Abstract
We describe the properties of a novel 252-kDa protein (P252) isolated from brush border membranes of Bombyx mori. P252 was found in a Triton X-100-soluble brush border membrane vesicle fraction, suggesting that it may be a component of the midgut epithelial cell membrane. P252 was purified to homogeneity, and the amino acid sequence of two internal peptides was determined, but neither of the peptides matched protein sequences in the available databases. The apparent molecular mass of the purified protein was estimated by denaturing gel electrophoresis to be 252 kDa, and it migrated as a single band on native gels. However, gel filtration chromatography indicated an apparent mass of 985 kDa, suggesting that P252 may exist as a homo-oligomer. The associations of P252 with Cry1Aa, Cry1Ab, and Cry1Ac were specific, and Kd constants were determined to be 28.9, 178.5, and 20.0 nM, respectively. A heterologous competition assay was also done. P252 did not exhibit Leu-pNA hydrolysis activity, and binding to the Cry1A toxins was not inhibited by GalNAc. Binding assays of P252 with various lectins indicated the presence of three antennal N-linked high-mannose-type as well as O-linked mucin-type sugar side chains. While the function of P252 is not yet clear, we propose that it may function with Cry1A toxins during the insecticidal response and/or Cry toxin resistance mechanism.
Cadherin-like proteins (CadLP) with molecular masses from 170 to 180 kDa or 210 kDa are suggested to be receptors for Cry1A toxins in Manduca sexta (40) and Bombyx mori (31, 32, 33). Recently, Gahan et al. (6) found that a Cry1Ac-resistant strain of Heliothis virescens carried a mutated gene encoding CadLP. Hara et al. (12) demonstrated that an antibody to CadLP (BtR175) inhibited the Cry1A toxin-mediated release of lactate dehydrogenase from midgut cells, thereby supporting the idea that CadLP may be a receptor for Cry1A toxins. Similarly, Gomez et al. (9) reported that specific binding between CadLP and Cry1Ab was inhibited by a peptide identified by phage display and that this peptide did not disrupt the binding between aminopeptidase N (APN) and Cry1Ab.
Although several reports lend support to the “CadLP theory” cited above, there is still uncertainty regarding the identity of the appropriate receptor(s). Prior to the proposal of the CadLP theory, several studies suggested that APN may be the receptor for Cry1A toxins (7, 21, 28, 29, 37, 42). In support of this hypothesis, the aminopeptidase activity of APN can modify the N termini of toxins to expose a recognition site for receptor binding. This is consistent with the observation that the N-terminal residues of Cry toxins are trimmed prior to insertion of domain I into the membrane (30). Thus, the activity of APN makes it an attractive candidate as a Cry toxin receptor. Furthermore, glycosylphosphatidylinositol (GPI) anchoring of APN may facilitate the clustering of toxin molecules on the plasma membrane. Based on these data, we propose that if CadLP is the Cry toxin receptor, then APN or another appropriate peptidase localized adjacent to the receptor must be present to modify domain I of Cry1A. Indeed, this concept was suggested from data published by Gomez et al. that demonstrated the existence of a protease activity (10).
We recently found that solubilized brush border membrane vesicle (BBMV) proteins from midguts of Cry1Ac-susceptible or highly resistant Plutella xylostella showed almost the same binding pattern with Cry1Ac in ligand blot analysis (24). Surface plasmon resonance analysis also demonstrated this phenomenon. Thus, Cry1Ac was shown to bind to BBMV proteins from both susceptible and resistant strains with almost identical binding kinetics (13). These data suggest that binding between Cry1Ac and BBMV proteins from highly Cry1Ac-resistant strains may confer noninsecticidal activity. The identification of these binding interactions may help to elucidate not only the mechanism of Cry toxin resistance but also that of Cry toxin lethality.
In the course of our research to understand toxin resistance, we discovered a novel 252-kDa protein (P252) that binds to Cry1A toxins but is distinct from the 120-kDa APN or 175-kDa CadLP. Furthermore, the association of P252 with Cry1A is not inhibited by GalNAc. Here we describe the characterization of P252 and show that like the 120-kDa APN, it can bind to all three Cry1A toxins. We discuss the properties of P252 in the context of the insecticidal mechanism of Cry toxins.
MATERIALS AND METHODS
Insects.
The silkworm B. mori hybrid Shunrei × Shogetsu was reared on the artificial diet Silkmate (Nosan Kogyo, Yokohama, Japan), and fifth-instar larvae were used in all experiments. Larvae ranging from neonate to fourth instar were used to check the occurrence of P252 in the midgut.
Bacterial culture.
Bacillus thuringiensis was cultured at 300 rpm in a rotary shaker for 3 days at 30°C using 500-ml Erlenmeyer flasks with baffle containing 100 ml of NYS medium (38). Escherichia coli JM109 harboring pYD4.0 (encoding the active portion of cry1Ab) was cultured for 48 h at 37°C in 100 ml of Luria-Bertani broth containing 50 μg of ampicillin/ml (19). The E. coli was a generous gift from K. Kanda, Saga University.
Preparation of Cry1A toxins.
Cry1Aa was prepared from B. thuringiensis subsp. sotto strain T84A1 (a generous gift from M. Ohba, Kyushu University). B. thuringiensis subsp. kurstaki strain HD-73 was used to generate Cry1Ac. Cry1Ab was generated by using the transformed E. coli JM109 described above. To purify Cry toxins, 100 ml of B. thuringiensis cultures was centrifuged at 10,000 × g for 10 min at 4°C, and the pellet was resuspended twice in 1.0 M NaCl and three times with water (each resuspension was centrifuged at 10,000 × g for 10 min). The final pellets were washed with water three times and resuspended in 10 ml of water. This suspension was adjusted to pH 10.0 with 50 mM Na2CO3 containing 10 mM dithiothreitol and allowed to stand for 2 h at 37°C. The alkali-soluble fraction was recovered by centrifugation at 15,000 × g for 10 min at 4°C. A crystalline product from the transformed E. coli strain was obtained as described previously (14, 26) and solubilized as indicated above for the B. thuringiensis materials.
Activation of Cry1A toxins.
The 130-kDa protoxins were activated with trypsin immobilized on Sepharose 4B as described previously (16). The activated 60-kDa Cry1A toxins were fractionated by DEAE-Sepharose column chromatography (30-ml bed volume) in Tris-HCl (pH 8.3) using a linear gradient of 50 to 400 mM NaCl over 300 ml at 4°C.
Preparation of BBMV from B. mori.
A total of 800 2-day-old fifth-instar larvae of B. mori were dissected, and isolated midguts were cut longitudinally to remove the peritrophic membrane. Each midgut was immersed in 0.1 ml of prechilled buffer A (50 mM Tris-HCl [pH 8.0] containing 5 mM EGTA and 300 mM mannitol). The midguts were then transferred to a total of 80 ml of fresh cold buffer A containing 0.1 μM phenylmethylsulfonyl fluoride and 6.3 μM leupeptin and homogenized with a Polytron homogenizer (DIAX-900; Heidolph Instruments) at 11,000 rpm (dial 3.5) for 4 min at 2-min intervals at 4°C. The homogenate was centrifuged at 1,000 × g for 10 min at 4°C, and the supernatant was recovered. The pellet was rehomogenized and centrifuged as described above. The two resulting supernatants were pooled and used to prepare BBMV as previously described (43).
Other instar larvae were also dissected for preparation of BBMV as described above to check for P252. For neonates, however, the whole midgut was homogenized prior to the analysis, since it was difficult to prepare a large amount of BBMV from these larvae.
Solubilization of BBMV.
BBMV (700 ml) were solubilized at 4°C for 1 h in buffer A containing both of the protease inhibitors (see above) and Triton X-100 (1% [vol/vol] final concentration) with gentle rotation. Approximately 700 mg of soluble protein was recovered after centrifugation at 100,000 × g for 1 h at 4°C.
Toyopearl HW-65F and Sephacryl S-300 column chromatography.
Solubilized BBMV proteins were fractionated using a Toyopearl HW-65 gel filtration column (4.5 cm d by 180 cm h) and a Sephacryl S-300 column (2.5 cm d by 100 cm h). Both resins were equilibrated with 100 mM Tris-HCl (pH 8.0) containing 100 mM NaCl and 1% (vol/vol) Triton X-100 (Toyopearl HW-65) or 0.05% Triton X-100 (Sephacryl S-300). For Toyopearl HW-65, 250 fractions of 8 ml each were collected at a flow rate of 80 ml/h. For Sephacryl S-300 chromatography, 2-ml fractions were collected at a flow rate of 40 ml/h. The S-300 fractions of interest were dialyzed against 20 mM Tris buffer (pH 8.0) containing 0.05% Triton X-100.
DEAE-Sepharose column chromatography.
Subsequent to the Toyopearl HW65 chromatography, samples were further fractionated using a DEAE-Sepharose column (2.5 cm d by 20 cm h). Samples were applied to the column, and nonadsorbed materials were removed using 20 mM Tris-HCl (pH 8.0) containing 0.05% (vol/vol) Triton X-100. Sample elution was performed using the same buffer with a 300-ml linear gradient of 0 to 0.3 M NaCl for each 150 ml, collecting 2-ml samples at a flow rate of 60 ml/h. The collected sample fraction was dialyzed and concentrated in 100 mM Tris buffer (pH 8.0) containing 100 mM NaCl and 0.05% Triton X-100. DEAE column chromatography was repeated three times, and after each of the first two times the sample was dialyzed against 20 mM Tris buffer (pH 8.0) containing 0.05% Triton X-100. After the third column, the collected fraction was dialyzed against 100 mM Tris (pH 8.0) containing 100 mM NaCl and 0.05% Triton X-100 and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
SDS-PAGE purification of a 252-kDa BBMV protein.
The final Sephacryl S-300 fractions containing P252 were subjected to SDS-PAGE on a 5% gel, and the gel slice containing P252 was excised and placed into a cellulose bag containing Tris-glycine buffer (pH 8.3). The proteins were electroeluted for 2 h and recovered from the buffer by membrane filtration (Ultra Filter 20,000-molecular-weight-cutoff UP-20; Advantec Toyo, Tokyo, Japan).
Aminopeptidase (AP) activity assay.
AP activity was assayed in a reaction mixture composed of 85 μl of 100 mM Tris-HCl (pH 8.0), 5 μl of 25 mM leucine-p-nitroanilide (L-pNA). The 10-min reaction was initiated by adding 10 μl of enzyme sample. Reactions were terminated by adding 400 μl of 0.5 M glycine-HCl (pH 3.0), and optical density was measured at 405 nm. One unit of AP activity was defined as the level of activity required to hydrolyze 1.0 μmol of L-pNA/min.
Gel electrophoresis.
SDS-PAGE was performed as described previously (25, 16), and the proteins were visualized by staining with silver or Coomassie brilliant blue (CBB). Native PAGE was performed as described by Kishimoto et al. (20).
Ligand blot analysis with Cry1A toxins.
Proteins resolved by SDS-PAGE were transferred to a polyvinylidene difluoride (PVDF) membrane at 2 mA/cm2 of membrane for 2 h using an electric blotter (AE-W675; ATTO, Tokyo, Japan). Membranes were blocked overnight using 1% skim milk in PBST (8.1 mM Na2HPO4, 1.5 mM KH2PO4, 0.13 M NaCl, 2.7 mM KCl, 0.05% [vol/vol] Tween 20).
Ligand blot analysis was performed to assess Cry1Aa, Cry1Ab, and Cry1Ac binding proteins. Membrane-bound Cry1Aa and Cry1Ab were detected using rabbit anti-Cry1Aa, and Cry1Ac was detected using anti-Cry1Ac. Toxin-bound antibodies were visualized using peroxidase-conjugated goat-anti rabbit immunoglobulin G (IgG) (Amersham Pharmacia Biosciences) and visualized using ECL reagents (Amersham Pharmacia Biosciences) and a BAS-3000 detector (Fujifilm Co. Ltd., Kanagawa, Japan).
Cy3 labeling of Cry1A.
Activated Cry1A toxin (1 mg) was labeled with the fluorescent dye Cy3 according to the manufacturer's protocol (Amersham Pharmacia Biosciences). Unreacted Cy3 was separated from labeled Cry 1A toxins by DEAE column chromatography (2.6 by 8 cm; bed volume, ∼2.5 ml). Free Cy3 flowed through the column, and the bound labeled toxin was eluted with 6 ml of 50 mM Tris-HCl (pH 8.0) containing 300 mM NaCl. The purified labeled toxin was quantified using the Bradford method (3). Approximately 85% of the activated Cry1A starting material was labeled with Cy3.
The protein concentrations of the labeled toxins were determined from a calibration curve constructed by using fluorescence intensities measured at 550 (excitation) and 570 (emission) nm. Because of their tendency for quenching, the labeled toxins were used within 1 week. A new calibration curve was plotted for each batch of labeled toxin.
Binding assay.
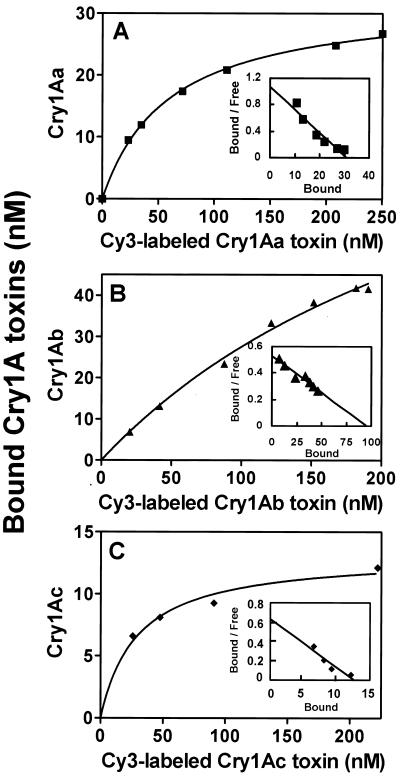
Purified P252 was covalently linked to Sepharose 4B (Amersham Pharmacia Biosciences) as described by the supplier, and 1.7 ml of the resin containing P252 was packed into a small glass column. Various amounts of each labeled toxin were applied to the column to determine the amount of toxin bound. Binding capacity of Cy3-labeled Cry1A was determined by measuring the fluorescence intensity of the bound toxin (550-nm excitation and 570-nm emission) using the calibration curve. To estimate Kd, various amounts of each Cy3-labeled Cry1A toxin were applied to the affinity column and incubated for 1 h at 20°C. After the column was washed with 50 mM Tris-HCl (pH 8.0), the bound labeled toxin was eluted with this buffer, containing 300 mM NaCl, and was quantitated using the calibration curve.
For the heterologous competition assay, Cy3-labeled Cry1A toxin solutions containing various concentrations of unlabeled Cry1A toxins or bovine serum albumin (BSA) were applied to the affinity columns and incubated as described above. Bound toxins were quantified as described above. The amount of bound material was expressed as a percentage of the maximum Cry1A toxin binding capacity of the matrix in the heterologous competition assay.
GraphPad Prism version 3.02 Windows was used to analyze binding parameters (GraphPad Software, San Diego, Calif.).
Nonspecific binding of Cy3-labeled Cry1A toxins was determined as described previously (11), with slight modification, adding 100-fold excess of unlabeled toxin together with the labeled one. And each labeled toxin exhibited only about 2% nonspecific binding.
Western blot analysis of P252.
Western blot analysis of P252 was performed using three antibodies raised against the four types of APN from B. mori (34) and CadLP of B. mori (BtR175). Detection of antibodies bound to P252 was achieved using peroxidase-conjugated goat anti-rabbit IgG or goat anti-mouse IgG, and bands were visualized using ECL. Recombinant APN-120K and BTR175 served as positive controls.
Lectin binding assay.
Peroxidase-conjugated Concanavalin A (ConA), soybean agglutinin (SBA), Lens culinaris agglutinin (LCA), wheat germ agglutinin (WGA), Phaseolus vulgaris agglutinin (PHA-E4), and peanut agglutinin (PNA) were purchased from Seikagaku Corporation (Tokyo, Japan). P252 isolated by SDS-PAGE was transferred to PVDF membranes by using an electric blotter as described above, and the membranes were blocked for 2 h with 1% BSA in TBST (20 mM Tris-HCl [pH 8.0], 150 mM NaCl, 0.05% [vol/vol] Tween 20) with the exception of WGA, for which the buffer contained 500 mM NaCl. After washing with water, membranes were incubated in blocking buffer containing various lectins. The bound lectins were detected using ECL as described for Western blotting.
Inhibition of P252-Cry1Ac binding by sugars.
To test the effects of sugars, such as GalNAc, N-acetyl-glucosamine (GlcNAc), mannose (Man), fucose (Fuc), and galactose (Gal), a 100 mM concentration of each sugar was added to the P252 on PVDF membranes in ligand blot binding solution 1 h prior to the reaction with Cry1Ac toxin, and the effects were evaluated by ligand blot assay as described above.
Protein quantification.
Solubilized or chromatographed BBMV proteins were monitored by optical density at 280 nm and quantified by the Bradford method (Bio-Rad) (3).
RESULTS
Purification of a 252-kDa Cry1A binding protein and SDS-PAGE analysis.
Our previous ligand blot analyses of BBMV proteins from B. mori often detected a protein that migrated at ∼252 kDa by SDS-PAGE. This protein was recognized in ligand blots with Cry1A toxins but not by antibodies to APN-120K or BtR175. Based on these unique properties, we performed a more detailed evaluation of Cry toxin binding proteins derived from preparations of midgut epithelial cell membranes.
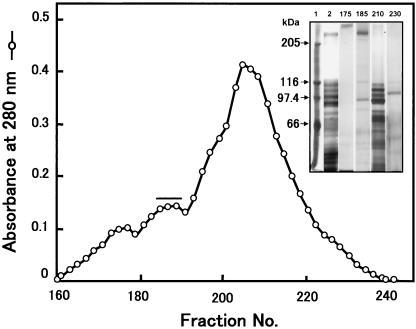
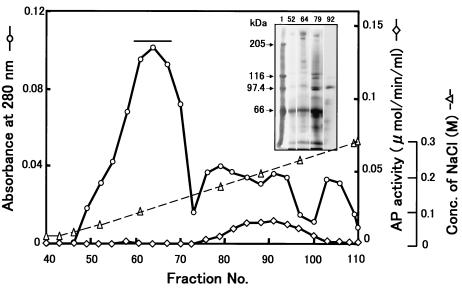
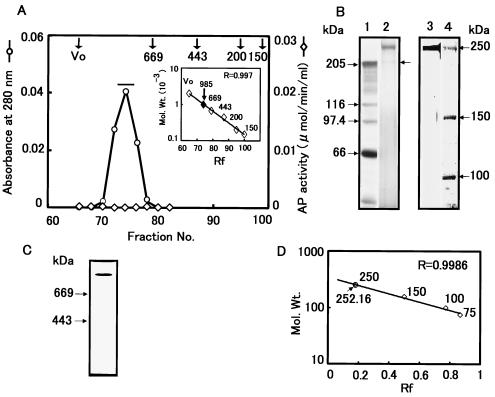
We fractionated soluble BBMV proteins using a Toyopearl HW-65F column (4.5 cm d by 180 cm h) as a first step because the initial use of DEAE ion-exchange chromatography did not effectively separate proteins in the 200- to 300-kDa range from proteins with molecular masses of 90 to 130 kDa. The Toyopearl column was effective in this regard (Fig. 1). The Toyopearl fractions containing the 252-kDa protein (P252) were then applied to a DEAE-Sepharose column that was eluted with an NaCl gradient (Fig. 2). P252 was eluted at ∼100 mM NaCl, and these fractions contained only residual APN activity. After two more identical rounds of DEAE chromatography, two rounds of Sephacryl S-300 chromatography were performed. The second Sephacryl S-300 column yielded a single, symmetrical absorbance peak that contained P252 (Fig. 3). SDS-PAGE/CBB staining showed a single band migrating at ∼250 kDa. Silver staining, however, showed a low level of protein contaminants (<0.1%) (Fig. 3B, lane 2) that were removed upon subsequent electrophoresis of this fraction via SDS-PAGE. The band corresponding to P252 was excised from the gel, and P252 was electroeluted from the gel slice. SDS-PAGE/silver staining of the electroeluted P252 showed a single band, demonstrating that we had recovered a homogeneous, highly purified preparation of this protein (Fig. 3B, lane 3). Native PAGE analysis with silver staining also showed a single band (Fig. 3C). Native PAGE does not necessarily indicate molecular mass, but the P252 band migrated to a position that was apparently larger than the 669-kDa marker.
FIG. 1.
Toyopearl HW-65F gel filtration chromatography. Chromatography of BBMV proteins prepared from fifth-instar larvae of B. mori. Each 8 ml of each column fraction was collected at a flow rate of 80 ml/h, and the proteins were resolved by SDS-PAGE (superimposed gel). Lane 1, protein molecular mass markers; lane 2, Triton X-100-soluble protein from BBMV; lane 3, fraction 175; lane 4, fraction 185; lane 5, fraction 210; lane 6, fraction 230. Fractions 185 to 190 (horizontal bar) were pooled and further purified by using DEAE-Sepharose chromatography.
FIG. 2.
DEAE-Sepharose chromatography of P252. Chromatogram of DEAE column fractionation of Toyopearl HW-65F gel filtration fractions containing P252. Each 2-ml sample was collected at a flow rate of 60 ml/h from the DEAE column and resolved by SDS-PAGE with silver staining. Lane 1, protein molecular mass markers; lanes 2, fraction 52; lane 3, fraction 64; lane 4, fraction 79; lane 5, fraction 92. Fractions 60 to 68 (horizontal bar) were pooled and further fractionated by DEAE column chromatography and gel filtration chromatography.
FIG. 3.
Sephacryl S-300 gel filtration chromatography of P252. (A) Fractions 52 to 64 collected from the DEAE chromatography shown in Fig. 2 were further chromatographed using a Sephacryl S-300 column. The elution profile represents the third of three sequential S-300 fractionation steps. A molecular size calibration standard curve derived using thyroglobulin (669 kDa), apoferritin (443 kDa), β-amylase (200 kDa), and alcohol dehydrogenase (150 kDa) is shown as an inset of panel A. (B) SDS-PAGE of P252 derived from Sephacryl S-300 chromatography from panel A. Lane 1, protein molecular mass markers; lane 2, sample from a pool of fractions 73 to 76 from the absorbance peak designated by the horizontal bar in panel A; lane 3, sample from lane 2 following gel purification; lane 4, protein molecular mass markers. Proteins were visualized by silver staining. (C) Native PAGE of purified P252. The gel was silver stained to determine the purity of the P252 preparation. (D) Determination of molecular weight of P252 by SDS-PAGE. A calibration curve derived from SDS-PAGE molecular weight standards run in parallel with the S-300 fractionated product was used to determine the molecular weight of P252.
The molecular mass of the protein was estimated to be 985 ± 55 kDa (n = 5) as measured by standardized Sephacryl S-300 column chromatography (Fig. 3A, superimposed graph) although a molecular mass of 252 ± 3.16 kDa (n = 8) was estimated by SDS-5% PAGE (Fig. 3D). The final yield of purified P252 was 0.08%. No measurable L-pNA hydrolysis activity was evident either in the fractions containing P252 in each purification step or in SDS-PAGE-purified P252.
Ligand blot and direct binding analysis of Cry1A toxins with P252.
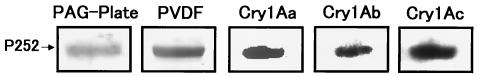
The toxin-binding characteristics of P252 were determined by ligand blot analysis using purified Cry1Aa, Cry1Ab, and Cry1Ac toxins (Fig. 4). Each of these toxins bound to P252, although the signal obtained for Cry1Ab was relatively low. The ratio of the amounts of Cry1Aa, Cry1Ab, and Cry1Ac bound under these conditions was 1.7:1.0:2.0, respectively, determined using a computer-aided densitometer (data not shown).
FIG. 4.
Ligand blot of P252 binding to Cry1A toxins. P252 was resolved by SDS-PAGE and transferred to a PVDF membrane. Samples from the SDS-PAGE gel (PAG-Plate) and the PVDF membrane (PVDF) were stained with CBB. Immobilized P252 was incubated with Cry1Aa, Cry1Ab, and Cry1Ac, and toxin binding was visualized using anti-Cry1Aa and anti-Cry1Ac.
The affinity of the toxins Cry1Aa, Cry1Ab, and Cry1Ac for P252 was determined using Cy3-labeled Cry1A toxins and a P252 affinity column. First, direct binding was analyzed using the Cry1A toxins (Fig. 5), and the data were used to construct Scatchard plots (Fig. 5, superimposed graphs). The experimental Kd values for Cry1Aa, Cry1Ab, and Cry1Ac were 28.9, 178.5, and 20.0 nM, respectively.
FIG. 5.
Total binding assay of Cy3-labeled Cry1A toxins with P252. Binding constants for Cy3-labeled Cry1Aa (A), Cry1Ab (B), and Cry1Ac (C) were determined using a P252 affinity column. Various amounts of Cy3-labeled proteins were incubated on the column and washed. The bound Cry toxin was eluted and quantitated by fluorescence, using a calibration curve. The superimposed graphs show Scatchard plots to determine each kd of Cy3-labeled Cry1A toxin.
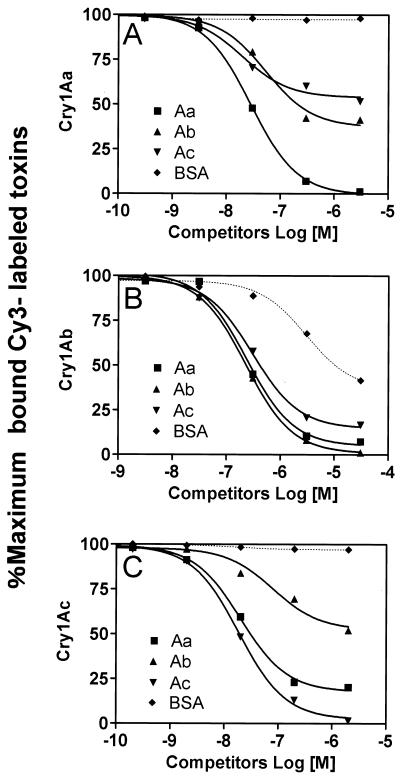
Heterologous assay for Cry1A toxin binding to P252.
A heterologous competitive binding assay was performed using labeled Cry1A toxins at a 30 nM concentration of Cry1Aa, an 180 nM concentration of Cry1Ab, and a 20 nM concentration of Cry1Ac (Fig. 6). First we demonstrated that the addition of BSA at 10-fold excess over the amount of labeled Cry1A toxins did not affect the binding of Cry1Aa and Cry1Ac to the affinity column. However, BSA strongly inhibited the binding between Cry1Ab and P252 (Fig. 6). Unlabeled Cry1Aa at 1 μM inhibited Cy3-labeled Cry1Aa binding by 95%, but 1 μM unlabeled Cry1Ab and Cry1Ac inhibited the binding by about 50% (Fig. 6A). Unlabeled Cry1Ab at a 1 μM concentration inhibited the binding of Cy3-labeled Cry1Ab to P252 by 95%, and 1 μM Cry1Aa and Cry1Ac inhibited the binding by 80 and ∼90%, respectively. Binding between Cy3-labeled Cry1Ac and P252 was inhibited by 90% in the presence of 1 μM unlabeled Cry1Ac, whereas 1 μM Cry1Aa inhibited the binding by 75% and Cry1Ab inhibited the binding by 40% (Fig. 6C). Thus, these studies indicated that the Cry1Ab binding site on P252 was shared by Cry1Aa, Cry1Ac, and partially even BSA. In contrast, the site for Cry1Ac binding to P252 was shared by Cry1Aa, but the Cry1Aa site was shared only partially by Cry1Ab and Cry1Ac.
FIG. 6.
Heterologous competition binding assay between Cry1A toxins and P252. The same procedure was used as for Fig. 5, except that the Cy3-labeled Cry1A toxin solution contained various amounts of competing unlabeled Cry1Aa, Cry1Ab, Cry1Ac, or BSA. The amount of bound Cy3-labeled Cry1A toxin was expressed as a percentage of the maximum labeled Cry1A toxins bound.
Effect of sugars on P252-Cry1A toxin binding.
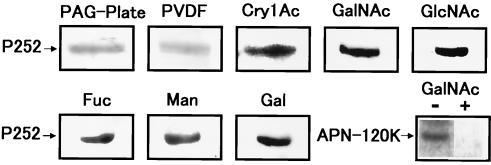
The binding between APN-120K and Cry1Ac is inhibited by GalNAc (21). Therefore, we examined the effect of the sugars GalNAc, GlcNAc, Man, Fuc, and Gal (each 100 mM) on P252-Cry1Ac binding (Fig. 7). None of these sugars inhibited binding. Furthermore, these sugars did not affect the binding of either Cry1Aa or Cry1Ab to P252 (data not shown). In parallel, the inhibitory effect of GalNAc on the binding of partially purified APN-120K and Cry1Ac was demonstrated (Fig. 7, bottom row, right panel).
FIG. 7.
Inhibition of binding between P252 and Cry1Ac by sugars, using ligand blot analysis. P252 was resolved by SDS-PAGE and transferred to a PVDF membrane, and samples from the SDS-PAGE gel (PAG-Plate) and the PDVF membrane (PVDF) were stained with CBB. P252 on the PVDF membrane was preincubated with the sugar GalNAc, GlcNAc, Gal, Fuc, or Man at a 100 mM concentration for an hour and then incubated with Cry1Ac. The sugars used in the blotting assays are shown above each panel. As a positive control, the inhibition of binding between partially purified APN-120K and Cry1Ac was demonstrated with (+) and without (−) GalNAc (bottom-right panel).
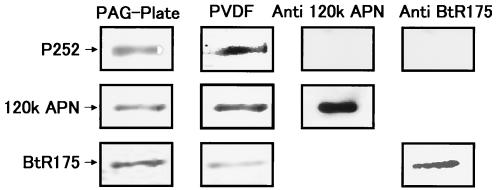
Western blot analysis of P252 using antibodies to APN-120K and BtR175.
Both APN-120K and BtR175 have been proposed to be receptors for Cry1A toxins. To determine whether P252 is antigenically related to either of these proteins, Western blot analysis of P252 (Fig. 8) was performed using antibodies against APN-120K (group 1 APN) (34) and BtR175. While APN-120K and BtR175 were detected by their respective antibodies, neither antibody detected purified P252. The PVDF membrane was also stained with CBB to verify that a sufficient quantity of P252 was transferred to the membrane to enable detection. In addition to the APN-120K (group 1) antibody, we also tested antisera against APN of groups 2, 3 and 4 and found that none recognized P252 (data not shown).
FIG. 8.
Western blots of P252 using anti-APN-120K and anti-BtR175. P252 was resolved by SDS-PAGE and transferred to a PVDF membrane, and samples from the SDS-PAGE gel (PAG-Plate) and the PDVF membrane (PVDF) were stained with CBB. Western blotting was performed using anti-APN-120K or anti-BtR175, as indicated (top row). As controls, partially purified APN-120K (middle row) and BtR175 (bottom row) were also analyzed using the appropriate antibodies.
Binding of various lectins to P252.
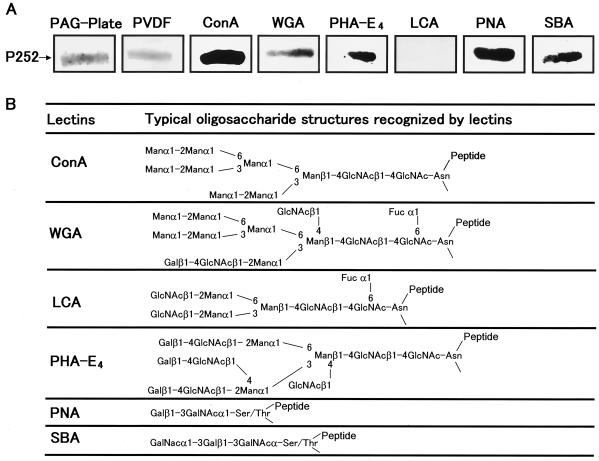
To determine the composition of the sugar side chains that may be present on P252, we assessed the binding of P252 to various lectins (ConA, WGA, LCA, PHA-E4, PNA, and SBA). ConA, WGA, PHA-E4, PNA, and SBA all bound to P252, but LCA did not (Fig. 9A), suggesting that P252 may contain both N- and O-linked sugar side chains.
FIG. 9.
Lectin binding analysis of P252. (A) P252 was resolved by SDS-PAGE and transferred to a PVDF membrane, and samples from the SDS-PAGE gel (PAG-Plate) and the PDVF membrane (PVDF) were stained with CBB. Lectins (ConA, WGA, PHA-E4, LCA, PNA, and SBA) conjugated to peroxidase were incubated with the immobilized P252. (B) Schematic of typical oligosaccharide structures recognized by various lectins used in this experiment.
Partial amino acid sequence.
To determine whether P252 is a novel protein, we attempted to ascertain the amino acid sequence from the N terminus but found that it was blocked. We therefore sequenced two internal peptides generated after Cleveland digestion (4) by using V8 protease. The resulting sequences were ATYLAGSGGVVPVCN and ATYTLNSDNTITVFN. Neither of these sequences matched any protein in the available databases.
DISCUSSION
Many detergent-soluble BBMV proteins from B. mori have been shown to bind Cry1Aa, Cry1Ab, and Cry1Ac in ligand blots as well as the toxin affinity columns, although the individual binding profiles differ among the three toxins (Y. Shitomi, Y. Nakano, T. Hayakawa, R. Kawai, T. Kishimoto, T. Mitui, K. Yaoi, R. Sato, and H. Hori, Abstr. Proc. 4th Pacific Rim Conf., p. 41, 2001). While the binding of these toxins has not been fully characterized, it is likely that many such toxin-protein interactions are necessary components of the insecticidal mechanism used by Cry1A toxins. Thus, a more precise and comprehensive investigation of BBMV proteins is a prerequisite to developing a full understanding of insecticidal mechanisms.
Here, we identified a 252-kDa protein, denoted P252, which may be an important toxin-binding component of BBMV proteins. The occurrence of P252 in BBMV and the binding of P252 to Cry1A toxins were comparable to that between the 110-kDa APN and Cry1Aa or APN-120K and Cry1Ac (Shitomi et al., unpublished data). Binding of P252 to all three toxins seems to be specific, since each labeled toxin was significantly inhibited by its respective unlabeled toxin. Because P252 did not react with either anti-APN-120K or anti-BtR175, we propose that P252 constitutes a unique Cry1A binding factor. This idea is supported by our internal amino acid sequence analysis, which indicates that P252 is a novel protein. Furthermore, the Kd values we determined indicate that P252 binds with high affinity to Cry1A toxins in midgut epithelial cells, suggesting that these interactions may occur under physiological conditions. Based on these analyses, we propose that that the binding between P252 and Cry1A toxins is specific and constitutes one of the strongest interactions known between Cry1A toxins and midgut epithelial membrane proteins, including both APN and cadherin-like proteins (27, 41, 17). These characteristics warrant further investigation of the functional significance of P252-Cry1A interactions.
The binding site for Cry1Aa in P252 seems to partially overlap with the binding sites for Cry1Ab and Cry1Ac. On the other hand, the Cry1Ab binding site in P252 appears to be shared by Cry1Aa, Cry1Ac, and even BSA. These unusual characteristics of Cry1Ab are very interesting and suggest that Cry1Ab may bind in a unique fashion to the BBM of the midgut.
GalNAc inhibits the binding of Cry1Ac to APN-120K (21). For this reason, the association between these molecules is thought to take place, at least in part, via a GalNAc residue. Our results suggest that P252 contains both N- and O-linked oligosaccharides that likely include GalNAc residues. However, toxin binding to P252 was not inhibited by GalNAc, Man, Gal, or GlcNAc, suggesting that the mechanism of P252 binding to Cry1Ac toxin may differ from that of APN-120K. However, at this time we cannot rule out the possibility that other P252 sugar side chains may be involved in the binding.
Our data suggest that P252 may exist as an oligomeric molecule in solution because its migration during gel filtration indicated a mass that is approximately four times larger than that measured by SDS-PAGE. Additionally, digestion of BBMV with phosphatidylinositol-specific phospholipase C (PIPLC) failed to release P252 from the plasma membrane (data not shown). The meaning of this experiment is not clear, since APN-120K, which is a GPI-anchored protein, was also not digested with PIPLC in control experiments (data not shown). However, if the GPI anchor of APN is modified with other lipid or dolichol derivatives, PIPLC may not cleave the GPI anchors (1). Thus, the failure of PIPLC to release P252 may be interpreted in two ways. First, P252 may not have a GPI anchor, and therefore it must be a transmembrane protein or one that is partially inserted into the membrane. Alternatively, the GPI anchor of P252 may be modified in a manner that prevents cleavage by PIPLC. Additionally, various APNs reportedly exist as dimeric or oligomeric proteins in mammalian cells (39), and oligomeric APN may lack a GPI anchor (2, 5). Also, it has been proposed that GPI-anchored proteins are localized in rafts in epithelial cell membranes, which are not thought to be detergent soluble. While we have not yet provided direct evidence that P252 is a transmembrane protein, its solubility in Triton X-100 (data not shown) suggests that it is not a GPI-anchored protein, in contrast to the 120-kDa APN that is primarily found in the Triton X-100-insoluble fraction (15). These data along with our preliminary data suggest that oligomeric P252 may be a transmembrane or membrane-anchored protein.
The lectins we used are known to recognize specific sugar constructs as indicated: ConA recognizes -Manα1-2Man1- in high-mannose- or complex-type oligosaccharides (8, 23); WGA recognizes the following oligosaccharides with the order cited, GlcNAcβ1-4GlcNAcβ1-4GlcNAc, GlcNAcβ1-4GlcNAc,and GlcNAc (45); LCA recognizes GlcNAcβ1-2Manα1-3(GlcNAcβ1-2Manα1-6)Manβ1-4GlcNAcβ1-4(Fucα1-6) GlcNAc-Asn (18, 45); PHA-E4, Galβ1-4GlcNAcβ1-2(Galβ1-4GlcNAcβ1-4)Manα1-3[(Galβ1-4GlcNAcβ1-2Manα1-6(GlcNAcβ1-4)]Manα1-4GlcNAcβ1-4GlcNAc-Asn (22); PNA recognizes Galβ1-3GalNAc (35); and SBA recognizesGalNAcα1-3Gal (36). These binding characteristics suggest that P252 contains N-linked complex and high-mannose-type oligosaccharides. These data also suggest that P252 may have the mucin-type sugar side chain, Galβ1-3GalNAc, or O-linked GalNAcα1-3Gal, since the protein reacted strongly with PNA and SBA. However, the lack of binding by LCA suggests that P252 is unlikely to have a fucosylated biantennal hybrid structure. PHA-E4 has specificity toward complex-type oligosaccharides having two or three antennal structures, and it also exhibits stronger binding to structures containing bisecting GalNAc residues. Therefore, since WGA bound to P252 and LCA did not, P252 may contain sugar side chains with triantennal-type structures, such as Galβ1-4GlcNAcβ1-2(Galβ1-4GlcNAcβ1-4)Manα1-3[Galβ1-4GlcNAcβ1-2Manα1-6(GlcNAcβ1-4)]Manα1-4GlcNAcβ1-4GlcNAc-Asn. O-linked sugar side chains, such as Galβ1-3GalNAc and GalNAcα1-3Gal-, may also be present on P252, since PNA and SBA bound to the protein (Fig. 9B).
While our present data enhance our understanding of the role of P252 in insecticidal mechanisms, the true function of this protein has not been definitively determined. However, we can assert that if P252 is a legitimate binding component involved in insecticidal mechanisms, then it may serve to assemble Cry toxins and adjacent proteases on the plasma membrane of host cells. Alternatively, if binding of P252 to Cry1A toxins bears no direct relationship to pore formation, then such binding may substantially reduce the toxicity of Cry1Ac to B. mori.
Interestingly, the B. mori strain we used in this experiment is very resistant to Cry1Ac. The 50% lethal concentrations of Cry1Ac and Cry1Aa were estimated, and the lethality of Cry1Aa was shown to be more than 3,000 times higher than that of Cry1Ac (Shitomi et al., unpublished data). Therefore, a large number of binding interactions between Cry1Ac and P252, whose Kd is comparable to that of Cry1Aa, may not exhibit lethal activity. Elucidating the role of the binding between P252 and the Cry1A toxins may ultimately contribute to a more complete understanding of the insecticidal mechanisms of Cry1A toxins and the sensitivity of B. mori to their action. P252 was shown to occur in midgut or BBMV from neonate and second- and third-instar larvae as well as fifth-instar larvae (data not shown).
Acknowledgments
This work was supported in part by research grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan (12558069 and 13306006 to H.H.). D.M.H. was supported by a scholarship from the Jin-nai Foundation.
REFERENCES
- 1.Baumann, N. A., and A. K. Menon. 2002. Lipid modification of proteins, p. 37-54. In D. E. Vance and J. E. Vance (ed.), Biochemistry of lipids, lipoproteins and membranes, 4th ed. Elsevier Sciences B. V., Amsterdam, The Netherlands.
- 2.Benajiba, A., and S. Maroux. 1981. Subunit structure of pig small-intestinal brush-border aminopeptidase N. Biochem. J. 197:573-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 4.Cleveland, D. W., S. G. Fischer, M. L. Kirschner, and U. K. Laemmli. 1977. Peptide mapping by limited proteolysis in sodium dodecylsulphate and analysis by gel electrophoresis. J. Biol. Chem. 252:1102-1106. [PubMed] [Google Scholar]
- 5.Danielsen, E. M. 1994. Dimeric assembly of enterocyte brush border enzymes. Biochemistry 33:1599-1605. [DOI] [PubMed] [Google Scholar]
- 6.Gahan, L. J., F. Gould, and D. G. Heckel. 2001. Identification of a gene associated with Bt resistance in Heliothis virescens. Science 293:857-860. [DOI] [PubMed] [Google Scholar]
- 7.Gill, S. S., E. A. Cowles, and V. Francis. 1995. Identification, isolation, and cloning of a Bacillus thuringiensis CryIAc toxin-binding protein from the midgut of the lepidopteran insect Heliothis virescens. J. Biol. Chem. 270:27277-27282. [DOI] [PubMed] [Google Scholar]
- 8.Goldstein, I. J., C. M. Reichert, and A. Masaki. 1974. Interaction of concanavalin A with model substrates. Ann. N. Y. Acad. Sci. 234:283-296. [DOI] [PubMed] [Google Scholar]
- 9.Gomez, I., D. I. Oltean, S. S. Gill, A. Bravo, and M. Soberon. 2001. Mapping the epitope in cadherin-like receptors involved in Bacillus thuringiensis Cry1A toxin interaction using phage display. J. Biol. Chem. 276:28906-28912. [DOI] [PubMed] [Google Scholar]
- 10.Gomez, I., J. Sanchez, R. Miranda, A. Bravo, and M. Soberon. 2002. Cadherin-like receptor binding facilitates proteolytic cleavage of helix α-1 in domain I and oligomer pre-pore formation of Bacillus thuringiensis Cry1Ab toxin. FEBS Lett. 513:242-246. [DOI] [PubMed] [Google Scholar]
- 11.Gonzalez-Cabrera, J., B. Escriche, B. El Tabashnik, and J. Ferre. 2003. Binding of Bacillus thuringiensis toxins in resistant and susceptible strains of pink bollworm (Pectinophora gossypiella). Insect Biochem. Mol. Biol. 33:929-935. [DOI] [PubMed] [Google Scholar]
- 12.Hara, H., S. Atsumi, K. Yaoi, K. Nakanishi, S. Higurashi, N. Miura, H. Tabunoki, and R. Sato. 2003. A cadherin-like protein functions as a receptor for Bacillus thuringiensis Cry1Aa and Cry1Ac toxins on midgut epithelial cells of Bombyx mori larvae. FEBS Lett. 538:29-34. [DOI] [PubMed] [Google Scholar]
- 13.Higuchi, M., N. S. Kumaraswami, T. Maruyama, T. Hayakawa, T. Mitsui, R. Sato, and H. Hori. 2001. Comparative study of proteins and neutral glycolipids of insect midgut brush border membrane in Cry1Ac susceptible and highly resistant Plutella xylostella, p. 199-208. In M. Ohba, O. Nakamura, E. Mizuki, and T. Akao (ed.), Proceedings of a centennial symposium commemorating Ishiwata's discovery of Bacillus thuringiensis. Kyushu University, Fukuoka, Japan.
- 14.Höfte, H., H. de Greve, J. Seurinck, S. Jansens, J. Mahillon, C. Ampe, J. Vandekerckhove, H. Vanderbruggen, M. van Montagu, M. Zabeau, and M. Vaeck. 1986. Structural and functional analysis of a cloned delta endotoxin of Bacillus thuringiensis berliner 1715. Eur. J. Biochem. 161:273-280. [DOI] [PubMed] [Google Scholar]
- 15.Hooper, N. M., and A. Bashir. 1991. Glycosyl-phosphatidylinositol-anchored membrane proteins can be distinguished from transmembrane polypeptide-anchored proteins by differential solubilization and temperature-induced phase separation in Triton X-114. Biochem. J. 280:745-751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Indrasith, L. S., K. Ogiwara, M. Minami, T. Iwasa, T. Maruyama, N. Suzuki, S. Asano, K. Sakanaka, and H. Hori. 1991. Processing of delta endotoxin from Bacillus thuringiensis subspp. Kurstaki HD-1 and HD-73 by immobilized trypsin and chymotrypsin. Appl. Entomol. Zool. 26:485-492. [Google Scholar]
- 17.Jenkins, J. L., and D. H. Dean. 2001. Binding specificity of Bacillus thuringiensis Cry1Aa for purified, native Bombyx mori aminopeptidase N and cadherin-like receptors. BMC Biochem. 2:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaifu, R., T. Osawa, and R. W. Jeanloz. 1975. Synthesis of 2-O-(2-acetamido-2-deoxy-beta-d-glucopyranosyl)-d-mannose, and its interaction with d-mannose-specific lectins. Carbohydr. Res. 40:111-117. [DOI] [PubMed] [Google Scholar]
- 19.Kim, Y. S., K. Kanda, F. Kato, and A. Murata. 1998. Effect of the carboxyl-terminal portion of Cry1Ab in Bacillus thuringiensis on toxicity against the silkworm, Bombyx mori. Appl. Entomol. Zool. 33:473-477. [Google Scholar]
- 20.Kishimoto, T., H. Hori, D. Takano, Y. Nakano, M. Watanabe, and T. Mitsui. 2001. Rice α-mannosidase digesting the high mannose glycopeptide of glutelin. Physiol. Plant. 112:15-24. [DOI] [PubMed] [Google Scholar]
- 21.Knight, P. J., N. Crickmore, and D. J. Ellar. 1994. The receptor for Bacillus thuringiensis CryIA(c) δ-endotoxin in the brush border membrane of the lepidopteran Manduca sexta is aminopeptidase N. Mol. Microbiol. 11:429-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kobata, A., and K. Yamashita. 1989. Affinity chromatography of oligosaccharides on E4-phytohemagglutinin-agarose column. Methods Enzymol. 179:46-54. [DOI] [PubMed] [Google Scholar]
- 23.Kornfeld, R., and C. Ferris. 1975. Interaction of immunoglobulin glycopeptides with concanavalin A. J. Biol. Chem. 252:2614-2619. [PubMed] [Google Scholar]
- 24.Kumaraswami, N. S., T. Maruyama, S. Kurabe, T. Kishimoto, T. Mitsui, and H. Hori. 2001. Lipids of brush border membrane vesicle (BBMV) from Plutella xylostella resistant and susceptible to Cry1Ac δ-endotoxin of Bacillus thuringiensis. Comp. Biochem. Physiol. B 129:173-183. [DOI] [PubMed] [Google Scholar]
- 25.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 26.Lee, M. K., R. E. Milne, A. Z. Ge, and D. H. Dean. 1992. Location of a Bombyx mori receptor binding region on a Bacillus thuringiensis δ-endotoxin. J. Biol. Chem. 267:3115-3121. [PubMed] [Google Scholar]
- 27.Luo, K.-E., S. Sangadala, L. Masson, A. Mazz, R. Brousseau, and M. J. Adang. 1997. The Heliothis virescens 170 kDa aminopeptidase function as “receptor A” by mediating specific Bacillus thuringiensis Cry1A δ-endotoxin binding and pore formation. Insect Biochem. Mol. Biol. 27:735-743. [DOI] [PubMed] [Google Scholar]
- 28.Martinez-Ramirez, A. C., S. Gonzalez-Nebauer, B. Esriche, and M. D. Real. 1994. Ligand blot identification of a Manduca sexta midgut binding protein specific to three Bacillus thuringiensis Cry1A-type ICPs. Biochem. Biophys. Res. Commun. 201:782-787. [DOI] [PubMed] [Google Scholar]
- 29.Masson, L., J. Y. Yang, Y. Lu, A. Mazza, R. Brousseau, and M. J. Adang. 1995. The CryIA(c) receptor purified from Manduca sexta displays multiple specificities. J. Biol. Chem. 270:20309-20315. [DOI] [PubMed] [Google Scholar]
- 30.Morse, R. J., T. Yamamoto, and R. M. Stroud. 2001. Structure of Cry2Aa suggests an unexpected receptor binding epitope. Structure 9:409-417. [DOI] [PubMed] [Google Scholar]
- 31.Nagamatsu, Y., S. Toda, F. Yamaguchi, M. Ogo, M. Kogure, M. Nakamura, Y. Shibata, and T. Katsumoto. 1998. Identification of Bombyx mori midgut receptor for Bacillus thuringiensis insecticidal CryIA(a) toxin. Biosci. Biotechnol. Biochem. 62:718-726. [DOI] [PubMed] [Google Scholar]
- 32.Nagamatsu, Y., S. Toda, T. Koike, Y. Miyoshi, S. Shigematsu, and M. Kogure. 1998. Cloning, sequencing, and expression of the Bombyx mori receptor for Bacillus thuringiensis insecticidal CryIA(a) toxin. Biosci. Biotechnol. Biochem. 62:727-734. [DOI] [PubMed] [Google Scholar]
- 33.Nagamatsu, Y., T. Koike, K. Sasaki, A. Yoshimoto, and Y. Furukawa. 1999. The cadherin-like protein is essential to specificity determination and cytotoxic action of the Bacillus thuringiensis insecticidal CryIAa toxin. FEBS Lett. 460:385-390. [DOI] [PubMed] [Google Scholar]
- 34.Nakanishi, K., K. Yaoi, Y. Nagino, H. Hara, M. Kitami, S. Atsumi, N. Miura, and R. Sato. 2002. Aminopeptidase N isoforms from the midgut of Bombyx mori and Plutella xylostella—their classification and the factors that determine their binding specificity to Bacillus thuringiensis Cry1A toxin. FEBS Lett. 519:215-220. [DOI] [PubMed] [Google Scholar]
- 35.Pereira, M. E., and E. A. Kabat. 1976. Immunochemical studies on blood groups LXII. Fractionation of hog and human A, H, and AH blood group active substance on insoluble immunoadsorbents of Dolichos and Lotus lectins. J. Exp. Med. 143:422-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pereira, M. E., E. A. Kabat, and N. Sharon. 1974. Immunochemical studies on the specificity of soybean agglutinin. Carbohydr. Res. 37:89-102. [DOI] [PubMed] [Google Scholar]
- 37.Sangadala, S., F. S. Walters, L. H. English, and M. J. Adang. 1994. A mixture of Manduca Sexta aminopeptidase and phosphatase enhances Bacillus thuringiensis insecticidal CryIA(c) toxin binding and 86Rb(+)-K(+) efflux in vitro. J. Biol. Chem. 269:10088-10092. [PubMed] [Google Scholar]
- 38.Suzuki, M., H. Hori, K. Ogiwara, S. Asano, R. Sato, M. Ohba, and H. Iwahana. 1992. Insecticidal spectrum of a novel isolate of Bacillus thuringiensis serovar japonensis. Biol. Control 2:138-142. [Google Scholar]
- 39.Taylor, A. 1993. Aminopeptidase: Structure and function. FASEB J. 7:290-298. [DOI] [PubMed] [Google Scholar]
- 40.Vadlamudi, R. K., E. Weber, I. Ji, T. H. Ji, and L. A. Bulla, Jr. 1995. Cloning and expression of a receptor for an insecticidal toxin of Bacillus thuringiensis. J. Biol. Chem. 270:5490-5494. [DOI] [PubMed] [Google Scholar]
- 41.Valaitis, A. P., J. L. Jenkins, M. K. Lee, D. H. Dean, and K. J. Garner. 2001. Isolation and partial characterization of Gypsy moth BTR-270, an anionic brush border membrane glycoconjugate that binds Bacillus thuringiensis Cry1A toxins with high affinity. Arch. Insect Biochem. Physiol. 46:186-200. [DOI] [PubMed] [Google Scholar]
- 42.Valaitis, A. P., M. K. Lee, F. Rajamohan, and D. H. Dean. 1995. Brush border membrane aminopeptidase-N in the midgut of the gypsy moth serves as the receptor for the CryIA(c) δ-endotoxin of Bacillus thuringiensis. Insect Biochem. Mol. Biol. 25:1143-1151. [DOI] [PubMed] [Google Scholar]
- 43.Wolfersberger, M., P. Luethy, A. Maurer, P. Parenti, F. V. Sacchi, B. Giordana, and G. M. Hanozet. 1987. Preparation and partial characterization of amino acid transporting brush border membrane vesicles from the larval midgut of the cabbage butterfly (Pieris brassicae). Comp. Biochem. Physiol. 86A:301-308. [Google Scholar]
- 44.Yamamoto, K., T. Tsuji, and T. Osawa. 1982. Requirement of the core structure of a complex-type glycopeptide for the binding to immobilized lentil- and pea-lectins. Carbohydr. Res. 110:283-289. [DOI] [PubMed] [Google Scholar]
- 45.Yamamoto, K., T. Tsuji, I. Matsumoto, and T. Osawa. 1981. Structural requirements for the binding of oligosaccharides and glycopeptides to immobilized wheat germ agglutinin. Biochemistry 20:5894-5899. [DOI] [PubMed] [Google Scholar]