Abstract
Methanogenic flowthrough aquifer columns were used to investigate the potential of bioaugmentation to enhance anaerobic benzene-toluene-ethylbenzene-xylene (BTEX) degradation in groundwater contaminated with ethanol-blended gasoline. Two different methanogenic consortia (enriched with benzene or toluene and o-xylene) were used as inocula. Toluene was the only hydrocarbon degraded within 3 years in columns that were not bioaugmented, although anaerobic toluene degradation was observed after only 2 years of acclimation. Significant benzene biodegradation (up to 88%) was observed only in a column bioaugmented with the benzene-enriched methanogenic consortium, and this removal efficiency was sustained for 1 year with no significant decrease in permeability due to bioaugmentation. Benzene removal was hindered by the presence of toluene, which is a more labile substrate under anaerobic conditions. Real-time quantitative PCR analysis showed that the highest numbers of bssA gene copies (coding for benzylsuccinate synthase) occurred in aquifer samples exhibiting the highest rate of toluene degradation, which suggests that this gene could be a useful biomarker for environmental forensic analysis of anaerobic toluene bioremediation potential. bssA continued to be detected in the columns 1 year after column feeding ceased, indicating the robustness of the added catabolic potential. Overall, these results suggest that anaerobic bioaugmentation might enhance the natural attenuation of BTEX in groundwater contaminated with ethanol-blended gasoline, although field trials would be needed to demonstrate its feasibility. This approach may be especially attractive for removing benzene, which is the most toxic and commonly the most persistent BTEX compound under anaerobic conditions.
The widespread contamination of surface and groundwater resources by the gasoline oxygenate methyl tert-butyl ether (MTBE) is leading to its phaseout. Ethanol, a likely candidate to substitute MTBE, is increasingly being used as a gasoline additive to meet renewable fuel and Clean Air Act requirements (48). Therefore, the presence of ethanol in groundwater contaminated with the gasoline constituents benzene, toluene, ethylbenzene, and xylenes (BTEX) is expected to increase in the near future.
Previous studies have shown that the preferential degradation of ethanol and the accelerated depletion of nutrients and electron acceptors that would otherwise be available for hydrocarbon biodegradation are likely to hinder BTEX removal (20, 50). These conditions could contribute to longer BTEX plumes, increasing the probability that a potential downgradient receptor will be exposed (47, 51). Thus, ethanol could deter the acceptability of the natural attenuation for controlling the migration of BTEX plumes at some sites, leaving a margin for alternative bioremediation approaches to solve the problem.
Aerobic bioremediation typically exhibits broader catabolic range and faster BTEX degradation kinetics than anaerobic systems (20, 50). However, aerobic processes are not universally applicable, and anaerobic strategies might be more appropriate to treat some ethanol-blended gasoline releases. Specifically, the high biochemical oxygen demand exerted by ethanol, typically exceeding 4,000 mg liter−1 near the source (41, 42), could make the delivery of sufficient oxygen (which has a relatively low solubility) a technically difficult if not prohibitively expensive task. Furthermore, oxygen addition could lead to clogging problems due to the precipitation of metal oxides (30, 61), particularly when the dissolved Fe(II) concentration exceeds 20 mg liter−1 (55). Thus, reductive dissolution of iron (possibly exacerbated by the anaerobic degradation of ethanol) would increase the difficulty to distribute oxygen and nutrients required to stimulate aerobic BTEX degradation.
Enhanced anaerobic BTEX biodegradation has been reported following the addition of nitrate (5, 17, 19), chelated Fe(III) (37, 39), and sulfate (3, 38). Nevertheless, anaerobic biostimulation may not be sufficient to ensure BTEX degradation if the aquifer material does not contain specific degraders in sufficient numbers to exert measurable degradation rates. In such cases, the addition of anaerobic microorganisms with the desired catabolic capacity directly into the contaminated zone should be evaluated for its ability to enhance the natural attenuation of BTEX and ethanol mixtures.
The benefits of bioaugmentation have been demonstrated in field tests for a wide variety of contaminants, including MTBE (52) and chlorinated solvents (23, 24, 26, 40). Similarly, benzene degradation in a sulfate-reducing zone of a petroleum-contaminated aquifer was observed only after the inoculation of a benzene-oxidizing, sulfate-reducing enrichment from aquatic sediments (59). These studies, however, did not deal with the high electron acceptor demand that is exerted during ethanol degradation, which is likely to drive the system rapidly to methanogenic conditions (22). To date, the ubiquity of methanogenic consortia capable of degrading benzene has not been established (36), and no previous studies have addressed how to enhance BTEX biodegradation under methanogenic conditions.
This paper addresses the potential of bioaugmentation to enhance the anaerobic degradation of BTEX-ethanol mixtures under methanogenic conditions that are characteristic of sites contaminated by ethanol-blended gasoline. An emphasis was placed on quantifying the presence of bssA and determining whether its concentration was correlated to toluene degradation activity. This gene codes for benzylsuccinate synthase (BSSA), a ubiquitous enzyme that initiates the anaerobic degradation of toluene and xylenes by catalyzing the addition of fumarate to the methyl group (1, 6, 9, 10, 12, 33, 49). The bssA gene is expressed in phototrophic (62), denitrifying (8), sulfate-reducing (9), and iron-reducing (31) bacteria. However, no previous studies have determined whether higher numbers of bssA gene copies correspond to higher toluene degradation rates under methanogenic conditions, which is important for the evaluation of the usefulness of this gene as a biomarker for environmental forensic analysis of anaerobic bioremediation potential.
MATERIALS AND METHODS
Aquifer columns.
Flowthrough aquifer columns (120-cm length, 5-cm diameter) equipped with eight sampling ports (at 2.5, 7.6, 14, 20, 40, 60, 80, and 100 cm from the inlet) were used to investigate the effect of ethanol on BTEX natural attenuation (22) and their enhanced biodegradation through anaerobic bioaugmentation. The columns were packed with aquifer material collected from the Northwest Terminal in Tigard, Oregon, and were fed continuously over 3 years in an up-flow mode with ethanol (∼1,000 mg liter−1) and BTEX (total of 13 to 26 mg liter−1) dissolved in a carbonate-buffered mineral medium. The substrates were fed by using a syringe pump (Harvard Apparatus model 22) and the carbonate medium was fed by using a peristaltic pump (Masterflex model 7519-15). The ratio of peristaltic pump to syringe pump was set at 20:1. The mineral medium composition (in milligrams per liter) was as follows: NO3− (5.0 to 17.0); SO42− (4.0); CaCO3 (1,000); NH4+ (5.5); Mg2+ (1.5); PO43− (0.06); and Ni(II), Cu(II), Zn(II), Co(II), and Mo(IV) (0.002 each). The medium was constantly purged with N2-CO2 (95:5, vol/vol) gas to remove dissolved oxygen. The hydraulic characteristics of the columns were estimated by fitting bromide tracer data to the one-dimensional advection-dispersion equation as described previously (22), and the hydraulic parameters (with values in parentheses) are as follows: flow (6.7 to 7.5 ml h−1), effective porosity (0.37), dispersion (5 cm2 h−1), and seepage velocity (0.9 cm h−1).
Cultures for bioaugmentation.
Two methanogenic BTEX-enriched consortia capable of mineralizing BTEX to CO2 and CH4 (27, 54) were utilized as inocula. The toluene- and o-xylene-enriched consortium was obtained from a site contaminated with creosote and maintained in the laboratory for 10 years (27). The benzene-enriched consortium was obtained from an oil refinery site (54). This consortium was derived from sulfate-reducing microcosms that became methanogenic after 1.5 years and was maintained in the lab for six subsequent years.
Previous molecular characterization of the toluene- and o-xylene-enriched consortium showed the presence of two archaea (a Methanosaeta sp. and a Methanospirillum sp.), one sulfate-reducing bacterium (a Desulfotomaculum sp.), and one bacterium not related to any known genus (27). The Methanosaeta sp. is an obligatory aceticlastic methanogen, oxidizing the carboxylic group of acetate to CO2 and reducing the methyl group to methane. The Methanospirillum sp. uses formate and hydrogen as electron donors to reduce CO2 to methane. The Desulfotomaculum sp. is capable of growing acetogenically on ethanol, propionate, butyrate, benzoate, and other metabolites. The limited substrate range of methanogens and the observed inhibitory effect of sulfate on toluene degradation implied that the unknown bacterium might be responsible for initiating toluene degradation. BSSA, which is the only enzyme known to initiate anaerobic toluene degradation, was previously detected in this consortium (6).
Molecular analysis of the methanogenic, benzene-enriched consortium revealed four archaeal clones (that grouped with acetoclastic and hydrogenotrophic methanogens) and two predominant bacterial clones (54). One of these bacteria grouped with the Desulfosporosinus sp. (which typically utilizes lactate, pyruvate, ethanol, or certain fatty acids as electron donors, reducing sulfate to hydrogen sulfide), and the second bacterium grouped with Desulfobacterium aniline (which can utilize aniline and phenol as substrates). Based on the phylogenetic association of D. aniline with a clone found in a sulfate-reducing culture capable of benzene degradation (45), and recognizing that sulfate-reducing bacteria can grow fermentatively in the absence of sulfate as an electron acceptor (16), Ulrich and Edwards (54) postulated that the latter microorganism initiates benzene degradation in this consortium.
To date, no toluene-, o-xylene-, or benzene-degrading organism has been isolated from either of these two consortia.
Column inoculation.
Two columns exposed to BTEX and ethanol, which had not exhibited BTEX degradation within 1 year were inoculated with methanogenic consortia. One column was bioagumented with the toluene- and o-xylene-enriched consortium, and another was inoculated with the benzene-enriched culture. About 40 ml of the cell stock solution (∼107 cells ml−1) was injected into a port vial located 20 cm from the column inlet. This location was selected to determine whether specific BTEX degraders would migrate towards the column's inlet, where BTEX and ethanol concentrations are higher, or away from the source, where ethanol no longer remained (22). The third column, which was not bioaugmented, served as a control to discern the benefits of bioaugmentation. The fourth column was poisoned with 0.01% of Kathon CG/ICP biocide (5-chloro-2-methyl-3(2H)-isothiazolone and 2-methyl-3(2H)-isothiazolone solution; Sigma-Aldrich) to distinguish biodegradation from abiotic losses (i.e., volatilization). All columns were operated in the dark and at an average temperature of 22°C.
Soil DNA extraction.
Column aquifer material was collected for DNA analyses. About 5 to 10 g of soil was taken from the port vials located 2.5, 14, 20, and 60 cm from the columns' inlet. Soil samples were dried overnight at room temperature (22°C), and 0.5 g of the dry soil was then transferred into a lysing matrix tube for DNA extraction by using a FastDNA SPIN kit according to the manufacturer's protocols. A bead-beating device (MINI Beadbeater) was utilized for soil lysis. A 50-μl soil DNA sample was collected in a 1.5-ml Eppendorf vial and stored in a freezer (ScienTemp) at −44°C.
RTQ-PCR.
The total numbers of bacteria, archaea, and bssA gene copies were estimated by using real-time quantitative PCR (RTQ-PCR) analysis with primers and probes (Integrated DNA Technologies, Inc.) described in Table 1. Bacteriophage λ (500 bp) was used as an internal standard for the determination of DNA efficiency recovery. When the recovery was lower than 100%, gene copy numbers were normalized to the fraction recovered. No correction was made when DNA recoveries exceeded 100%. Recoveries for the internal standard in the column bioaugmented with benzene-enriched consortium were 115% (at 2.5 cm), 14% (at 14 cm), and 4% (at 60 cm). Recoveries for the internal standard in the column bioaugmented with toluene- and o-xylene-enriched consortium were 148% (at 2.5 cm), 4% (at 14 cm), 6% (at 20 cm), and 15% (at 60 cm). Such wide variations in DNA recoveries (e.g., 0.6 to 126%) are commonly reported (44, 63) and are probably due to the binding of sample impurities (e.g., humic acids) that interfere with the activity of Taq polymerase during PCR analysis (46).
TABLE 1.
Primers and probe sequences used in RTQ-PCR
| Target | Forward primer | Reverse primer | Probed |
|---|---|---|---|
| bssAa | 5′ACGACGGYGGCATTTCTC3′ | 5′GCATGATSGGYACCGACA3′ | FAM-5′CTTCTGGTTCTTCTGCACCTTGGACACC3′-TAMRA |
| Bacteriab | 5′CGGTGAATACGTTCYCGG3′ | 5′GGWTACCTTGTTACGACTT3′ | FAM-5′CTTGTACACACCGCCCGTC3′-BHQ-1 |
| Archeaec | 5′CGGTGAATACGTCCCTGC3′ | 5′AAGGAGGTGATCCTGCCGCA3′ | FAM-5′CTTGTACACACCGCCCGTC3′-BHQ-1 |
| 5′CGGTGAATATGCCCCTGC3′ | |||
| Phage (λ)a | 5′ACGCCACGCGGGATG3′ | 5′AGAGACACGAAACGCCGTTC3′ | TET-5′ACCTGTGGCATTTGTGCTGCCG3′-TAMRA |
The forward and reverse primer as well as the probe were designed by Beller et al. (7).
The forward primer BACT1369F, reverse primer PROK1492R, and probe TM1389F were developed by Suzuki et al. (53).
The forward primer ARCHMIX1369F (ARCH1-1369F and ARCH2-1369F), reverse primer PROK1541R, and probe TM1389F were developed by Suzuki et al. (53).
The reporter dye used was FAM (6-carboxyfluorescein) or TET (tetrachloro-6-carboxyfluorescein), and the quencher dye was either TAMRA (6-carboxy- tetramethyl rhodamine) or BlackHole Quencher-1.
The PCR mixture contained a 0.9 μM concentration of each primer (a 0.45 μM concentration of each forward primer for archaea), a 0.25 μM concentration of the probe, 1× TaqMan Universal PCR Master mix (Applied Biosystems), 2.5 μl of undiluted DNA, and nuclease-free sterile water (AMRESO-E476) to a final volume of 25 μl. The RTQ-PCR was conducted with an ABI PRISM 7000 sequence detection system (Applied Biosystems) with the following temperature conditions: 50°C for 2 min, followed by 95°C for 10 min and 40 cycles at 95°C for 15 s, and 60°C for 1 min. The initial concentration of DNA in the standards was measured by using a Beckman DU-600 fluorometer (Amersham Pharmacia, Piscataway, N.J.).
The number of bssA gene copies in each sample was estimated based on the following equation (7):
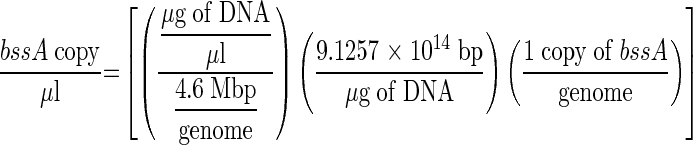 |
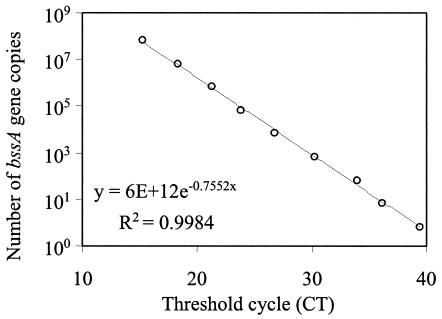
This equation is based on the following assumptions: (i) the bssA primer and probe set designed on different denitrifying bacteria (i.e., Azoarcus sp. strain T, Thauera aromatica strains T1 and K172, and Azoarcus tolulyticus strain Tol-4) was representative of all other bacteria containing bssA; (ii) the approximate size of the strain T1 genome used as the standard in the calibration curves was 4.6 Mbp (and there are approximately 9.12576 × 1014 bp μg of DNA−1), which is equivalent to the size of the Escherichia coli genome (13); and (iii) there is one copy of bssA per genome. These assumptions were also used to quantify bacteria and archaea but not bacteriophage λ, because, in this case, the solutions contained DNA fragments of identical length as those used in the standards. Figure 1 shows an example of a calibration curve prepared for bssA. Similar calibration curves were also prepared for bacteria (T. aromatica strain T1 [102 to 108 copies; r2 = 0.988]), archaea (Methanococcus maripaludis [103 to 108 copies; r2 = 0.990]), and bacteriophage λ (101 to 107 copies; r2 = 0.986).
FIG. 1.
Example of calibration curve used for RTQ-PCR. Data show the standardization of the bssA gene by using genomic DNA of T. aromatica strain T1 (ATCC 700265D). The primers and probes used are shown in Table 1.
Analytical procedures.
Samples for BTEX, ethanol, and methane analyses were collected with a 1-ml gas-tight syringe directly from the columns' port vials and injected into 5-ml gas chromatography (GC) vials previously capped with a 20-mm Teflon-coated septum and aluminum crimps. The vials were placed in a water bath (Buchi 461) at 85°F for 30 min. A 1-ml headspace sample was then injected directly into a Hewlett-Packard model 5890 series II GC equipped with a DB-wax column (30 m, 0.53-mm diameter; J & W Scientific) and a flame ionization detector. GC operational temperatures were set at 175°C for the injector, 250°C for the detector, and 150°C for the oven.
Bromide, acetate, nitrate, and sulfate were analyzed in an Alcott 728 auto sampler equipped with a Gilson 307 pump and a Dionex IonPac AS4A 4-mm column. A Dionex ASRS-I 4-mm anion self-regenerating suppressor and a Dionex conductivity detector ion chromatograph were connected to an interface (HP35900E) and a conductivity detector (Ionpac AS4A column). The eluent solution consisted of 250 mg of sodium bicarbonate liter−1 plus 933 mg of sodium carbonate liter−1 in deionized water. The regenerant solution consisted of deionized water with 2.68 ml of concentrated sulfuric acid (18 M) added per liter. The pH of the samples was measured by using a Fisher Scientific AB15 pH meter.
RESULTS AND DISCUSSION
Column operation history.
Flowthrough aquifer columns were used to simulate the bioattenuation of BTEX and ethanol mixtures over 3 years. The removal of BTEX or ethanol was attributable to biodegradation, since negligible abiotic losses for BTEX and ethanol (<8%) were observed in the poisoned control column (data not shown). Methane concentrations measured in these columns (18 to 23 mg liter−1) were close to the solubility level (24 mg liter−1 at 1 atm and 20°C) (Fig. 2B and 3B), confirming that methanogenic conditions prevailed.
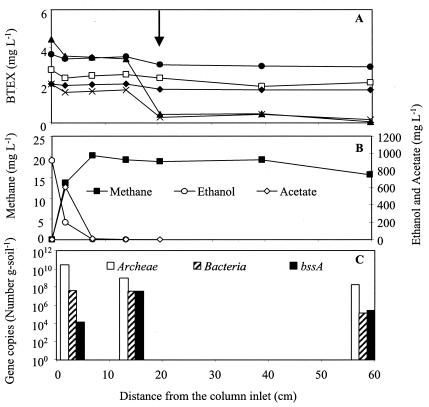
FIG. 2.
Concentration profiles in the column bioaugmented with the toluene- and o-xylene-enriched methanogenic consortium, taken 1 year after inoculation. Concentrations of BTEX (A) and of ethanol, acetate, and methane (B) and the bacterial gene distribution (C) are depicted along the length of the column. The arrow shows the inoculation port. Symbols: □, benzene; ▴, toluene; ⧫, ethylbenzene; ×, o-xylene; •, m-p-xylene.
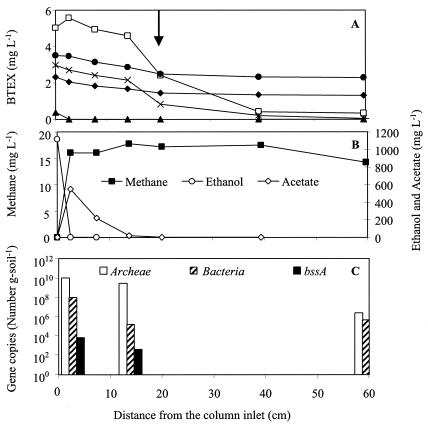
FIG. 3.
Concentration profiles of the column bioaugmented with the benzene-enriched methanogenic consortium, taken 1 year after inoculation. Concentrations of BTEX (A) and ethanol, acetate, and methane (B) and the bacterial gene distribution (C) are depicted along the length of the column. The arrow shows the inoculation port. Symbols: □, benzene; ▴, toluene; ⧫, ethylbenzene; ×, o-xylene; •, m-p-xylene.
Toluene was the only hydrocarbon that was biodegraded in the column that was not bioaugmented. However, its degradation occurred after only 2 years of acclimation (∼137 pore volumes exchanged) (data not shown). On the other hand, benzene degradation was not observed within 3 years. The overall persistence of BTEX compounds was attributed to the development of strongly anaerobic (methanogenic) conditions as result of the high electron acceptor demand exerted during ethanol degradation, which depleted the influent nitrate and sulfate. The low oxidation-reduction found in these columns (Eh = −300 mV) (22) represents decreased thermodynamic feasibility for BTEX bio-oxidation. The recalcitrance of benzene, which is the most toxic and the least frequently degraded of the BTEX compounds under anaerobic conditions (2, 29), motivated us to investigate whether anaerobic bioaugmentation could enhance its natural attenuation in the presence of ethanol.
Effect of bioaugmentation.
Major concerns about the use of bioaugmentation include the survival of the added microorganisms (34, 43) and/or low bacterial transport through the aquifer material, which acts as a filter (24). However, bioaugmentation has a greater probability of success when the added microorganisms fill a metabolic niche that is not being exploited by the indigenous microfloras (58). Since aerobic BTEX-degrading organisms are ubiquitous (28), bioaugmentation is generally considered to be unnecessary to enhance aerobic BTEX bioremediation. Nonetheless, the common persistence of benzene in strongly anaerobic environments (exacerbated by the presence of ethanol) suggests that indigenous microorganisms do not readily exploit benzene degradation as an ecological niche under methanogenic conditions. Therefore, bioaugmentation with competent methanogenic consortia may be justified in such cases to shorten long acclimation periods and enhance degradation rates.
Figure 2A and B shows BTEX and ethanol concentration profiles, respectively, in the column bioaugmented with the toluene- and o-xylene-enriched methanogenic consortium. No BTEX removal had been observed prior to bioaugmentation. The removal rate of toluene and o-xylene occurred within 30 days after bioaugmentation and was most pronounced near the inoculation port (located 20 cm from the inlet). BSSA, which is the enzyme responsible for initiating toluene degradation by the added consortium (6), was probably responsible for the degradation of o-xylene during bacterial growth on toluene, possibly cometabolically as discussed previously (11). Toluene-dependent degradation of o-xylene appears to be a common occurrence in anaerobic systems (2). Interestingly, m- and p-xylene degradation was not observed. Whereas the initial biotransformations of m-, p-, and o-xylene have been reported to be analogous to the biotransformation of toluene by BSSA (8, 11, 12, 33), differences in substrate specificity among BSSA enzymes expressed in different organisms are not well understood. For example, all of the m-xylene-anaerobic degrading bacteria isolated thus far can degrade toluene, but it is unclear why the reverse is not always observed (29). Apparently, the BSSA expressed in this consortium is selective towards toluene and o-xylene degradation.
Benzene and ethylbenzene were not degraded in this column (Fig. 2A). The persistence of these compounds corroborates previous studies showing that this consortium is unable to degrade them (25). Benzene (which lacks a methyl group) and ethylbenzene (which is transformed by the dehydrogenation of the ethyl group) are degraded by different anaerobic pathways (4, 32, 57, 60).
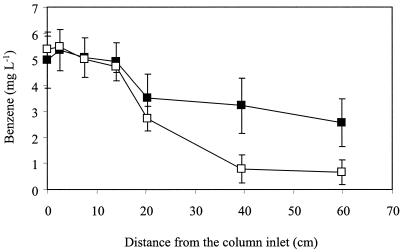
Significant benzene degradation (>49%) was found only in the column bioaugmented with benzene-enriched microorganisms (Fig. 3A). Interestingly, benzene removal efficiency increased from 49% ± 14% to 88% ± 7% after the influent toluene concentration was decreased from 6.8 to 1.0 mg liter−1 (Fig. 4). A subsequent increase in the influent toluene concentration to 2 mg liter−1 resulted in a decrease in removal efficiency to about 75% (data not shown). These results suggest that toluene inhibited anaerobic benzene degradation. Competitive inhibition is unlikely because anaerobic benzene degradation is not mediated by BSSA, which attacks methyl groups that benzene lacks. Rather, anaerobic benzene degradation has been reported to occur through oxidation of the aromatic ring to form phenol (57, 60) and, subsequently, benzoate and a variety of aliphatic acids (29). It is unknown if toluene was degraded by the same organisms that degraded benzene and, if so, whether toluene acted as a noncompetitive inhibitor, perhaps by being utilized preferentially or contributing to metabolic flux dilution (35). o-Xylene degradation was also observed in this column 90 days after inoculation, and its removal increased over time (up to 99%). Overall, the high BTEX removal efficiency after bioaugmentation was sustainable for over 1 year.
FIG. 4.
Effect of toluene on anaerobic benzene degradation. Benzene removal increased from 49 to 88% after the influent toluene concentration was decreased from 7 to 1 mg liter−1. Symbols: ▪, influent toluene of 6.75 ± 1.6 mg liter−1; □, influent toluene of 1.1 ± 1.0 mg liter−1.
Acetate, a by-product of ethanol anaerobic degradation, was observed in all columns (580 to 650 mg liter−1) near the inlet, where the highest ethanol degradation activity occurred (Fig. 2B and 3B). Acetate accumulation resulted in the reduction of pH from 8.3 to 6.3 (data not shown), even though the medium was well buffered with calcium carbonate (∼1 g liter−1). Edwards and Grbić-Galić (25) found that acetate, which is also a by-product of toluene degradation by the toluene- and o-xylene-enriched methanogenic consortium, inhibited toluene degradation. Characterizing and elucidating the inhibitory mechanisms of acetate on BTEX degradation was beyond the scope of this study. However, it did not escape our attention that high benzene, toluene, and o-xylene removal rates were continuously observed near the inoculation port where neither ethanol nor acetate was present (Fig. 2B and 3B).
The increase in biomass concentration due to bioaugmentation could potentially decrease soil permeability and affect nutrient and substrate transport to the growing cells (18, 21, 56). However, no clogging was observed within 1 year of inoculation, suggesting that bioaugmentation (with 40 ml at ∼1010 cells liter−1) had a negligible effect on column permeability. For example, a microbial concentration (x) on the order of 1010 cells g of soil−1, which was the highest measured value (Fig. 2C and 3C), would decrease soil porosity by only about 0.2%, assuming a dry cell weight (DCW) of 1.33 × 10−13 g (14), a soil bulk density (ρbulk) of 1,600 g liter−1, and a biomass density (ρcell) of 1,100 g liter−1 (15), i.e., the pore volume fraction occupied by the microorganisms is calculated as (x× DCW × ρbulk)/ρcell (18).
Molecular analysis.
Soil DNA analysis was performed in the aquifer material prior to inoculation and after 1 year of experimentation. RTQ-PCR was used to estimate the total number of archaea, bacteria, and specific toluene degraders harboring the bssA gene.
The total numbers of bacteria (∼108 cells g of soil−1) and archaea (e.g., methanogens) (∼1010 cells g of soil−1) were higher near the column inlet and decreased along the column length as the substrate concentration decreased (Fig. 2 and 3). Total bacterial concentrations were also estimated independently for these samples by using phospholipid fatty acid analysis by Microbial Insights, Inc. Its results (5 × 107 to 7 × 107 cells g of soil−1) were in good agreement with our RTQ-PCR results.
Little is known about the diversity of the bssA gene in different bacteria. Therefore, it is unclear whether the bssA primer-probe combination used in this study (which was designed based on the genome sequence of denitrifying bacteria) might require alterations to enhance hybridization with the bssA sequences in the methanogenic consortia present in the columns. A suboptimal primer-probe sequence could have resulted in an underestimation of the total number of bssA copies. Nevertheless, the primer-probe set used in this work reproductively detected bssA (detection limit of ≥10 bssA copies g of soil−1) in both columns, and the highest number of bssA gene copies was measured near the location exhibiting the highest toluene degradation activity (Fig. 2). Therefore, this gene might be a useful biomarker for environmental forensic analyses to evaluate anaerobic toluene biodegradation potential. The detection of bssA near the inlet, albeit at levels 4 orders of magnitude lower than that at the point of highest toluene degradation activity (Fig. 2C), suggests chemotaxis of anaerobic toluene degraders toward areas of higher substrate concentration.
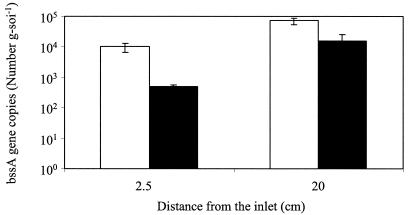
To investigate the persistence of the bssA gene, RTQ-PCR analysis was performed up to 1 year after column feeding was stopped (and thus, exposure to BTEX and ethanol had ceased) (Fig. 5). The results showed that bssA genes were still present in the aquifer material (although at lower concentrations, decreasing by 92% near the inlet and 65% at the inoculation port). This detection suggests the robustness of anaerobic bioaugmentation, which may offer long-term enhanced biodegradation potential.
FIG. 5.
Persistence of the bssA gene in soil samples from the column bioaugmented with toluene- and o-xylene-enriched methanogenic consortium. Open bars, 1 year after inoculation; filled bars, 1 year after exposure to BTEX and ethanol (EtOH) ceased.
In conclusion, the increasing likelihood of encountering ethanol as a cocontaminant in groundwater contaminated with BTEX may require the reconsideration of current remediation practices and the development of novel remediation approaches. This work supports the notion that anaerobic bioaugmentation could be a robust and sustainable approach to accelerate the natural attenuation of BTEX in some aquifers contaminated with ethanol-blended gasoline. This approach may be especially attractive for removing benzene, which is the most toxic and commonly the most persistent BTEX compound under anaerobic conditions. Nevertheless, anaerobic bioaugmentation is unlikely to be universally applicable, and pilot studies should be conducted to identify potential critical limitations associated with scale-up issues, including the required inoculum size and cost, the need for pH control, and performance at lower temperatures in the presence of potential inhibitory compounds.
Acknowledgments
We thank Elizabeth Edwards and Ania Ulrich (University of Toronto, Toronto, Canada) for donating the bacterial consortia utilized in this work, Tim Buschek for donating the aquifer material, Harry Beller and Stacy Kane from Lawrence Livermore National Laboratory for their comments and suggestions regarding RTQ-PCR, Craig Just and Collin Just (University of Iowa) for their analytical assistance, and the DNA Facility (University of Iowa) for its help with PCR analysis.
This work was funded by a grant from the U.S. Environmental Protection Agency STAR program. M.L.B.D.S. was also supported by a fellowship from CAPES-Brazil.
REFERENCES
- 1.Achong, G. R., A. M. Rodriguez, and A. M. Spormann. 2001. Benzylsuccinate synthase of Azoarcus sp. strain T: cloning, sequencing, transcriptional organization, and its role in anaerobic toluene and m-xylene mineralization. J. Bacteriol. 183:6763-6770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alvarez, P. J. J., and T. M. Vogel. 1995. Degradation of BTEX and their aerobic metabolites by indigenous microorganisms under nitrate reducing conditions. Water Sci. Technol. 31:15-28. [Google Scholar]
- 3.Anderson, R. T., and D. R. Lovley. 2000. Anaerobic bioremediation of benzene under sulfate-reducing conditions in a petroleum-contaminated aquifer. Environ. Sci. Technol. 34:2261-2266. [Google Scholar]
- 4.Ball, H. A., H. A. Johnson, M. Reinhard, and A. M. Spormann. 1996. Initial reactions in anaerobic ethylbenzene oxidation by a denitrifying bacterium, strain EB1. J. Bacteriol. 178:5755-5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ball, H. A., and M. Reinhard. 1996. Monoaromatic hydrocarbon transformation under anaerobic conditions at Seal Beach, California: laboratory studies. Environ. Toxicol. Chem. 15:114-122. [Google Scholar]
- 6.Beller, H. R., and E. A. Edwards. 2000. Anaerobic toluene activation by benzylsuccinate synthase in a highly enriched methanogenic culture. Appl. Environ. Microbiol. 66:5503-5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beller, H. R., S. R. Kane, T. C. Legler, and P. J. J. Alvarez. 2002. A real-time polymerase chain reaction method for monitoring anaerobic, hydrocarbon-degrading bacteria based on a catabolic gene. Environ. Sci. Technol. 36:3977-3984. [DOI] [PubMed] [Google Scholar]
- 8.Beller, H. R., and A. M. Spormann. 1997. Anaerobic activation of toluene and o-xylene by addition to fumarate in denitrifying strain T. J. Bacteriol. 179:670-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beller, H. R., and A. M. Spormann. 1997. Benzylsuccinate formation as a means of anaerobic toluene activation by sulfate-reducing strain PRTOL1. Appl. Environ. Microbiol. 63:3729-3731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beller, H. R., and A. M. Spormann. 1999. Substrate range of benzylsuccinate synthase from Azoarcus sp. strain T. FEMS Microbiol. Lett. 178:147-153. [DOI] [PubMed] [Google Scholar]
- 11.Beller, H. R., A. M. Spormann, P. K. Sharma, J. R. Cole, and M. Reinhard. 1996. Isolation and characterization of a novel toluene-degrading, sulfate-reducing bacterium. App. Environ. Microbiol. 62:1188-1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Biegert, T., G. Fuchs, and J. Heider. 1996. Evidence that anaerobic oxidation of toluene in the denitrifying bacterium Thauera aromatica is initiated by formation of benzylsuccinate from toluene and fumarate. Eur. J. Biochem. 238:661-668. [DOI] [PubMed] [Google Scholar]
- 13.Blattner, F. R., G. Plunkett, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 14.Bratbak, G. 1985. Bacterial biovolume and biomass estimations. Appl. Environ. Microbiol. 49:1488-1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bratbak, G., and I. Dundas. 1984. Bacterial dry-matter content and biomass estimations. Appl. Environ. Microbiol. 48:755-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bryant, M., L. Campbell, C. Reddy, and M. Crabill. 1977. 1977. Growth of Desulfovibrio in lactate or ethanol media low in sulfate in association with H2-utilizing methanogenic bacteria. Appl. Environ. Microbiol. 33:1162-1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burland, S. M., and E. A. Edwards. 1999. Anaerobic benzene biodegradation linked to nitrate reduction. Appl. Environ. Microbiol. 65:529-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clement, T. P., B. S. Hooker, and R. S. Skeen. 1996. Macroscopic models for predicting changes in saturated porous media properties caused by microbial growth. Ground Water 34:934-942. [Google Scholar]
- 19.Coates, J. D., R. Chakraborty, J. G. Lack, S. M. O'Connor, K. A. Cole, K. S. Bender, and L. A. Achenbach. 2001. Anaerobic benzene oxidation coupled to nitrate reduction in pure culture by two strains of Dechloromonas. Nature 411:1039-1043. [DOI] [PubMed] [Google Scholar]
- 20.Corseuil, H. X., C. S. Hunt, R. C. F. Dos Santos, and P. J. J. Alvarez. 1998. The influence of the gasoline oxygenate ethanol on aerobic and anaerobic BTX biodegradation. Water Res. 32:2065-2072. [Google Scholar]
- 21.Cunningham, A. B., W. G. Characklis, F. Abedeen, and D. Crawford. 1991. Influence of biofilm accumulation on porous-media hydrodynamics. Environ. Sci. Technol. 25:1305-1311. [Google Scholar]
- 22.Da Silva, M. L. B., and P. J. J. Alvarez. 2002. Effects of ethanol versus MTBE on benzene, toluene, ethylbenzene, and xylene natural attenuation in aquifer columns. J. Environ. Eng. ASCE 128:862-867. [Google Scholar]
- 23.Duba, A. G., K. J. Jackson, M. C. Jovanovich, R. B. Knapp, and R. T. Taylor. 1996. TCE remediation using in situ, resting-state bioaugmentation. Environ. Sci. Technol. 30:1982-1989. [Google Scholar]
- 24.Dybas, M. J., M. Barcelona, S. Bezborodnikov, S. Davies, L. Forney, H. Heuer, O. Kawka, T. Mayotte, L. Sepulveda-Torres, K. Smalla, M. Sneathen, J. Tiedje, T. Voice, D. C. Wiggert, M. E. Witt, and C. S. Criddle. 1998. Pilot-scale evaluation of bioaugmentation for in-situ remediation of a carbon tetrachloride contaminated aquifer. Environ. Sci. Technol. 32:3598-3611. [Google Scholar]
- 25.Edwards, E. A., and D. Grbić-Galić. 1994. Anaerobic degradation of toluene and o-xylene by a methanogenic consortium. Appl. Environ. Microbiol. 60:313-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ellis, D. E., E. J. Lutz, J. M. Odom, R. J. Buchanan, C. L. Bartlett, M. D. Lee, M. R. Harkness, and K. A. Deweerd. 2000. Bioaugmentation for accelerated in situ anaerobic bioremediation. Environ. Sci. Technol. 34:2254-2260. [Google Scholar]
- 27.Ficker, M., K. Krastel, S. Orlicky, and E. Edwards. 1999. Molecular characterization of a toluene-degrading methanogenic consortium. Appl. Environ. Microbiol. 65:5576-5585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gibson, D. T., and V. Subramanian. 1984. Microbial degradation of aromatic hydrocarbons, p. 181-252. In D. T. Gibson (ed.), Microbial degradation of organic compounds. Marcel Dekker, Inc., New York, N.Y.
- 29.Heider, J., A. M. Spormann, H. R. Beller, and F. Widdel. 1998. Anaerobic bacterial metabolism of hydrocarbons. FEMS Microbiol. Rev. 22:459-473. [Google Scholar]
- 30.Hinchee, R. E., and D. C. Downey. 1988. The role of hydrogen peroxide in enhanced bioreclamation, p. 931-948. In National Water Well Association (ed.), Proceedings of the NWWA/API Conference on Petroleum Hydrocarbons and Organic Chemicals in Ground Water: Prevention, Detection and Restoration. American Petroleum Institute, Houston, Tex.
- 31.Kane, S. R., H. R. Beller, T. C. Legler, and R. T. Anderson. 2002. Biochemical and genetic evidence of benzylsuccinate synthase in toluene-degrading, ferric iron-reducing Geobacter metallireducens. Biodegradation 13:149-154. [DOI] [PubMed] [Google Scholar]
- 32.Kniemeyer, O., and J. Heider. 2001. (S)-1-phenylethanol dehydrogenase of Azoarcus sp. strain EbN1, an enzyme of anaerobic ethylbenzene catabolism. Arch. Microbiol. 129:129-135. [DOI] [PubMed] [Google Scholar]
- 33.Krieger, C. J., H. R. Beller, M. Reinhard, and A. M. Spormann. 1999. Initial reactions in anaerobic oxidation of m-xylene by the denitrifying bacterium Azoarcus sp. strain T. J. Bacteriol. 181:6403-6410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krumme, M. L., R. L. Smith, J. Egestorff, S. M. Thiem, J. M. Tiedje, K. N. Timmis, and D. F. Dwyer. 1994. Behavior of pollutant-degrading microorganisms in aquifers—predictions for genetically-engineered organisms. Environ. Sci. Technol. 28:1134-1138. [DOI] [PubMed] [Google Scholar]
- 35.Lovanh, N., and P. J. J. Alvarez. 2004. Effect of ethanol, acetate, and phenol on toluene degradation activity and tod-lux expression in Pseudomonas putida TOD102: evaluation of the metabolic flux dilution model. Biotechnol. Bioeng. 86:801-808. [DOI] [PubMed] [Google Scholar]
- 36.Lovley, D. R. 2000. Anaerobic benzene degradation. Biodegradation 11:107-116. [DOI] [PubMed] [Google Scholar]
- 37.Lovley, D. R. 1991. Dissimilatory Fe(III) and Mn(IV) Reduction. Microbiol. Rev. 55:259-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lovley, D. R., J. D. Coates, J. C. Woodward, and E. J. P. Phillips. 1995. Benzene oxidation coupled to sulfate reduction. Appl. Environ. Microbiol. 61:953-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lovley, D. R., J. C. Woodward, and F. H. Chapelle. 1994. Stimulated anoxic biodegradation of aromatic-hydrocarbons using Fe(III) ligands. Nature 370:128-131. [DOI] [PubMed] [Google Scholar]
- 40.Major, D. W., M. L. McMaster, E. E. Cox, E. A. Edwards, S. M. Dworatzek, E. R. Hendrickson, M. G. Starr, J. A. Payne, and L. W. Buonamici. 2002. Field demonstration of successful bioaugmentation to achieve dechlorination of tetrachloroethene to ethene. Environ. Sci. Technol. 36:5106-5116. [DOI] [PubMed] [Google Scholar]
- 41.Malcolm Pirnie, Inc. 1998. Evaluation of the fate and transport of ethanol in the environment. Report prepared for the American Methanol Institute. Malcolm Pirnie, Inc., Oakland, Calif.
- 42.Molson, J. W., J. F. Barker, E. O. Frind, and M. Schirmer. 2002. Modeling the impact of ethanol on the persistence of benzene in gasoline-contaminated groundwater. Water Resour. Res. 38:1003. [Google Scholar]
- 43.Munakata-Marr, J., P. L. McCarty, M. S. Shields, M. Reagin, and S. C. Francesconi. 1996. Enhancement of trichloroethylene degradation in aquifer microcosms bioaugmented with wild type and genetically altered Burkholderia (Pseudomonas) cepacia G4 and PR1. Environ. Sci. Technol. 30:2045-2052. [Google Scholar]
- 44.Mygind, T., L. Ostergaard, S. Birkelund, J. S. Lindholt, and G. Christiansen. 2003. Evaluation of five DNA extraction methods for purification of DNA from atherosclerotic tissue and estimation of prevalence of Chlamydia pneumoniae in tissue from a Danish population undergoing vascular repair. BMC Microbiol. 3:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Phelps, C. D., L. J. Kerkhof, and L. Y. Young. 1998. Molecular characterization of a sulfate-reducing consortium which mineralizes benzene. FEMS Microbiol. Ecol. 27:269-279. [Google Scholar]
- 46.Porteous, L. A., R. J. Seidler, and L. S. Watrud. 1997. An improved method for purifying DNA from soil for polymerase chain reaction amplification and molecular ecology applications. Mol. Ecol. 6:787-791. [Google Scholar]
- 47.Powers, S. E., C. S. Hunt, S. E. Heermann, H. X. Corseuil, D. Rice, and P. J. J. Alvarez. 2001. The transport and fate of ethanol and BTEX in groundwater contaminated by gasohol. Crit. Rev. Environ. Sci. Technol. 31:79-123. [Google Scholar]
- 48.Powers, S. E., D. Rice, B. Dooher, and P. J. J. Alvarez. 2001. Will ethanol-blended gasoline affect groundwater quality? Using ethanol instead of MTBE as a gasoline oxygenate could be less harmful to the environment. Environ. Sci. Technol. 35:24A-30A. [DOI] [PubMed] [Google Scholar]
- 49.Rabus, R., and J. Heider. 1998. Initial reactions of anaerobic metabolism of alkylbenzenes in denitrifying and sulfate reducing bacteria. Arch. Microbiol. 170:377-384. [Google Scholar]
- 50.Ruiz-Aguilar, G. M. L., J. M. Fernandez-Sanchez, S. R. Kane, D. Kim, and P. J. J. Alvarez. 2002. Effect of ethanol and methyl-tert-butyl ether on monoaromatic hydrocarbon biodegradation: response variability for different aquifer materials under various electron-accepting conditions. Environ. Toxicol. Chem. 21:2631-2639. [PubMed] [Google Scholar]
- 51.Ruiz-Aguilar, G. M. L., K. O'Reilly, and P. J. J. Alvarez. 2002. A comparison of benzene and toluene plume lengths for sites contaminated with regular vs. ethanol-amended gasoline. Ground Water Monit. Res. 23:48-53. [Google Scholar]
- 52.Salanitro, J. P., P. C. Johnson, G. E. Spinnler, P. M. Maner, H. L. Wisniewski, and C. Bruce. 2000. Field scale demonstration of enhanced MTBE bioremediation through aquifer bioaugmentation and oxygenation. Environ. Sci. Technol. 34:4152-4162. [Google Scholar]
- 53.Suzuki, M. T., L. T. Taylor, and E. F. DeLong. 2000. Quantitative analysis of small-subunit rRNA genes in mixed microbial populations via 5 ′-nuclease assays. Appl. Environ. Microbiol. 66:4605-4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ulrich, A. C., and E. A. Edwards. 2003. Physiological and molecular characterization of anaerobic benzene-degrading mixed cultures. Environ. Microbiol. 5:92-102. [DOI] [PubMed] [Google Scholar]
- 55.U.S. Environmental Protection Agency. 1985. How to evaluate alternative cleanup technologies for underground storage tank sites: a guide for corrective action plan reviewers. U.S. Environmental protection agency document no. 510-B-94-003 and 510-B-95-007. U.S. Environmental Protection Agency, Washington, D.C.
- 56.Vandevivere, P., and P. Baveye. 1992. Relationship between transport of bacteria and their clogging efficiency in sand columns. Appl. Environ. Microbiol. 58:2523-2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vogel, T. M., and D. Grbicgalic. 1986. Incorporation of oxygen from water into toluene and benzene during anaerobic fermentative transformation. Appl. Environ. Microbiol. 52:200-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vogel, T. M., and M. V. Water. 2003. Bioaugmentation, p. 952-959. In C. J. Hurst, R. L. Crawford, G. R. Knudsen, M. J. McInerney, and L. D. Stetzenbach (ed.), Manual of environmental microbiology, 2nd ed. ASM Press, Washington, D.C.
- 59.Weiner, J. M., and D. R. Lovley. 1998. Anaerobic benzene degradation in petroleum-contaminated aquifer sediments after inoculation with a benzene-oxidizing enrichment. Appl. Environ. Microbiol. 64:775-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weiner, J. M., and D. R. Lovley. 1998. Rapid benzene degradation in methanogenic sediments from a petroleum-contaminated aquifer. Appl. Environ. Microbiol. 64:1937-1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wiesner, M. R., M. C. Grant, and S. R. Hutchins. 1996. Reduced permeability in groundwater remediation systems: Role of mobilized colloids and injected chemicals. Environ. Sci. Technol. 30:3184-3191. [Google Scholar]
- 62.Zengler, K., J. Heider, R. Rossello-Mora, and F. Widdel. 1999. Phototrophic utilization of toluene under anoxic conditions by a new strain of Blastochloris sulfoviridis. Arch. Microbiol. 172:204-212. [DOI] [PubMed] [Google Scholar]
- 63.Zhou, J. Z., M. A. Bruns, and J. M. Tiedje. 1996. DNA recovery from soils of diverse composition. Appl. Environ. Microbiol. 62:316-322. [DOI] [PMC free article] [PubMed] [Google Scholar]