Abstract
It was long been noted that secondary metabolism is associated with fungal development. In Aspergillus nidulans, conidiation and mycotoxin production are linked by a G protein signaling pathway. Also in A. nidulans, cleistothecial development and mycotoxin production are controlled by a gene called veA. Here we report the characterization of a veA ortholog in the aflatoxin-producing fungus A. parasiticus. Cleistothecia are not produced by Aspergillus parasiticus; instead, this fungus produces spherical structures called sclerotia that allow for survival under adverse conditions. Deletion of veA from A. parasiticus resulted in the blockage of sclerotial formation as well as a blockage in the production of aflatoxin intermediates. Our results indicate that A. parasiticus veA is required for the expression of aflR and aflJ, which regulate the activation of the aflatoxin gene cluster. In addition to these findings, we observed that deletion of veA reduced conidiation both on the culture medium and on peanut seed. The fact that veA is necessary for conidiation, production of resistant structures, and aflatoxin biosynthesis makes veA a good candidate gene to control aflatoxin biosynthesis or fungal development and in this way to greatly decrease its devastating impact on health and the economy.
Among the most common toxigenic fungi affecting the human food chain are Aspergillus spp., infecting major agricultural commodities such as corn, peanuts, cotton, sorghum, and other oil-seed sources. Aspergillus parasiticus produces the polyketide mycotoxin aflatoxin (AF) (26, 30, 31), one of the most mutagenic and carcinogenic natural compounds described to date. Ingestion of food or feed contaminated with AF has been associated with hepatotoxicity, teratogenicity, immunotoxicity, and even death (10, 31). For these reasons, efforts are focused on finding methods to control the production of AF or to prevent the dissemination or survival of the fungi that produce this type of mycotoxin.
A common biosynthetic pathway leads to production of both AF and a mycotoxin called sterigmatocystin (ST); ST is the penultimate intermediate in the AF pathway. One of the best genetically characterized eukaryotic systems, that of the fungus Aspergillus nidulans, produces ST and is especially productive for the study of the AF-ST gene clusters and signal transduction pathways governing mycotoxin production and fungal development (5, 14, 18, 19, 34). For both A. parasiticus and A. nidulans fungi, a correlation between conidiation and secondary metabolism has been demonstrated. Kale et al. (16) showed that nonaflatoxigenic variants of A. parasiticus had abundant vegetative mycelium and reduced conidiation. Hicks et al. (14) reported that ST biosynthesis is genetically linked with the production of asexual spores (conidia) in A. nidulans through the FadA G protein signaling pathway. This same study showed the conservation of this genetic linkage in A. parasiticus. Interestingly, certain mutations in genes involved in the FadA pathway also alter cleistothecial development (27, 28), suggesting the existence of an association not only with conidiation and secondary metabolism but also with processes leading to the development of the sexual stage.
Only a few genes are known to be involved in sexual development; one example is veA, the velvet gene. The veA gene product does not present homology with any other protein of known function; however, it is known that veA mediates a developmental light response in A. nidulans (33). In A. nidulans strains containing a wild-type allele of the velvet gene (referred to as AnveA in this work), light reduces and delays cleistothecial formation and the fungus develops asexually forming conidia, whereas in the dark, fungal development is directed towards the formation of cleistothecia. In a study focused on morphogenesis, it was reported that deletion of AnveA blocks cleistothecial production in A. nidulans, demonstrating that AnveA is necessary for sexual development (20). Recently, we reported that deletion of AnveA also blocks ST production by repression of aflR, a specific transcription factor that activates the ST and AF gene clusters (6, 9, 17, 25, 32, 35).
We have found a veA homolog in the AF producer A. parasiticus. Our results indicate that the A. parasiticus veA deletion strain is unable to produce sclerotia or the AF intermediate versicolorin A. We also show that the blockage in the production of AF intermediates in the veA deletion strain is related to the absence of expression of both aflR and the adjacent gene aflJ, also involved in the expression of AF structural genes (7, 23). Additionally, the veA deletion strain also has reduced growth and conidial production when grown on the natural substrate peanut seed. Because veA is required for both mycotoxin production and morphological development, we propose veA as a potential target gene for the implementation of a control strategy to prevent the health and economic effects of these carcinogenic mycotoxins.
MATERIALS AND METHODS
Strains and growth conditions.
The A. parasiticus strains used in this study were ATCC 36537 (wA; ver1), CSS10 (wA; ver1; pyrG−), provided by John Linz, and also TJW41.21 (wA; ver1; pyrG−; veA::pyrG), generated in this study. The strains were inoculated on YGT medium (0.5% [wt/vol] yeast extract, 2% [wt/vol] glucose, and 1 ml of trace element solution per liter of medium) (15). Appropriate supplements corresponding to the auxotrophic markers were added to the medium (15). Agar (15 g/liter) was added in the case of solid medium. All strains were maintained as glycerol stocks.
Identification and sequencing of veA.
The identification of A. parasiticus veA was achieved by hybridizing an A. parasiticus genomic library (kindly provided by John Linz) with the A. nidulans AnveA gene as a heterologous probe. The AnveA gene was contained in a 4-kb XhoI fragment obtained from the plasmid pPK11 (kindly provided by Larry Yager). The screening of the A. parasiticus library yielded a cosmid containing veA (GenBank accession number AY445513). This cosmid, which we named pAMC10, was subcloned using a shotgun method and sequenced using synthetic primers and an ABI PRISM DNA sequencing kit (Perkin Elmer Life Science). This sequence information was then used for the construction of the deletion vector.
Construction of the veA deletion strain.
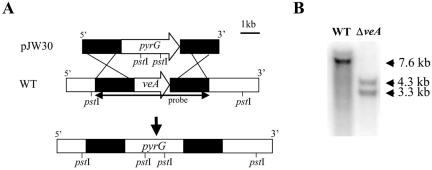
The transformation vector, which was called pJW30 and was utilized to delete veA, was constructed as follows. First, a 3-kb EcoRI-BamHI fragment containing A. parasiticus pyrG from pBZ5 (29) was subcloned into pBluescript SK(-) to obtain pJW24. DNA fragments corresponding to the upstream and downstream regions of veA were generated by PCR from pAMC10 by the use of Pfu Turbo polymerase (Stratagene Co.) and primers 5′AGAGATGTCAAGTTCGAGTCGAG3′ and 5′CAGAATTCGACGGGCCGTAAATTGAC3′ and primers 5′TTGGATCCTGCTTTTCCTCCCGCTCA3′ and 5′CTCGGCACCCAGCGTCATCC3′, respectively. A 2-kb upstream region fragment was inserted into the EcoRI site in pJW24, and a 2-kb downstream region fragment was ligated into the BamHI site in pJW24 after blunt ending the vector in both steps. The finished deletion vector, pJW30, was used to transform A. parasiticus CSS10 (wA; ver1; pyrG−) to create TJW41.21. The veA deletion strain (ΔveA) was obtained by a double-crossover event, exchanging the pyrG selectable marker gene for the veA coding region (Fig. 1A). Southern analysis was used to confirm a single gene replacement event in TJW41.21 (Fig. 1B). On the basis of the DNA sequence, in this Southern analysis we predicted a hybridization band corresponding to a PstI fragment of 7,560 bp for the wild type and two bands corresponding to two PstI fragments of 3,381 and 4,256 bp in the deletion strain when the probe indicated in Fig. 1 was used. This pattern can only be obtained by a double-recombination event in the flanking regions of veA that results in veA gene replacement. Fungal transformation essentially followed that of Miller et al. (24), with the modification of embedding the protoplasts in top agar (0.75%) rather than spreading them with a glass rod on solid medium.
FIG. 1.
Deletion of the veA gene in A. parasiticus. (A) Diagram of the veA open reading frame and how it was replaced with the pyrG selectable marker from pJW30 by homologous recombination to generate the ΔveA mutant, TJW41.21. Solid bars indicate the 5′ and 3′ flanking regions of veA. (B) Southern analysis of wild-type (WT) strain and the ΔveA mutant strain. Genomic DNA was digested by PstI. A 6.1-kb PCR product containing veA was obtained with the primers 5′-AGA GAT GTC AAG TTC GAG TCG AG-3′ and 5′ CTC GGC ACC CAG CGT CAT CC 3′ and was used as a veA probe (indicated by arrows in panel A). Expected hybridization band patterns: wild-type strain, 7.6-kb band; ΔveA strain, 3.3- and 4.3-kb bands.
Physiological studies.
The studies on fungal development were performed with the ΔveA strain (wA; ver1; pyrG−; veA::pyrG) and the corresponding control strain (wA; ver1). Plates containing 30 ml of solid YGT medium were spread with 100 μl of water containing 105 spores. The cultures were incubated at 30°C in continuous light (25 microeinsteins/m2/s) or in the dark. After 7 days, a 16-mm-diameter core was removed from each spread-plate culture and homogenized in water to release spores. Conidia were counted using a hemacytometer. Identical cores were taken to examine sclerotial production. The cores were spread with 95% ethanol to enhance visualization of sclerotia. Sclerotial production was examined under a stereo-zoom microscope. Colony growth was recorded as colony diameter measurements in point inoculation cultures on YGT medium. Experiments were carried out with four replicates.
Mycotoxin analysis.
Four cores (16-mm diameter) or, alternatively, 2 g of fungal biomass (specified in each experiment described below) was collected from each replicate of veA and ΔveA cultures and placed in a 50-ml Falcon tube. AF intermediates were extracted from these samples by adding 5 ml of CHCl3 three consecutive times. Extracts were allowed to dry and then resuspended in 500 μl of CHCl3 before fractionating 15 μl of each extract on a silica gel thin-layer chromatography (TLC) plate by the use of a benzene:glacial acetic acid (95:5 [vol/vol]) solvent system. The TLC plates were sprayed with aluminum chloride (15% in ethanol) to intensify fluorescence upon exposure to long-wave (365-nm) UV light and baked for 10 min at 80°C prior to viewing.
mRNA studies.
Cultures of 250 ml of YGT liquid medium containing 5 × 105 spores ml−1 of either veA or ΔveA strains were incubated (300 rpm and 30°C) for 24 h. At that time the mycelia from these cultures were recovered by filtration. Approximately 9 g of mycelium from either veA or ΔveA strains was then shifted and spread on each petri dish (9-cm diameter) containing solid YGT medium and allowed to grow in a light incubator at 30°C. At the time of the shift (t = 0 h) onto solid medium and 12 and 24 h after the shift, the mycelial samples were then harvested for RNA extraction. Total RNA was isolated from mycelia by using Trizol as described by the supplier (Invitrogen). Approximately 20 μg of total RNA was used for RNA blot analysis. PCR products amplified from A. parasiticus genomic DNA with the following primer pairs were used as specific probes: for aflR, 5′-CACCTCCACGATGGTTG-3′ and 5′-GTATAGACTGTTCAAGATTCG-3′; for aflJ, 5′-GCATGCTTGCGGTGGCTG-3′ and 5′-CCGTTGCCATTCCCAAG-3′; for pksL1, 5′-CTGCAGAGCATGAACACG-3′ and 5′-GAATGGTACGATGGTGAC-3′; and for brlA, 5′-ATGAGAGCTCAAAACAACC-3′ and 5′-TTAATCGTCTAACCCATC-3′. A ver1-specific 2.2-kb EcoRI-HindIII fragment was obtained from pBVER-1 (provided by John Linz). The identity of the probes was confirmed by sequencing. ImageQuant 5.2 software was used to perform the densitometry analysis of brlA expression. The results were plotted as relative band intensity values normalized to rRNA and to the highest band intensity (considered 1 U).
Infection studies.
Seeds of the near-isogenic Florunner, Southern Runner, and 72cc peanut lines were kindly provided to us by C. Holbrook (U.S. Department of Agriculture, Atlanta, Georgia). Prior to infection by A. parasiticus strains, the seeds were shelled, the cotyledons were separated, and the embryos were removed. Seeds from both peanut lines were weighed so that all the peanuts used were between 0.4 and 0.6 g. The cotyledons were surfaced sterilized by immersion in 10% Clorox bleach for 1 min followed by immersion in sterile distilled water for 1 min. The seeds were inoculated as follows. For both veA and ΔveA strains, four peanut cotyledons were placed in a 50-ml Falcon tube, to which was added 500 μl of sterile distilled water containing 106 spores of the appropriate strain. Each tube was vortexed for 1 min, the caps were loosened, and the samples were placed in the light or in the dark at 30°C. These experiments were performed with four replicates. After 7 days, spores from the infected seeds were released by adding 5 ml of Tween 80 water to each Falcon tube, vortexing for 1 min, and using an aliquot of this suspension for counting with a hemocytometer.
RESULTS
Deletion of veA affects growth and morphological development in A. parasiticus.
Fig. 1 shows a diagram of the strategy followed to delete veA as well as the results of the Southern analysis hybridization confirming the veA deletion. Loss of this gene resulted in reduction of colony growth (estimated as colony diameter) under either light or dark conditions (Fig. 2A). In the light the reduction was 25%, and in the dark the reduction was approximately 50%. Growth was also evaluated in liquid culture in an experiment in which 250 ml of YGT medium was inoculated with either veA or ΔveA strains (5 × 105 spores ml−1) and incubated at 30°C and 300 rpm. After 24 h, the mycelium mass produced in the ΔveA strain was 25% with respect to that obtained in the control strain (data not shown).
FIG. 2.
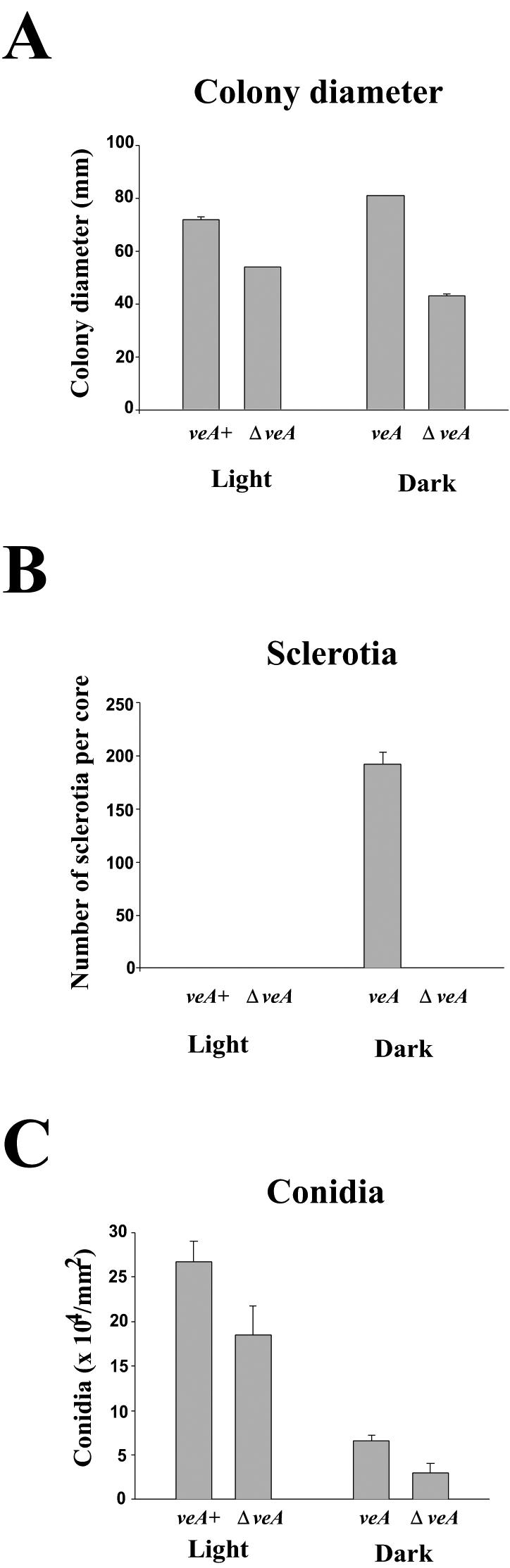
veA is required for normal growth and morphological development in A. parasiticus. Wild-type and ΔveA cultures were grown on YGT medium in the light or in the dark for 7 days. The experiment was carried out with four replicates. (A) Fungal growth, measured as colony diameter. (B) Resistant structure production, measured as number of sclerotia per core (core = 200 mm2). (C) Asexual stage, measured as number of conidia per surface area.
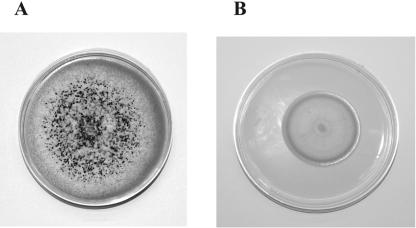
Figure 2B shows that sclerotia production only occurred in the dark in the control strain and that the veA deletion completely prevented the formation of sclerotia (Fig. 2B and 3). To test whether the absence of sclerotia could be a consequence of reduction in growth, we cultured the veA and ΔveA strains (5 × 105 spores ml−1) in YGT medium at 30°C and 300 rpm for 24 h. Then, the mycelia were harvested by filtration and equal amounts of mycelium (4 g/9-cm-diameter plate) were collected and spread on solid YGT medium and cultured in the dark at the same temperature for 10 days. The results concurred with those of the experiment shown in Fig. 2B and 3, where sclerotial production occurred in the control strain but was completely abolished in the ΔveA strain. Later examination of these cultures at 14 days of growth from the moment of the shift to solid medium showed the same results; sclerotial production was absent from the deletion strain.
FIG. 3.
A. parasiticus veA (A) and ΔveA (B) strains cultured on YGT medium for 7 days in the dark. Both strains produce white conidia. The black structures observed only in the veA strain are sclerotia.
The A. parasiticus strain with a wild-type veA produced more conidia (75%) in the light than in the dark (Fig. 2C). Deletion of veA also resulted in a reduction of conidial production in A. parasiticus (30% in the light and 46% in the dark) (Fig. 2C). The effect of veA deletion on A. parasiticus conidiation was further evaluated by mRNA studies (see below).
Role of veA in the production of versicolorin A in A. parasiticus.
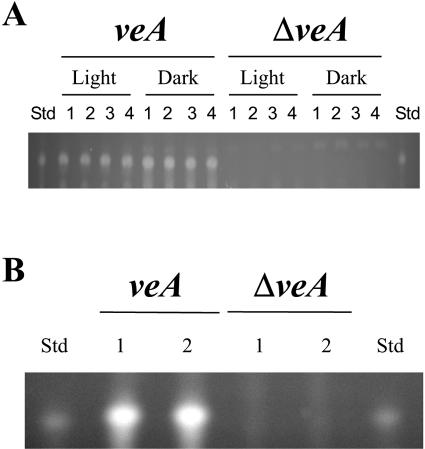
We evaluated the effect of veA deletion on AF biosynthesis in A. parasiticus. To facilitate the detection of cluster activation and AF pathway activity, we used the visible orange AF intermediate versicolorin A. Strains of A. parasiticus that produce versicolorin A have a mutation in a gene (ver1, encoding a ketoreductase) involved in the multistep AF pathway (21, 22). As a result of this mutation, these strains have an interrupted AF pathway in which versicolorin A accumulates instead of AF. AF biosynthetic clustered genes are simultaneously activated or deactivated; therefore, accumulation of versicolorin A is a good indicator of said activation and of AF biosynthetic pathway activity. Our TLC analysis showed that the A. parasiticus ΔveA strain does not produce versicolorin A whereas it was detected in the light and in the dark in the veA control strain when grown on solid YGT medium for 7 days (Fig. 4A). An additional experiment was carried out to test whether the blockage of versicolorin A production was caused by a reduction in growth. The veA and ΔveA strains (5 × 105 spores ml−1) were cultured in YGT medium at 30°C and 300 rpm for 24 h, and then equal amounts of mycelium (6 g/9-cm-diameter plate) were shifted into solid medium and allowed to grow for 96 h. A total of 2 g of mycelium from veA and ΔveA cultures was extracted, and versicolorin A levels were analyzed. The results of this experiment, shown in Fig. 4B, indicate that while versicolorin A was present in the control strain, it was completely absent from the ΔveA strain. The effect of veA deletion on AF biosynthesis was also further analyzed in mRNA studies (see below).
FIG. 4.
TLC analysis of versicolorin A production in A. parasiticus. (A) The veA and ΔveA spread plate cultures were grown in the light or in the dark on solid YGT medium for 7 days. The numbers 1 to 4 represent replicates. (B) In a shift experiment, the veA and ΔveA cultures were incubated in liquid YGT for 24 h and then equal amounts of mycelium were shifted into solid medium (6 g/9-cm-diameter plate) and allowed to grow for an additional 96 h. The experiment was repeated twice with similar results (indicated as lanes 1 and 2). Versicolorin A standards (Std) are shown on both sides of the TLC plate.
mRNA analysis.
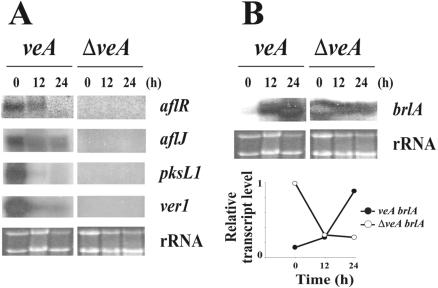
In this study we evaluated the effect of veA deletion on regulatory genes controlling AF biosynthesis and conidiation. We looked at the expression of the regulatory AF genes aflR and aflJ and of the two structural genes pksL1 (encoding a polyketide synthase) and ver1 as indicators of cluster activation (Fig. 5A). Transcripts corresponding to aflR and aflJ as well as to pksL1 and ver1 were detected in the wild-type strain while they were completely absent from the ΔveA strain.
FIG. 5.
Effects of the veA deletion on the transcription of genes required for AF biosynthesis and morphological development in A. parasiticus. The veA and ΔveA strains were cultured in shaken YGT liquid medium for 24 h and then shifted onto solid YGT medium. Total RNAs of the veA and ΔveA strains were isolated at the time of the shift (t = 0) and 12 and 24 h after the shift to solid culture. (A) Transcriptional patterns of the AF genes aflR, aflJ, pksL1, and ver1. rRNA stained with ethidium bromide is shown to indicate RNA loading. (B) Transcriptional pattern of the developmental gene brlA. brlA mRNA, present in both veA and ΔveA, was quantified by densitometry using ImageQuant 5.2 software and plotted as relative band intensity normalized to rRNA and to the highest band intensity (considered 1 U). Two separate repetitions of this experiment yielded similar results.
With respect to conidiation, we also studied the effect of the veA deletion on the expression of A. parasiticus brlA, a key transcription factor first described in A. nidulans (1) and known to regulate conidiophore formation. Besides eliminating the problem that reduction of growth could cause in our analysis, shift experiments are particularly useful in the study of regulation of conidiation in Aspergillus species. As reviewed by Adams et al. (2), conidiophore formation is an event that is temporally regulated. The vegetative stage can be continuously maintained by growing hyphae in submerged liquid medium, a trait that provides a means for synchronous development induction by harvesting hyphae from the submerged culture and exposing them to air. During the time of vegetative growth in the submerged culture, the mycelium becomes competent for conidiation induction occurring after the shift to solid medium. The kinetics of conidiation under these conditions have been found to be very reproducible.
In our studies, A. parasiticus brlA transcripts were observed in both the control strain and the ΔveA strain (Fig. 5B). Only one brlA transcript band was detected in the Northern analysis. At t = 0 h (corresponding to the moment of the shift from liquid to solid culture) brlA transcripts were present in the ΔveA strain while brlA was detected only at very low levels (6-fold lower) in the wild-type control (Fig. 5B). The densitometry analysis indicated that after the shift into solid culture brlA levels increased in the wild type while they decreased in the ΔveA strain. Although brlA transcript was present in the liquid YGT culture after 24 h in the ΔveA strain, no conidiophore formation was observed using the light microscope under this condition. Furthermore, microscopic observations of these cultures revealed that after the shift to solid medium the production of conidia in ΔveA was also notably lower (approximately 10%) with respect to that seen with the control.
Role of veA in A. parasiticus conidiation on seeds.
After chemical surface sterilization, three lines of peanut seeds (Florunner, Southern Runner, and 72cc) were inoculated with A. parasiticus veA and ΔveA strains to evaluate the effect of veA in the production of conidia when the fungus is interacting with live seeds. The control strain produced the highest number of conidia when it was grown on Florunner peanut seeds, while it produced less conidia (40% reduction with respect to the results obtained with Florunner in both the light and the dark) when growing on 72cc peanut line seeds (Fig. 6). Conidial production in veA was higher in the light than in the dark (35% lower in Florunner, 55% lower in Southern Runner, and 38% lower in 72cc peanut lines). Regardless of the peanut line, deletion of veA resulted in a notable reduction of conidial production in both the light and the dark (Fig. 6).
FIG. 6.
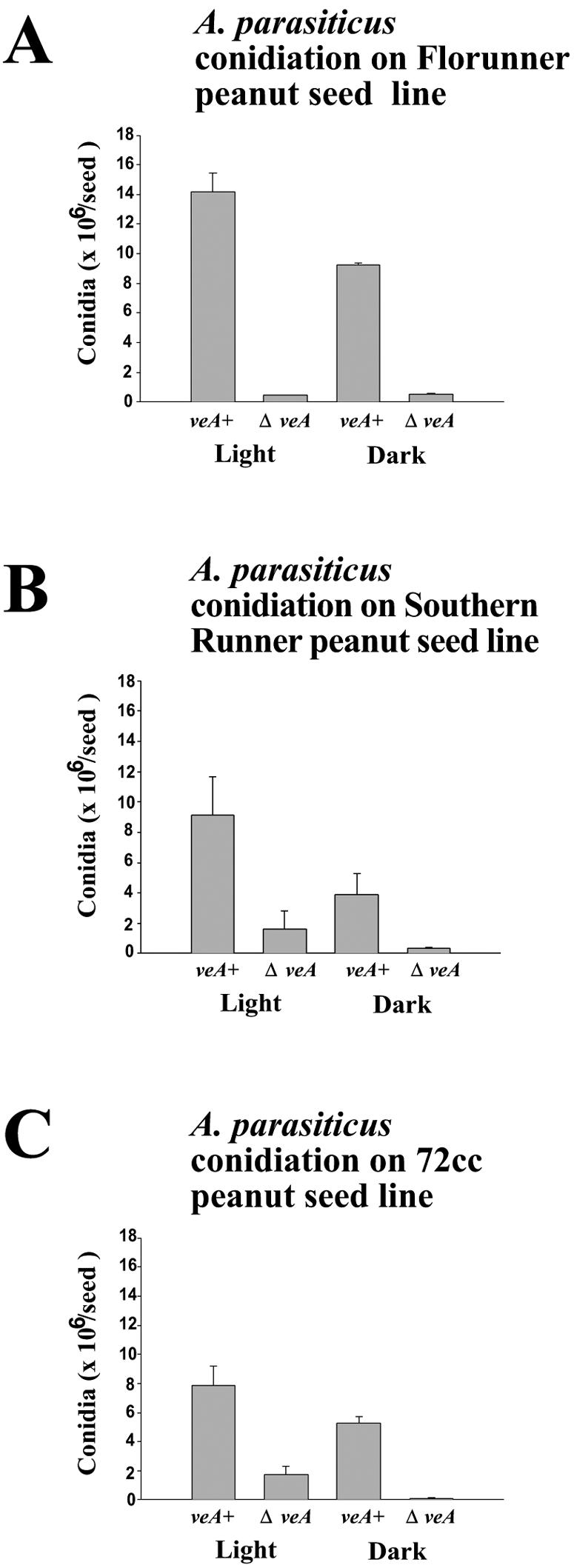
Conidiation of A. parasiticus veA and ΔveA strains on peanut seeds. The cultures were grown in the light or in the dark on peanut seeds for 7 days. The experiment was carried out with four replicates. (A) Florunner line; (B) Southern Runner line; (C) 72cc line.
DISCUSSION
Current control strategies fail to effectively eliminate AF contamination, which continues to be a major economic problem in the United States and a serious health threat in developing countries. Researching the viability of regulation of mycotoxin biosynthesis and/or fungal development by targeting regulatory genes may help to control a worldwide threat to food supplies and human health by Aspergillus mycotoxin. We have found a homolog of the A. nidulans AnveA in the AF-producing species A. parasiticus. The AnveA gene has been described as necessary for both development (3, 4, 17, 20, 33) and mycotoxin production (17) in this model system. Here we find that veA is required for sclerotial and AF production in A. parasiticus.
We found that veA is relevant in A. parasiticus colony growth. The reduction in growth in the veA deletion strain is more pronounced than the slight reduction reported for the A. nidulans AnveA deletion strain (17). We also found that conidiation is reduced in the A. parasiticus veA deletion strain (Fig. 2C). This result contrasts with the increase in conidial production in the AnveA deletion strain (17, 20). It is possible that veA regulates conidiation in A. parasiticus in a different manner than in A. nidulans. Because the reduction in conidiation was still observed when equal amounts of fungal biomass from veA deletion and control cultures were used, we do not think it likely that the decrease in conidiation is a consequence of a reduction in growth. Furthermore, brlA, encoding a transcription factor required for conidiophore formation, was found to be expressed in both strains (Fig. 5B). Under the experimental condition assayed, brlA was highly expressed in 24-h YGT submerged cultures in the ΔveA strain; however, conidiophore formation was not observed. brlA expression was still detected after the cultures were shifted into solid medium, decreasing brlA levels in the ΔveA strain and increasing levels in the control strain, in which conidiation was approximately 10-fold higher than in the deletion strain. One possibility that could explain the inverted brlA expression pattern observed in veA deletion and control strains is that veA lack of function could result in alteration of the sporulation temporal program (2) such that the sporulation program occurs early on, activating brlA prematurely and decreasing brlA levels when brlA expression is increasing in the wild type. Although the lack of an air interphase did not prevent brlA expression in the ΔveA strain growing in submerged cultures, brlA expression was not sufficient to initiate conidiation in A. parasiticus with a veA deletion background under these conditions.
In A. nidulans, brlA generated two types of transcripts, α and β transcripts, (13) and we have previously reported that AnveA affects the α/β ratio that controls conidiation in this fungus. In contrast with brlA in A. nidulans, which has been studied in depth (12, 13), little is known about the A. parasiticus brlA gene. Our A. parasiticus brlA mRNA study showed only one transcript band. It is unknown whether this band corresponds to one transcript or to two or more transcripts of similar sizes. Further characterization of A. parasiticus brlA is needed to understand its functionality. Besides brlA, there could also be additional factors affected by the absence of veA that reduce conidiation in A. parasiticus. We also investigated whether this reduction in conidia number holds true when the fungus is growing on peanut seed, a natural substrate for A. parasiticus. Our results with three different lines of peanut seeds indicated that veA is essential for normal conidiation in this plant pathogen.
One interesting observation was the regulation of sclerotial production by veA. In the A. parasiticus control strain, sclerotia formed only in the dark (Fig. 2B and 3), a condition that also favored the formation of cleistothecia in A. nidulans (33). Additionally, deletion of veA results in a complete blockage of sclerotial production. It is interesting that some researchers consider sclerotia as possible vestiges of cleistothecia (11, 33), taking into account that deletion of AnveA in A. nidulans results in a blockage of cleistothecial formation (17, 20).
The blockage of sclerotial production persisted when equal amounts of fungal biomass from both deletion and control strains were used, indicating that the effect of the veA deletion on sclerotial development is independent of the reduction of growth observed.
Besides regulating morphological differentiation, veA also regulates secondary metabolism. An association between sclerotial formation and AF biosynthesis in A. parasiticus has been reported by Chang et al. (8). These authors introduced aflR and aflJ into an O-methylsterigmatocystin-accumulating strain. The increase in toxin production coincided with a decrease in sclerotial size and an alteration in sclerotial shape and suggested that these alterations could be caused by competition for a common substrate such as acetate. They also described an increase in sclerotial numbers in some cases; however, this effect was medium dependent. In our studies, besides blocking sclerotial production, deletion of veA prevented the accumulation of versicolorin A, an intermediate indicator of AF pathway activity (21, 22, 29) (Fig. 4). We also demonstrated that this blockage is independent of the reduction in growth in the veA deletion strain. Furthermore, our mRNA analysis indicated that the absence of veA prevents the expression of the regulatory genes aflR and aflJ and consequently the expression of structural AF genes such as pksL1 and ver1 (Fig. 5A). These results are consistent with the fact that in A. nidulans, AnveA is necessary for ST production (17), a related polyketide mycotoxin synthesized through a highly conserved biosynthetic pathway. Because of this conservation, it is likely that a veA homolog also exists in A. flavus, another important seed-infecting fungus, and that veA might have a similar role in regulating AF production in A. flavus.
Since veA is required for morphological development and mycotoxin biosynthesis, veA is a promising potential target for a control strategy to reduce the impact of AF on health worldwide and on the economy. Further studies will include the elucidation of possible connections of veA with genetic elements of signal transduction pathways known to regulate morphological development and toxin production. Interestingly, the conservation of veA extends beyond the genus Aspergillus. As mentioned in Kato et al. (17), homologs of veA were found in other filamentous fungi, some of them plant pathogens, across genera. Importantly, veA has not been found in plants or animals. Due to the conserved nature of the regulatory networks among fungal species, a broader control strategy involving veA may well be effective against Aspergillus and other pathogenic fungi.
Acknowledgments
We thank C. Holbrook (U.S. Department of Agriculture, Atlanta, Ga.) for providing us with the peanut lines; J. Linz for providing the cosmid library, ATCC 36537 and CSS10 strains, and pBVER clone; L. Yager for providing the pPK11 clone; and L. Smith for her technical support.
This study was financed by Northern Illinois University and the Plant Molecular Biology Center at the same university and by a U.S. Department of Agriculture grant (NRI 2001-35319-10996).
REFERENCES
- 1.Adams, T. H., M. T. Boylan, and W. E. Timberlake. 1988. brlA is necessary and sufficient to direct conidiophore development in Aspergillus nidulans. Cell 54:353-362. [DOI] [PubMed] [Google Scholar]
- 2.Adams, T. H., J. W. Wieser, and J-H. Yu. 1998. Asexual sporulation in Aspergillus nidulans. Microbiol. Mol. Biol. Rev. 62:35-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Calvo, A. M., L. L. Hinze, H. W. Gardner, and N. P. Keller. 1999. Sporogenic effect of polyunsaturated fatty acids on development of Aspergillus spp. Appl. Environ. Microbiol. 65:3668-3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calvo, A. M., H. W. Gardner, and N. P. Keller. 2001. Genetic connection between fatty acid metabolism and sporulation in Aspergillus nidulans. J. Biol. Chem. 276:25766-25774. [DOI] [PubMed] [Google Scholar]
- 5.Calvo, A. M., R. A. Wilson, J. W. Bok, and N. P. Keller. 2002. Relationship between secondary metabolism and fungal development. Microbiol. Mol. Biol. Rev. 66:447-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cary, J. W., K. C. Ehrlich, M. Wright, P. K. Chang, and D. Bhatnagar. 2000. Generation of aflR disruption mutants of Aspergillus parasiticus. Appl. Microbiol. Biotechnol. 53:680-684. [DOI] [PubMed] [Google Scholar]
- 7.Chang, P. K. 2003. The Aspergillus parasiticus protein AFLJ interacts with the aflatoxin pathway-specific regulator AFLR. Mol. Genet. Genom. 268:711-719. [DOI] [PubMed] [Google Scholar]
- 8.Chang, P. K., J. W. Bennett, and P. J. Cotty. 2001. Association of aflatoxin biosynthesis and sclerotial development in Aspergillus parasiticus. Mycopathologia 153:41-48. [DOI] [PubMed] [Google Scholar]
- 9.Chang, P.-K., J. W. Cary, D. Bhatnagar, T. E. Cleveland, J. W. Bennett, J. E. Linz, C. P. Woloshuk, and G. A. Payne. 1993. Cloning of the Aspergillus parasiticus apa-2 gene associated with the regulation of aflatoxin biosynthesis. Appl. Environ. Microbiol. 59:3273-3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dvorackova, I., and V. Kusak. 1990. Hepatocellular carcinoma (a 28-year necropsy review). J. Environ. Pathol. Toxicol. Oncol. 10:220-224. [PubMed] [Google Scholar]
- 11.Geiser, D. M., W. E. Timberlake, and M. L. Arnold. 1996. Loss of meiosis in Aspergillus. Mol. Biol. Evol. 13:809-817. [DOI] [PubMed] [Google Scholar]
- 12.Han, S., and T. H. Adams. 2001. Complex control of the developmental regulatory locus brlA in Aspergillus nidulans. Mol. Genet. Genom. 266:260-270. [DOI] [PubMed] [Google Scholar]
- 13.Han, S., J. Navarro, R. A. Greve, and T. H. Adams. 1993. Translational repression of brlA expression prevents premature development in Aspergillus. EMBO J. 12:2449-2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hicks, J. K., J. H. Yu, N. P. Keller, and T. H. Adams. 1997. Aspergillus sporulation and mycotoxin production both require inactivation of the FadA Gα protein-dependent signaling pathway. EMBO J. 16:4916-4923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Käfer, E. 1977. Meiotic and mitotic recombination in Aspergillus and its chromosomal aberrations. Adv. Genet. 19:33-131. [DOI] [PubMed] [Google Scholar]
- 16.Kale, S. P., J. W. Cary, D. Bhatnagar, and J. W. Bennett. 1996. Characterization of experimentally induced, nonaflatoxigenic variant strains of Aspergillus parasiticus. Appl. Environ. Microbiol. 62:3399-3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kato, N., W. Brooks, and A. M. Calvo. 2003. The expression of sterigmatocystin and penicillin genes in Aspergillus nidulans is controlled by veA, a gene required for sexual development. Eukaryot. Cell 2:1178-1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keller, N. P., and T. H. Adams. 1995. Analysis of a mycotoxin gene cluster in Aspergillus nidulans. SAAS Bull. Biochem. Biotechnol. 8:14-21. [PubMed] [Google Scholar]
- 19.Keller, N. P., and T. M. Hohn. 1996. Metabolic pathway gene clusters in filamentous fungi. Fungal Genet. Biol. 21:17-29. [PubMed] [Google Scholar]
- 20.Kim, H., K. Han, K. Kim, D. Han, K. Jahng, and K. Chae. 2002. The veA gene activates sexual development in Aspergillus nidulans. Fungal Genet. Biol. 37:72-80. [DOI] [PubMed] [Google Scholar]
- 21.Liang, S.-H., C. D. Skory, and J. E. Linz. 1996. Characterization of the function of the ver-1A and ver-1B genes, involved in aflatoxin biosynthesis in Aspergillus parasiticus. Appl. Environ. Microbiol. 62:4568-4575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liang, S.-H., T.-S. Wu, R. Lee, F. S. Chu, and J. E. Linz. 1997. Analysis of mechanisms regulating expression of the ver-1 gene, involved in aflatoxin biosynthesis. Appl. Environ. Microbiol. 63:1058-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meyers, D. M., G. Obrian, W. L. Du, D. Bhatnagar, and G. A. Payne. 1998. Characterization of aflJ, a gene required for conversion of pathway intermediates to aflatoxin. Appl. Environ. Microbiol. 64:3713-3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller, B., K. Miller, and W. Timberlake. 1985. Direct and indirect gene replacements in Aspergillus nidulans. Mol. Cell. Biol. 5:1714-1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Payne, G. A., G. J. Nystrom, D. Bhatnagar, T. E. Cleveland, and C. P. Woloshuk. 1993. Cloning of the afl-2 gene involved in aflatoxin biosynthesis from Aspergillus flavus. Appl. Environ. Microbiol. 59:156-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Payne, G. A., and M. P. Brown. 1998. Genetics and physiology of aflatoxin biosynthesis. Annu. Rev. Phytopathol. 36:329-362. [DOI] [PubMed] [Google Scholar]
- 27.Rosen, S., J. H. Yu, and T. H Adams. 1999. The Aspergillus nidulans sfaD gene encodes a G protein beta subunit that is required for normal growth and repression of sporulation. EMBO J. 18:5592-5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shimizu, K., and N. P. Keller. 2001. Genetic involvement of a cAMP-dependent protein kinase in a G protein signaling pathway regulating morphological and chemical transitions in Aspergillus nidulans. Genetics 157:591-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Skory, C. D., P.-K. Chang, J. Cary, and J. E. Linz. 1992. Isolation and characterization of a gene from Aspergillus parasiticus associated with the conversion of versicolorin A to sterigmatocystin in aflatoxin biosynthesis. Appl. Environ. Microbiol. 58:3527-3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sweeney, M. J., and A. D. Dobson. 1999. Molecular biology of mycotoxin biosynthesis. FEMS Microbiol. Lett. 175:149-163. [DOI] [PubMed] [Google Scholar]
- 31.Trail, F., N. Mahanti, and J. Linz. 1995. Molecular biology of aflatoxin biosynthesis. Microbiology 141:755-765. [DOI] [PubMed] [Google Scholar]
- 32.Woloshuck, C. P., Foutz, K. R., Brewer, J. F., Bhatnagar, D., Cleveland, T. E., and G. A. Payne. 1994. Molecular characterization of aflR, a regulatory locus for aflatoxin biosynthesis. Appl. Environ. Microbiol. 60:2408-2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yager, L. N. 1992. Early developmental events during asexual and sexual sporulation in Aspergillus nidulans. Biotechnology 23:19-41. [PubMed] [Google Scholar]
- 34.Yu, J.-H., and T. J. Leonard. 1995. Sterigmatocystin biosynthesis in Aspergillus nidulans requires a novel type I polyketide synthase. J. Bacteriol. 177:4792-4800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu, J. H., R. A. Butchko, M. Fernandes, N. P. Keller, T. J. Leonard, and T. H. Adams. 1996. Conservation of structure and function of the aflatoxin regulatory gene aflR from Aspergillus nidulans and Aspergillus flavus. Curr. Genet. 29:549-555. [DOI] [PubMed] [Google Scholar]