DNA restriction and modification enzymes were obtained from New England Biolabs. DNase I grade II was obtained from Boehringer Mannheim. Chaperonins 60 and 10 from O. antarctica RB8T were expressed in and purified from E. coli XLOLR harboring phagemid pPst26 with the cpn operon, as described previously (7, 8). GroEL and GroES chaperonins from E. coli were provided by Novagen. ATP, chromatographic media, and an LMW calibration kit for native electrophoresis were obtained from Amersham Pharmacia Biotech (Little Chalfont, United Kingdom). For immunodetection, antiserum against a synthetic peptide deduced from N-terminal sequencing of EstRB8 (MKIRPLHDRIVVRRKE) was purchased from SEQLAB (Sequence Laboratories Göttingen GmbH).
Detection and cloning of EstRB8 in O. antarctica RB8T lambda expression library.
The genome library of O. antarctica RB8T in bacteriophage lambda was established by using a ZAP Express kit (Stratagene) with E. coli XL1-Blue MRF′. Esterase-positive clones were detected as follows. After infection of E. coli XL1-Blue MRF′ and subsequent incubation, plates (22.5 by 22.5 cm) containing about 10,000 phage clones per plate were overlaid with 20 ml of a water solution containing 320 μl of α-naphthyl acetate (20 mg/ml in acetone), 5 mM isopropyl-β-d-thiogalactopyranoside (IPTG), and 320 μl of Fast Blue RR (80 mg/ml in dimethyl sulfoxide). Positive clones exhibiting a brown halo after about 2 h of incubation were picked, and the separate positive clones were isolated after phage particle dilution, E. coli infection, and halo detection. From one of the selected phage colonies, the pBK1Est phagemid was derived by using the helper phage excision procedure (Stratagene) and was transferred to E. coli XLOLR cells.
Construction of the gene fusions of cpn and estRB8.
A DNA fragment containing genes for chaperonin 60 (cpn60) and its cochaperonin (cpn10) from O. antarctica RB8T was amplified from the genomic DNA of O. antarctica RB8T. Primers Cpn F SacI (5′-AGA GCT CCT AAT ACT TGG GAT CCA ACA GTT G) and CpnR SalI (5′-AGT CGA CAC GGT AAG CAG ATC AGG ACA ATG) contained sites for the corresponding endonucleases (underlined). The PCR was performed by using Taq polymerase (QIAGEN) and 27 cycles consisting of 95°C for 1 min, 50°C for 45 s, and 72°C for 2 min; the resulting amplicons were gel purified (QiaExII; QIAGEN) and cloned into plasmid pCR2.1 (Invitrogen). A plasmid designated pCRWT derived from the PCR amplicon from the template was extracted and digested with endonucleases SacI and SalI, and the approximately 2-kbp DNA fragments were extracted from the agarose gel, ligated with plasmid pBK1Est that was initially linearized with the same endonucleases (the SacI site was located in the phagemid pBK-CMV multiple-cloning site; SalI cut upstream of the initial Met of EstRB8), dephosphorylated with shrimp alkaline phosphatase (Roche) for 1 h at 37°C, and purified from the agarose gel. The ligation products were electroporated into E. coli TOP10 (Invitrogen) as recommended by the supplier, and plasmid pBK1CpnEst harboring the cpn10::cpn60::estRB8 gene fusion was obtained.
Growth conditions.
E. coli strains containing plasmids pBK1Est and pBK1CpnEst were electroporated into E. coli TOP10 cells and were grown at the desired temperature in Luria-Bertani medium containing 1 mM IPTG and 50 μg of kanamycin per ml.
Purification of EstRB8 after expression in cpn+ and cpn-negative E. coli strains.
Once the cultures (see above) exhibited the optimal esterase activity, as determined by titrating free fatty acids released by hydrolysis of tributyrin (12, 127, 504, and 1,560 μmol min−1 g−1 for lyophilized cpn-negative E. coli cells grown at 37, 30, 20, and 15°C, respectively, and 12, 127, 768, 2,040, 2,304, 2,400, and 2,400 μmol min−1 g−1 for lyophilized cpn+ E. coli cells grown at 37, 30, 20, 15, 10, 8, and 4°C, respectively), bacterial cells were harvested and resuspended in buffer A (50 mM Tris-HCl, pH 9.0) containing one protease inhibitor cocktail tablet (Roche) and DNase I grade II, incubated on ice for 30 to 45 min, and then sonicated for 4 min (total time). The soluble fraction was separated from insoluble debris by centrifugation (10,000 × g, 30 min, 4°C), dialyzed overnight against buffer A, concentrated by ultrafiltration on a Centricon YM-10 membrane (Amicon, Millipore) to obtain a total volume of 2,000 μl, and applied to an ion-exchange chromatography Mono-Q HR 10/10 column that was preequilibrated with buffer A. Proteins were eluted with an increasing NaCl gradient (0 to 1 M) in the same buffer for 200 min at a rate of 0.5 ml/min. Fractions that eluted at about 0.3 M NaCl were pooled and dialyzed against buffer B [50 mM Tris-HCl (pH 9.0), 1 M (NH4)2SO4]. Then 1,000 μl of a Centricon YM-10-concentrated and filtered (pore size, 0.22 μm) sample containing esterase was loaded on a Resource 15PHE hydrophobic chromatography column (PE 4.6/100) that was previously equilibrated with buffer B. The column was washed with a decreasing linear (NH4)2SO4 gradient (1.0 to 0 M in of 25 ml [total volume]), and desorption of the remaining bound esterase was performed with 10 ml of buffer C (10 mM Tris-HCl buffer, pH 9.0). A flow rate of 0.5 ml/min was used. The active fractions were pooled, dialyzed overnight at 4°C against buffer D (10 mM Tris-HCl [pH 7.0], 150 mM NaCl), and concentrated by ultrafiltration on a Centricon YM-10 membrane to obtain a total volume of 1 ml. The sample was purified further by using a Superose 12 HR 10/30 gel filtration column preequilibrated with buffer D at a flow rate of 0.4 ml/min. The purified recombinant esterase was dialyzed against buffer A and stored at −20°C at a concentration of 50 μM until it was used; 0.5 to 1.2 mg of EstRB8 (depending on the culture temperature) was obtained from 94 mg of crude E. coli extract, and the level of recovery was about 25.6% (wt/wt). When SDS-PAGE and native PAGE were used, a single component with a molecular mass of ca. 35,000 Da was detected. The N-terminal amino acid sequence of the purified esterase was MKIRPLHDRIVVRRKE, which is identical to amino acid residues 1 to 16 in the deduced sequence of EstRB8.
Level of expression of estRB8.
For quantification of protein expression, cell extracts, prepared as described above, were examined by using SDS-PAGE with 12 to 15% (vol/vol) acrylamide. Proteins in the gel were stained with Coomassie brilliant blue R-250, and the gel region corresponding to the EstRB8 size was examined for rough estimation of the quantity of the expressed protein in the total protein fraction. A Molecular Dynamics densitometer was used to scan the Coomassie brilliant blue-stained gel, and ImageQuant software was used to quantify the intensity of the bands by volume integration. When necessary, esterase-specific antibodies were used for immunodetection after Western blotting.
Esterase assay.
Esterase activity was assayed spectrophotometrically by using p-nitrophenyl esters ranging from acetate to palmitate as substrates as described elsewhere (13), with small modifications. Briefly, the enzyme activity was assayed by addition of 5 μl of a solution containing esterase (50 μM) to 150 μl of a 16 mM p-nitrophenyl ester (Sigma) stock solution (in isopropanol) in 2,850 μl of a mixture containing the appropriate buffer (0.1 M citrate [pH 5.5], 0.1 M morpholineethanesulfonic acid [MES] [pH 5.5 to 7.0], 0.1 M HEPES [pH 7.0 to 8.0], 0.1 M Tris-HCl [pH 8.0 to 9.0], or 0.1 M glycine-NaOH [pH 9.0 to 10.5]), 15% acetonitrile, and 0.038 mM Triton X-100 at temperatures ranging from 4 to 50°C. The esterase reaction was monitored at 405 nm. One unit of enzymatic activity was defined as the amount of protein that released 1 μmol of p-nitrophenoxide per min from p-nitrophenyl ester at the temperature and pH used. All values were determined in triplicate and were corrected by taking into consideration the autohydrolysis of the substrate. Unless indicated otherwise, the standard esterase assay used in the present study was performed at 20°C in 100 mM Tris-HCl buffer (pH 9.0) with 0.8 mM p-nitrophenyl butyrate (p-NPB) as the substrate. Hydrolytic activity was also determined by using lyophilized cells by titrating free fatty acids released by hydrolysis of tributyrin by using the pH-stat method (17). The hydrolysis of tributyrin was assayed titrimetrically with a pH-stat (model DL50; Mettler) by using 0.01 M NaOH as the titrant. The reaction mixture (20 ml) contained 65.6 mM substrate, 0.15 M NaCl, and 0.09% (vol/vol) acetonitrile. One unit of lipase activity was defined as the amount of enzyme that liberated 1 μmol of free fatty acid per min.
The thermostability was estimated by preincubating the enzyme in 100 mM Tris-HCl (pH 9.0) at temperatures ranging from 4 to 50°C. Aliquots (100 μl) were removed at different times, and the remaining esterase activity was measured by the standard esterase assay after the substrate (p-NPB) was added. The kinetic parameter (kcat/KM) was determined at temperatures ranging from 4 to 70°C. The substrate was p-NPB at concentrations of 10, 25, 50, 75, 100, 150, 200, 250, 300, and 400 mM in 100 mM Tris-HCl buffer (pH 9.0). Initial velocity values were fitted to the Hanes transformation of the Michaelis-Menten equation.
Assays and other methods.
The protein concentration was determined by the Bradford dye-binding method with a Bio-Rad protein assay kit by using bovine serum albumin as the standard (3). SDS-PAGE and native electrophoresis were performed as described by Laemmli (11). The temperature-induced unfolding was monitored by measuring the loss of secondary structure by means of peptide ellipticity at 220 nm. Circular dichroism spectra of EstRB8 (0.3 mg/ml) in Tris-HCl buffer (100 mM, pH 9.0) were measured with a Jasco J-720 spectropolarimeter equipped with a constant-temperature cell holder at temperatures ranging from 4 to 50°C. Spectra were measured in standard buffer by using a 0.1-cm cell.
Accession numbers.
The nucleotide sequences of pBK1Est (harboring the esterase gene alone) and pBK1CpnEst (containing the fusion cpn10::cpn60::estRB8) have been deposited in the EMBL/GenBank/DDBJ databases under accession numbers AJ606098 and AJ606099, respectively.
RESULTS
Detection and subcloning of EstRB8 in lambda expression library of O. antarctica RB8T.
An intracellular esterase (EstRB8) was detected after screening of the genomic DNA library of O. antarctica RB8T, established in E. coli, by using α-naphthyl acetate and Fast Blue RR (for details see Materials and Methods). Sequence analysis of the positive clone revealed an open reading frame consisting of 1,450 nucleotides that encode a protein (EstRB8) that contains 333 amino acids and has a theoretical molecular mass of 36,666 Da and a deduced isoelectric point of 5.58. Sequencing of the cloned fragment confirmed that the levels of identity and similarity of the deduced amino acid sequence of the cloned enzyme to sequences of known ester hydrolases classified in different families were low. Thus, EstRB8 had 37% amino acid identity to the lipase from Pseudomonas aeruginosa PAO1 (accession no. AAG06337) (the amino acid similarity to this top hit was not greater than 50%), 36% amino acid identity to triacylglycerol lipase from Moraxella sp. (accession no. P24640), 37% amino acid identity to triacylglycerol lipase from Moritella marina (accession no. AAK07450), and 34% amino acid identity to triacylglycerol lipase from Psychrobacter immobilis (accession no. CAA47949). The esterase seemed to belong to the alpha/beta hydrolase superfamily, because it contained the sequence motif Gly-X-Ser-X-Gly from residues 151 to 155, which is found in most serine hydrolases belonging to this superfamily (10). The catalytic triad should be formed by Ser-119, Asp-248, and His-276 according to a comparison with the Ser-Asp-His catalytic triad of other esterases and lipases (2).
Construction of gene fusions of cpn and estRB8.
The objective of the present study was to test a previously engineered E. coli strain that contained cpn60 and cpn10 genes from O. antarctica RB8T and that was able to grow at 4°C (maximum growth rate [μmax], 0.28 h−1) (7, 8) as a host for gene expression at low temperatures. An E. coli cpn10::cpn60::estRB8 strain containing a single DNA fragment with cpn60, cpn10, and estRB8 from O. antarctica RB8T (for details see Materials and Methods) was grown at 37, 30, 20, 15, 10, 8, and 4°C (at these temperatures the μmax values were determined to be 1.42, 0.88, 0.49, 0.45, 0.36, 0.30, and 0.28 h−1, respectively), and the level of expression of estRB8 and the specific activity of purified EstRB8 for hydrolysis of p-NPB were measured. As shown in Table 2, the maximum esterase activity was observed after growth at 4 to 10°C (∼32,380 μmol min−1 g−1), whereas at 15, 20, 30, and 37°C the specific activities were 15, 32, 81, and 95% lower, respectively (27,523, 10,362, 1,714, and 162 μmol min−1 g−1). SDS-PAGE (Fig. 1) and immunoblotting with anti-EstRB8 antibody (data not shown) revealed that the levels of expression of estRB8 in cpn+ E. coli were similar at temperatures ranging from 4 to 37°C (25 to 50 μg/mg) (Table 2). As the data show, the same amount of protein exhibited to 200-fold more activity at 4 to 10°C, which may indicate the presence of more highly folded species in EstRB8 expressed at 4 to 10°C.
TABLE 2.
Summary of the biochemical and growth characteristics of clones containing estRB8
| Growth temp (°C) |
E. coli/pBK1Est
|
E. coli/pBKCpnEst without esterase inductiona
|
||||
|---|---|---|---|---|---|---|
| Without esterase inductiona
|
With esterase inductionb
|
|||||
| Expression of EstRB8 (μg/mg)c | Esterase sp actd | Expression of EstRB8 (μg/mg)c | Esterase sp actd | Expression of EstRB8 (μg/mg)c | Esterase sp actd | |
| 37 | 25-50 | 179 | 25-50 | 184 | 25-50 | 162 |
| 30 | 25-50 | 1,894 | 25-50 | 1,800 | 25-50 | 1,714 |
| 20 | 25-50 | 7,518 | 25-50 | 9,636 | 25-50 | 10,362 |
| 15 | 25-50 | 23,270e | 25-50 | 22,375 | 25-50 | 27,523f |
| 10 | NDg | ND | 12-20 | 1,188 | 25-50 | 31,488h |
| 8 | ND | ND | 1.2 | 850 | 25-50 | 32,380h |
| 4 | ND | ND | <0.1 | 239 | 25-50 | 32,380h |
An E. coli strain harboring pBK1Est (without cpn60 and cpn10) or pBKCpnEst (with cpn60 and cpn10) was grown directly at temperatures ranging from 4 to 37°C in the presence of IPTG (1 mM).
An E. coli strain harboring pBK1Est (without cpn60 and cpn10) was grown at 37°C (optical density, 1.0), and esterase activity was induced by addition of IPTG (1 mM) after incubation at suboptimal temperature (37 to 4°C).
Level of expression (in micrograms of EstRB8/per milligram of total protein) in E. coli cell extract (for details see Materials and Methods).
Specific activity (in micromoles per minute per gram of pure protein) in Tris-HCl buffers (pH 9.0) at 20°C when p-NPB was used as the substrate.
Growth rate, 0.15 h−1.
Growth rate, 0.45 h−1.
ND, no growth or growth rate lower than 0.02 h−1.
Growth rate, 0.28 to 0.36 h−1.
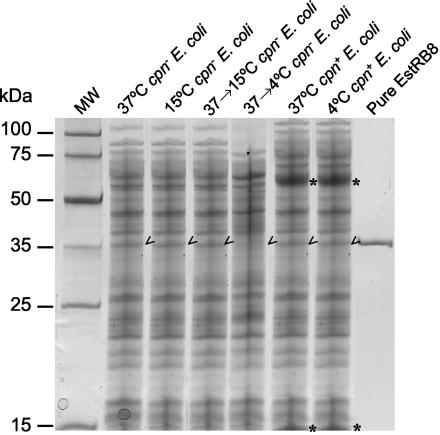
FIG. 1.
SDS-PAGE analysis of cell extracts of E. coli cpn and cpn+ strains. From left to right, the lanes contained molecular weight markers (MW), E. coli cpn cells grown at 37°C, E. coli cpn cells grown at 15°C, E. coli cpn cells induced at 37°C and then 15°C, E. coli cpn cells induced at 37°C and then 4°C, E. coli cpn cells grown at 37°C, E. coli cpn cells grown at 4°C, and pure esterase. The asterisks and arrowheads indicate the positions of Cpn60/Cpn10 and EstRB8, respectively. Each lane contained 10 μg of protein.
Comparative analysis of E. coli cpn+ and cpn-negative strains for production of active thermolabile esterase.
The results described above may have practical implications for developing additional expression capability in E. coli at low temperatures. To determine the advantage of using a E. coli cpn+ strain based on its ability to produce active thermolabile proteins, we examined the expression and levels of activity of EstRB8 in (i) E. coli expressing pBK1Est grown at 37°C and suboptimal temperatures (15 to 30°C) and (ii) E. coli expressing pBK1Est grown at 37°C and induced with IPTG at temperatures shifted from 4 to 20°C. These two methods, which are normally used when workers are trying to express psychrophilic genes, were compared by using the chaperonin-based system.
For expression of estRB8 in an E. coli cpn-negative strain, plasmid pBK1Est was transferred to E. coli TOP10 cells, which were grown at 37, 30, 20, and 15°C (the μmax values were determined to be 1.0, 0.90, 0.22, and 0.15 h−1, respectively) in Luria-Bertani medium containing 1 mM IPTG and 50 μg of kanamycin per ml. Temperatures of ≤10°C were not evaluated due to the inability of E. coli to grow (μmax, <0.02 h−1). Figure 1 shows that the E. coli cpn-negative strain grown at temperatures ranging from 15 to 37°C expressed EstRB8 at similar levels (25 to 50 μg/mg of proteins [Table 2]), like the cpn+ strain. In addition, as Table 2 shows, when EstRB8 was produced in the cpn strain, the specific activity was minimal after expression at 37°C (190 μmol min−1 g−1, 180-fold lower than the specific activity purified from the E. coli cpn+ strain grown at 4 to 10°C) and maximal at 15°C (23,270 μmol min−1 g−1). As the data show, the cpn+ strain grown at 4 to 10°C gave 28% more activity than the cpn-negative strain at 15°C (Table 2) and grew more effectively at low temperatures (the μmax values were 0.28 to 0.45 h−1 for the cpn+ strain grown at 4 to 10°C, compared to 0.15 h−1 for the cpn strain grown at 15°C).
In a second set of experiments, an E. coli strain harboring pBK1Est was grown at 37°C and then incubated at suboptimal temperatures (4, 8, 10, 15, 20, and 30°C), and esterase activity was induced by addition of IPTG (1 mM). As shown in Table 2, when cells pregrown at 37°C were transferred to 15°C, the esterase activity for hydrolysis of p-NPB was maximal (22,375 μmol min−1 g−1) and similar to that found for EstRB8 purified from E. coli cpn+ and cpn-negative cells cultured at 15°C (27,523 and 23,270 μmol min−1 g−1, respectively). Induction temperatures of ≤10°C correlated to low expression and activity levels; e. g., at 4°C the specific esterase activity was approximately 1% of that found at 15°C (239 versus 22,375 μmol min−1 g−1). This may correlate with the inability of E. coli to adequately express EstRB under these conditions (<0.1 μg/μg at 4°C after induction with IPTG) (Fig. 1), which seems to be negative in terms of the total esterase activity in the cells.
O. antarctica chaperonins do not assist with the proper folding of EstRB8.
It has been reported previously that coexpression of the GroEL/GroES molecular chaperones improves proper folding of the product proteins (1). Thus, to determine whether the high level of EstRB8 activity was caused by a lower expression temperature or chaperonin-mediated refolding, chemically denatured EstRB8 incubated in the presence of 6 M urea for 12 h at 4°C or purified esterase from cells grown at 37°C (∼180 μmol min−1 g−1) was incubated in vitro at temperatures ranging from 4 to 30°C with pure Cpn60 and Cpn10 from O. antarctica (7) and ATP in refolding buffer (50 mM morpholinepropanesulfonic acid [MOPS] [pH 7.0], 50 mM MgCl2, 50 mM KCl). It has been demonstrated previously that Oleispira chaperonins can efficiently refold chemically denatured polypeptides (i.e., bovine rhodanese and porcine lactate dehydrogenase), like other GroEL homologues (7, 8, 20). The esterase solution (50 μM purified enzyme in refolding buffer) was equilibrated with Cpn60 (100 μM) and different concentrations of Cpn10 (50 to 200 μM), and the recovery of esterase activity was measured after various incubation times (2 to 48 h) at 4 to 30°C after addition of 1 mM ATP. The activity was measured at 20°C by using the standard esterase assay and p-NPB as the substrate. The results of our experiments showed that the EstRB8 activity did not increase after incubation at a low temperature (4°C) or an intermediate temperature (30°C) with Oleispira chaperones and ATP, exhibiting the initial specific activity value (180 μmol min−1 g−1). Similar results were obtained when the refolding experiments were carried out in the presence of GroEL and GroES chaperonins from E. coli. We also observed that after dialysis of urea-denatured EstRB8 against buffer A overnight at 4°C, esterase could not be successfully folded, since the activity of EstRB8 did not improve.
Coexpression of Oleispira chaperonins did not affect the catalytic properties of EstRB8.
We also determined whether the coexpression of chaperonins affected the catalytic properties of EstRB8. To do this, the substrate specificity and biochemical properties of the esterases purified from (i) E. coli cells expressing pBK1Est grown at 37°C, (ii) E. coli cells expressing pBK1Est grown at 37°C and then transferred to 15°C, and (iii) E. coli cells expressing pBK1CpnEst grown at 4°C were compared. We found that esterases purified from the three E. coli variants were able to hydrolyze p-nitrophenyl esters (butyrate > caproate > propionate > acetate ≫ caprate ≫ laurate ≫ palmitate at a similar ratio [98:61:31:29:5:2:1]); however, esterase purified from cpn+ cells grown at 4°C was more efficient than the esterases purified from cpn-negative cells grown at 37 and 15°C (32,380 versus 190 and 22,375 μmol min−1 g−1 for hydrolysis of p-NPB, respectively). Short acylglycerols, triolein, and olive oil were not good substrates for O. antarctica esterase, suggesting that the enzyme is an esterase rather than a lipase. The best physiological efficiency of EstRB8 produced at 4°C in E. coli cpn10::cpn60::estRB8 for hydrolysis of p-NPB was observed at 4 to 10°C, similar to the enzyme purified at 37 or 15°C. However, EstRB8 produced at 4°C in the E. coli cpn+ strain was significantly more efficient and had kcat/Km values, measured at 4°C, that were approximately 110- and 2.4-fold higher than those of the enzyme produced at 37°C (0.6 s−1 mM−1 versus 65.9 s−1M−1) and 15°C (26.5 s−1 M−1versus 65.9 s−1 M−1), respectively. The pH and temperature optima (pH 8.5 to 9.5, 24°C), thermostability (<1 min at 50°C, 3 min at 40°C, 7 min at 35°C, 197 min at 20°C, and >600 min at temperatures below 18°C), and midpoint for thermal denaturation measured by circular dicroism (∼27°C) were similar independent of the growth temperature and the plasmid used for estRB8 expression.
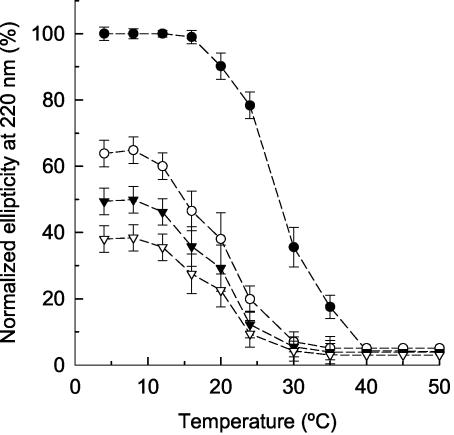
To assess whether esterase was folded more correctly at low temperatures, the secondary-structure content was determined by far-UV circular dicroism. Far-UV circular dicroism spectra at 4 to 50°C for each esterase were recorded. EstRB8 variants exhibited typical α/β mixed-type circular dichroism spectra with a negative peak at 210 to 220 nm (data not shown). As Fig. 2 shows, the relative peptide ellipticity at 220 nm, and thus the helical structure content, of EstRB8 purified from the E. coli cpn+ strain grown at 4°C was 1.6-, 2-, and 2.6-fold higher than the values obtained after cultivation at 20, 30, and 37°C, respectively, when either parental (cpn) or recombinant (cpn+) cells were used. Based on these observations and the findings of Yang et al. (22), we concluded that EstRB8 had a more helical structure and thus folded more efficiently when it was expressed at 4°C, and this may correlate with higher esterase activities (see above). This, together with the low activity if the enzyme was expressed at temperatures higher than 20°C, clearly indicated the cold-adapted phenotype of EstRB8.
FIG. 2.
Analysis of the secondary structure by far-UV circular dicroism. Far-UV circular dicroism spectra at 4 to 50°C for pure EstRB8 from E. coli bearing pBK1CpnEst grown at 4°C (•), 20°C (○), 30°C (▾), or 37°C (▿) were recorded, and the relative ellipticity at 220 nm was measured. The conditions used are described in Materials and Methods. The ellipticity at 220 nm obtained with the EstRB8 produced in E. coli grown at 4°C was assumed to be 100%. The three sets of data corresponding to EstRB8 purified from E. coli expressing plasmid pBK1CpnEst or pBK1Est at 20, 30, and 37°C were statistically equivalent, and only the data for pBK1CpnEst are shown.
DISCUSSION
The present study demonstrated for first time that a cold-resistant (due to psychrophilic chaperonins) E. coli strain might successfully be employed to express heat-labile recombinant proteins at temperatures below 15°C. We proved this by studying E. coli bearing a plasmid with a psychrophilic esterase-psychrophilic chaperonin gene fusion. This E. coli strain grew at temperatures ranging from 4 to 15°C at high rates (μmax, 0.28 to 0.45 h−1). The EstRB8 purified from this recombinant strain was up to 180-fold more active than the enzyme purified from a common E. coli strain grown at 37°C. In addition, the level of expression of the target protein (Fig. 1 and 2) and its biochemical properties were not affected by coexpression of cpn. Although it is well known that many proteins are more soluble in E. coli when the bacteria are cultured at a low temperature (i.e., 16 to 18°C) (6, 14), we demonstrated that the E. coli cpn+ strain was more efficient (in terms of rate of production and protein expression) than a common E. coli strain grown at suboptimal temperatures or after induction of 37°C-pregrown cultures at lower temperatures (Table 2 and Fig. 1 and 2). The use of this approach may be an attractive alternative to existing methodologies for expression of thermosensitive proteins in mesophilic hosts in order to improve the solubility and activity of thermosensitive recombinant proteins, which otherwise are produced in an insoluble form in E. coli at 37°C. The results of the refolding experiments also demonstrated that the high level of activation of EstRB8 directly correlated with the low growth temperature rather than with the chaperonin-mediated folding. However, this finding does not exclude the possibility that other thermosensitive polypeptides that require fold-catalyzing proteins, such as GroEL/ES homologues, to enhance their activities (1) may benefit from being expressed in the recombinant E. coli strains which we studied here. Although it was difficult to obtain sufficient quantities of native esterase for further biochemical comparisons with the recombinant variants due to the extremely low level of expression of EstRB8 in O. antarctica at 4°C, we suggest that the enzyme produced in E. coli grown at 4°C may have enzymatic activities that are very similar to those of the enzyme from O. antarctica RB8T thriving in its natural environment.
Finally, the principle which we demonstrated to be valid for E. coli may not be restricted to this organism and may also be used to establish new hosts previously known as mesophilic organisms for expression under cold conditions. The expression principle described here could also facilitate a better screening system to analyze (meta)genome expression libraries (e.g., bacteriophage lambda libraries) for thermosensitive enzymes that cannot normally be assessed by using the common E. coli host routinely used for library construction and screening. A host that bears a chromosomally integrated chaperonin cassette is now under construction, and its applicability in screening for other psychrophilic enzymes in, e.g., a genome library of O. antarctica RB8T and metagenomic libraries obtained from Arctic environmental DNA samples is being investigated. An E. coli cpn+ strain grown at ≤15°C may also be an important tool for expression of other bacterial, archaeal, and eukaryotic proteins, as well as antibody fragments in E. coli that are difficult to obtain in a soluble form with high yields at 37°C, so that the proteins or antibodies could fold more slowly and thus more properly at low temperatures.
Acknowledgments
M.F. thanks the European Commission for a Marie Curie postdoctoral fellowship. K.N.T. thanks Fonds der Chemischen Industrie. P.G. acknowledges the generous support of INTAS project Nr-2060 and European Union project EVK3-2002-00077 (COMMODE).
We also thank Rita Getzlaft for performing the protein sequence analysis and Nieves López and Dolores Reyes for determination of esterase activity towards tributyrin.
REFERENCES
- 1.Amrein, K. E., B. Takacs, M. Steiger, J. Molnos, N. A. Flint, and P. Burn. 1995. Purification and characterization of recombinant human p50csk protein-tyrosine kinase from an Escherichia coli expression system overproducing the bacterial chaperones GroES and GroEL. Proc. Natl. Acad. Sci. USA 92:1048-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arpigny, J. L., and K. E. Jaeger. 1999. Bacterial lipolytic enzymes: classification and properties. Biochem. J. 343:177-183. [PMC free article] [PubMed] [Google Scholar]
- 3.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 4.Cavicchioli, R., K. S. Siddiqui, D. Andrews, and K. R. Sowers. 2002. Low-temperature extremophiles and their applications. Curr. Opin. Biotechnol. 13:253-261. [DOI] [PubMed] [Google Scholar]
- 5.Feller, G., and C. Gerday. 2003. Psychrophilic enzymes: hot topics in cold adaptation. Nat. Rev. 1:200-208. [DOI] [PubMed] [Google Scholar]
- 6.Feller, G., O. Le Bussy, and C. Gerday. 1998. Expression of psychrophilic genes in mesophilic hosts: assessment of the folding state of a recombinant α-amylase. Appl. Environ. Microbiol. 64:1163-1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferrer, M., H. Lünsdorf, T. N. Chernikova, M. Yakimov, K. N. Timmis, and P. N. Golyshin. 2004. Functional consequences of single:double ring transition in chaperonins: life in the cold. Mol. Microbiol. 53:167-182. [DOI] [PubMed] [Google Scholar]
- 8.Ferrer, M., T. N. Chernikova, M. M. Yakimov, P. N. Golyshin, and K. N. Timmis. 2003. Chaperonins govern growth of E. coli at low temperatures. Nat. Biotechnol. 21:1266-1267. [DOI] [PubMed] [Google Scholar]
- 9.Hough, D. W., and M. J. Danson. 1999. Extremozymes. Curr. Opin. Chem. Biol. 3:39-46. [DOI] [PubMed] [Google Scholar]
- 10.Jaeger, K. E., S. Ransac, B. W. Dijkstra, C. Colson, M. van Heuvel, and O. Misset. 1994. Bacterial lipases. FEMS Microbiol. Rev. 15:29-63. [DOI] [PubMed] [Google Scholar]
- 11.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 12.Lorenz, P., K. Liebeton, F. Niehaus, and J. Eck. 2002. Screening for novel enzymes for biocatalytical processes: accessing the metagenome as a resource of novel functional sequence space. Curr. Opin. Biotechnol. 13:572-577. [DOI] [PubMed] [Google Scholar]
- 13.Manco, G., E. Adinolfi, F. M. Pisani, G. Ottolina, G. Carrea, and M. Rossi. 1998. Overexpression and properties of a new thermophilic and thermostable esterase from Bacillus acidocaldarius with sequence similarity to hormone-sensitive lipase subfamily. Biochem. J. 332:203-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mujacic, M., K. W. Cooper, and F. Baneyx. 1999. Cold-inducible cloning vectors for low-temperature protein expression in Escherichia coli: application to the production of a toxic and proteolytically sensitive protein. Gene 238:325-332. [DOI] [PubMed] [Google Scholar]
- 15.Remaut, E., C. Biliki, M. Iturriza-Gomara, and K. Keymeulen. 1999. Development of regulatable expression systems for cloned genes in cold-adapted bacteria, p. 1-16. In R. Margesin and F. Schinner (ed.), Biotechnological applications of cold-adapted organisms. Springer-Verlag, Heidelberg, Germany.
- 16.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 17.San Clemente, C. L., and D. V. Vadegra. 1967. Instrumental assay of microbial lipase at constant pH. Appl. Environ. Microbiol. 15:110-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmid, A., J. S. Dordick, B. Hauser, B. Kiener, M. Wubbolts, and B. Witholt. 2001. Industrial biocatalysis today and tomorrow. Nature 409:258-268. [DOI] [PubMed] [Google Scholar]
- 19.Tutino, M. L., A. Duilio, E. Parrilli, E. Remaut, G. Sannia, and G. Marino. 2001. A novel replication element from an Antarctic plasmid as a tool for the expression of proteins at low temperatures. Extremophiles 5:257-264. [DOI] [PubMed] [Google Scholar]
- 20.Walter, S., and J. Buchner. 2002. Molecular chaperones—cellular machines for protein folding. Angew. Chem. Int. Ed. Engl. 41:1098-1113. [DOI] [PubMed] [Google Scholar]
- 21.Yakimov, M. M., L. Giuliano, G. Gentile, E. Crisafi, T. N. Chernikova, W.-R. Abraham, H. Lunsdorf, K. N. Timmis, and P. N. Golyshin. 2003. Oleispira antarctica gen. nov., sp. nov., a novel hydrocarbonoclastic marine bacterium isolated from Antarctic coastal sea water. Int. J. Syst. Evol. Microbiol. 53:779-785. [DOI] [PubMed] [Google Scholar]
- 22.Yang, J. T., C.-S. C., Wu, and H. M. Martinez. 1986. Calculation of protein conformation from circular dichroism. Methods Enzymol. 130:208-269. [DOI] [PubMed] [Google Scholar]