Abstract
External addition of the β-lactam precursor α-aminoadipic acid to the filamentous fungus Penicillium chrysogenum leads to an increased intracellular α-aminoadipic acid concentration and an increase in penicillin production. The exact route for α-aminoadipic acid uptake is not known, although the general amino acid and acidic amino acid permeases have been implicated in this process. Their corresponding genes, PcGAP1 and PcDIP5, of P. chrysogenum were cloned and functionally expressed in a mutant of Saccharomyces cerevisiae (M4276) in which the acidic amino acid and general amino acid permease genes (DIP5 and GAP1, respectively) are disrupted. Transport assays show that both PcGap1 and PcDip5 mediated the uptake of α-aminoadipic acid, although PcGap1 showed a higher affinity for α-aminoadipic acid than PcDip5 (Km values, 230 and 800 μM, respectively). Leucine strongly inhibits α-aminoadipic acid transport via PcGap1 but not via PcDip5. This difference was exploited to estimate the relative contribution of each transport system to the α-aminoadipic acid flux in β-lactam-producing P. chrysogenum. The transport measurements demonstrate that both PcGap1 and PcDip5 contribute to the α-aminoadipic acid flux.
The production of penicillin, still the most widely used antibiotic together with its derivatives, is mainly based on an industrial fermentation utilizing the filamentous fungus Penicillium chrysogenum. Although the biochemistry of penicillin biosynthesis has been researched extensively, much less is known about the transport processes involved. These include, for instance, the transport of penicillin or its precursors across membranes of the intracellular compartments and its transport across the plasma membrane (43). The penicillin biosynthesis pathway starts in the cytosol with the condensation of three amino acids, l-α-aminoadipic acid (α-AAA), l-cysteine, and l-valine, to form the tripeptide l-α-amino-adipyl-l-cysteinyl-d-valine (ACV) (44). ACV is converted into isopenicillin N (IPN) by IPN synthase. In this step the characteristic β-lactam ring structure is formed by ring closure. IPN is subsequently transported into microbodies or peroxisomes, where the α-AAA moiety is replaced by a phenylacetic acid group involving a coenzyme A derivative of this aromatic acid. This process yields penicillin G, which subsequently must be transported across the microbody membrane and the plasma membrane in order to accumulate in the extracellular medium (1, 7, 29, 43). α-AAA is released in the microbody and can be recycled for either lysine or penicillin biosynthesis. However, a portion of the α-AAA is lost by the irreversible formation of 6-oxopiperidine-2-carboxylic acid (OPC), a cyclized form of α-AAA. The extent of OPC formation ranges from 6 to 60% relative to the formation of penicillin (on a molar basis), depending on the strain and cultivation conditions. A major fraction of the OPC seems to be released from the cell, but the mechanism of secretion is unknown (16).
The intracellular concentration of α-AAA, but not that of cysteine or valine, appears limiting for the synthesis of the tripeptide ACV, which also affects the overall penicillin biosynthesis rate in low-producing strains (13, 19). This has become apparent from experiments in which the mycelium is supplemented with α-AAA, whereupon the intracellular α-AAA concentration is elevated concomitantly with enhanced levels of ACV and penicillin production (13, 19). In addition, strains producing higher levels of penicillin have increased intracellular α-AAA levels (23). In P. chrysogenum, α-AAA is a branching intermediate of the lysine and penicillin biosynthesis pathways, which share common enzymatic steps starting from α-ketoglutarate. The routes diverge at the point where α-AAA can be converted into α-AAA-semialdehyde by α-AAA reductase (5, 12, 23). Disruption of the gene encoding this enzyme, LYS2, results in a higher intracellular α-AAA concentration and concomitantly higher levels of penicillin production (10). In Aspergillus nidulans, which also produces penicillin but is not used for industrial production, the relative amount of α-AAA entering either the penicillin synthesis pathway or the lysine synthesis pathway is tightly regulated (9). Another important aspect of the α-AAA biosynthesis route is that in the yeast Saccharomyces cerevisiae, some of the enzymes of the lysine biosynthesis pathway are localized in the microbodies instead of the mitochondria (8). It is unclear how this pathway is organized in filamentous fungi.
The observation that externally added α-AAA enhances penicillin biosynthesis strongly suggests that there is a mechanism of α-AAA uptake. Fungi have different amino acid permeases that differ in specificity. Some systems are specific only for a group of related amino acids, while the general amino acid permease can transport all amino acids (20, 32, 37). These systems all belong to one family, referred to as the amino acid permease family (3). Exceptions are the permease encoded by the mtr locus of Neurospora crassa and its homologue in P. chrysogenum, which are specific for neutral aliphatic and aromatic amino acids (26, 39; H. Trip et al., unpublished data). In general, amino acid transport occurs by proton symport and thus is driven by the proton motive force (18, 35). Although α-AAA is not one of the natural amino acids but an intermediate of amino acid metabolism, it shares the characteristics of glutamate and aspartate as a dicarboxylic amino acid. In S. cerevisiae, these amino acids can enter the cell via the general amino acid permease Gap1 and the acidic amino acid permease Dip5 (33). For P. chrysogenum, transport systems have mostly been described only on the basis of functional studies; these systems have not been genetically described. The general amino acid permease is a nonspecific system that is involved in the uptake of all amino acids (4), while the acidic amino acid permease mediates the uptake of glutamate and aspartate. Previous studies have shown that glutamate transport by the acidic amino acid permease can be inhibited by an excess of α-AAA, which suggests that this compound is a substrate for this transporter (21).
Here we have analyzed the uptake of α-AAA by P. chrysogenum in more detail, and we show that there are two major uptake routes: (i) via the acidic amino acid permease and (ii) via the general amino acid permease. The genes encoding these transporters were cloned into an S. cerevisiae mutant in which acidic amino acid uptake is defective, allowing the functional characterization of the P. chrysogenum Gap1 and Dip5 homologues. The results indicate that both systems contribute to the uptake of α-AAA by P. chrysogenum under conditions in which β-lactams are formed.
MATERIALS AND METHODS
Strains and culture conditions.
P. chrysogenum strain Wisconsin 54-1255 (kindly provided by DSM-Anti-Infectives, Delft, The Netherlands) was grown in a minimal medium based on a penicillin production medium as described previously (27). The medium contained 4% (wt/vol) glucose or lactose as the carbon source and either 0.4% (wt/vol) ammonium acetate, 0.4% (wt/vol) urea, 10 mM l-glutamate, 10 mM l-serine, or 10 mM l-lysine as the nitrogen source. Cultures were started on YPG medium (1% yeast extract, 2% peptone, 2% glucose), incubated for 16 to 20 h in a rotary shaker at 200 rpm and 25°C, and then diluted into minimal medium and incubated for 24 or 48 h.
S. cerevisiae strain M4276 (MATα ura3 Δgap1 Δdip5) (33) was grown either in minimal medium (1% [wt/vol] succinic acid, 0.6% [wt/vol] NaOH, 0.16% [wt/vol] yeast nitrogen base [Difco] without ammonium sulfate and amino acids, and 2% [wt/vol] d-glucose) (15), in MA medium (minimal medium containing 0.5% [wt/vol] ammonium sulfate as the sole nitrogen source), in MP medium (containing 0.1% [wt/vol] l-proline as the sole nitrogen source), or in MG medium (containing 0.5 g of l-glutamate/liter as the sole nitrogen source). Where indicated, these media were solidified with 1.5% agar.
Escherichia coli strain DH5α was used as the plasmid host, and DNA manipulations were carried out essentially as described elsewhere (34). E. coli XL10-Gold Ultracompetent cells (Stratagene, Amsterdam, The Netherlands) were used for amplification of the partial cDNA library.
Expression of a partial cDNA library of P. chrysogenum in S. cerevisiae M4276.
A cDNA library of P. chrysogenum (kindly provided by DSM-Anti-Infectives, Delft, The Netherlands) was transferred from pCMVSPORT 4.0 (Stratagene) to a suitable yeast expression vector. For this purpose, the ampicillin resistance marker of the yeast/E. coli shuttle vector yEP352 (17) was replaced by a kanamycin resistance marker, and the copper-inducible promoter of CUP1, pCUP1 (36), was introduced together with the terminator of CYC1 (45), yielding plasmid yEX-C. The SbfI and NotI sites between pCUP1 and tCYC1 were used for insertion of the cDNA library. The cDNA library was isolated from pCMVSPORT 4.0 by digestion with SbfI and NotI and was subjected to gel electrophoresis. Since the sizes of known fungal amino acid permease genes range from 1.4 to 2 kb, fragments ranging from 1.0 to 2.5 kb were isolated, purified, and ligated into yEX-C. To amplify the library, the ligation mixture was transformed to E. coli XL10-Gold Ultracompetent cells (Stratagene) for high transformation efficiency, and the cells were spread on Luria broth plates with kanamycin. Plasmid DNA was isolated by resuspending the colonies in water, followed by a standard plasmid isolation procedure. About 5 μg of plasmid DNA was transformed to S. cerevisiae M4276, a strain deficient in glutamate and aspartate uptake, as described previously (14). Cells were spread on MG plates containing 0.05 g of l-glutamate/liter as the sole nitrogen source and 0.2 mM CuSO4 for induction of the copper-inducible promoter pCUP1. At the same time, the transformation efficiency was checked by spreading transformed cells on MA plates, containing NH4+ as the nitrogen source; an efficiency of approximately 35,000 transformants per μg of DNA was obtained. After 6 days of incubation at 30°C, eight colonies appeared; they were transferred to liquid minimal medium containing 0.05 g of l-glutamate/liter as the sole nitrogen source. Only one of the colonies was able to grow on glutamate, and the plasmid DNA was isolated by following a standard plasmid rescue protocol. For propagation, plasmid DNA was transformed to E. coli DH5α. The plasmid was analyzed by restriction, revealing a 1.9-kb insert in the yEX-C vector; the DNA sequence of the insert was determined.
Cloning of PcGAP1.
The following degenerate primers were designed on the basis of conserved regions of known fungal (putative) general amino acid permease genes: primer Gap F (5′-CCCGGCGCCATCAARCARGTNTTYTGG-3′) and primer Gap R1 (3′-CCGGAGATGGCCAGNAGCCARTCRAA-5′). PCR was performed with genomic DNA of P. chrysogenum as a template. PCR products were analyzed by gel electrophoresis, and a clear, distinct fragment of 0.4 kb was isolated, purified, and ligated into pGEM-T Easy (Promega, Leiden, The Netherlands). Of the four clones analyzed by sequencing, three contained a fragment showing high homology (50% amino acid identity) with Gap1 of S. cerevisiae and one contained a fragment showing highest homology (45 to 48% amino acid identity) with yeast basic amino acid permeases. The Gap1 homologous fragment was used to make a radioactively labeled probe for the screening of a genomic library.
Isolation of genomic DNA of permease genes.
A genomic library of P. chrysogenum Wisconsin 1255-54 in phage λZAP II (Stratagene) was screened by plaque hybridization using a radioactively labeled 400-bp internal fragment of PcDIP5 amplified by PCR or the PcGAP1 gene fragment described above. Positive plaques were isolated, phages were released, and pBluescript, containing the genome fragments (average size, 8 kb), was rescued from the phage by incubation with helper phage M13. DNA sequencing of the PcDIP5 positive clone revealed a complete, 1,677-bp open reading frame, interrupted by five introns of approximately 60 bp. The PcGAP1 positive clone contained a 1,743-bp open reading frame interrupted by three introns of 63, 55, and 35 bp. Introns were identified on the basis of consensus sequences of introns of P. chrysogenum and, in the case of PcDIP5, by alignment of the cDNA and genomic DNA.
Cloning of the cDNA of PcGAP1 in the yeast expression vector yEX-C.
A forward primer with an EcoRI restriction site (5′-ATTCACATAGAATTCATGGAGGAGAAGAAGTTTGAGGC-3′) and a reverse primer with a NotI restriction site (3′-ACAGCGGCCGCTTTCCAGGTAAGATGC-5′) were used in a PCR to amplify the cDNA of PcGAP1 with the cDNA library as a template. The 1.8-kb product obtained was digested with EcoRI and NotI and ligated into the EcoRI- and NotI-digested vector yEX-C. The resulting plasmid, yEX-PcGAP1, was transformed to S. cerevisiae M4276.
α-AAA transport assays with the mycelium of P. chrysogenum.
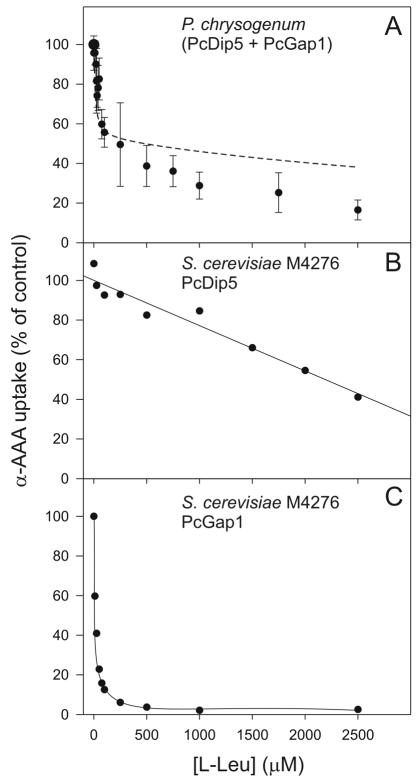
P. chrysogenum was precultured overnight as described above, and germinating spores were added to penicillin production media. After 48 h of growth, the mycelium was harvested by suction filtration, washed, and resuspended in ice-cold 50 mM KH2PO4—K2HPO4 (pH 6.0) to a density of approximately 15 mg (dry weight)/ml. This suspension was preincubated for 5 min at 25°C with aeration, and dl-[3H]α-aminoadipate was added to final concentrations of 10 μM to 2.5 mM. For inhibition studies, dl-[3H]α-aminoadipate was present at 25 μM, while the competitor l-leucine was used at a concentration range of 10 μM to 2.5 mM. After 3 min, uptake was stopped by addition of 2 ml of ice-cold 0.1 M LiCl and subsequent suction filtration through a 0.45-μM-pore-size nitrocellulose filter. The filter was washed with 2 ml of 0.1 M LiCl, and radioactivity was measured in a scintillation counter. Background values were determined by measuring uptake in the mycelium, which was pretreated with the protonophore carbamoyl cyanide m-chlorophenylhydrazone (CCCP). Uptake measurements with P. chrysogenum were made in four independent experiments, for which average values and standard deviations are shown (see Fig. 1A).
FIG. 1.
Uptake of α-AAA and inhibition by unlabeled l-leucine. (A) The uptake of 3H-α-AAA (25 μM) by the mycelium of P. chrysogenum grown under penicillin-producing conditions was assayed in the presence of increasing concentrations of unlabeled l-leucine. The dashed line represents the best-fitting curve after computer-aided regression and curve fitting of the equation given in Materials and Methods (see “Amino acid transport assays with S. cerevisiae” above) to the measured data. (B and C) Assays of uptake of [3H]α-AAA by S. cerevisiae M4276 cells expressing PcDIP5 and PcGAP1, respectively. Assay conditions were as described for panel A.
Amino acid transport assays with S. cerevisiae.
S. cerevisiae strains M4276 yEX-PcDIP5, M4276 yEX-PcGAP1, and M4276 yEX-C (empty vector) were grown overnight in MP medium at 30°C. Cultures were then diluted 10-fold in fresh MP medium, containing 0.2 mM CuSO4 for induction of transcription of PcDIP5 or PcGAP1, and were grown to an optical density at 600 nm (OD600) of 0.4 to 0.8. Cells were harvested by centrifugation, washed with minimal medium without a nitrogen source (MM), and resuspended to an OD600 of 10 in the same medium. Before the transport assay was started, cells were incubated for 10 min at 30°C with stirring. Subsequently, 100 μl of the cell suspension was added to 150 μl of MM with 25 μM 14C-labeled l-amino acid (0.05 μCi), and incubation was continued at 30°C with stirring. Uptake reactions were stopped either immediately after addition of the substrate or after 30, 60, or 120 s by the addition of 2 ml of ice-cold 0.1 M LiCl and subsequent suction filtration through a 0.45-μM-pore-size nitrocellulose filter. Radioactivity retained on the filters was measured in a scintillation counter. For Km determinations, uptake of glutamate and aspartate at concentrations varying from 10 to 2,500 μM was measured after 30 s to ensure linearity with time. For Km determinations of α-AAA transport, uptake was assessed after 3 min. Ki values of leucine inhibition were determined at α-AAA concentrations of 10 to 2,500 μM, in the presence or absence of l-leucine at 1 or 2.5 mM in PcDIP5-expressing cells and at 10 μM in PcGAP1-expressing cells. All uptake measurements with S. cerevisiae M4276 were made in at least two independent experiments, and average values are given.
The Km values for α-AAA transport via PcDip5 and PcGap1 and the respective Ki values for the competitive inhibition by leucine were used to assess the relative contributions of the two transporters to the uptake of α-AAA by P. chrysogenum in the presence of increasing concentrations of leucine. The data were analyzed by regression according to Michaelis-Menten kinetics by using the following equation:
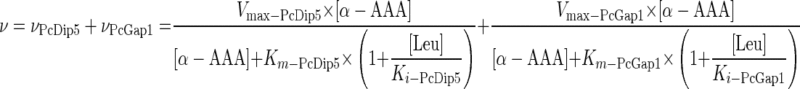 |
where v is the observed rate of α-AAA uptake; vPcDip5 and vPcGap1 are the rates of uptake via PcDip5 and PcGap1, respectively; Km-PcDip5 and Km-PcGap1 are the Michaelis constants and Vmax-PcDip5 and Vmax-PcGap1 are the maximal velocities for α-AAA uptake via PcDip5 and PcGap1, respectively; Ki-PcDip5 and Ki-PcGap1 are the inhibition constants for competitive inhibition by leucine of α-AAA uptake via PcDip5 and PcGap1, respectively; and [α-AAA] and [Leu] are the concentrations of α-AAA and leucine, respectively.
Semiquantitative RT-PCR.
Total RNA extracts were prepared by a Trizol-based method (Invitrogen, Breda, The Netherlands). The mycelium was frozen in liquid nitrogen and ground. Approximately 5 volumes of Trizol were added, and samples were mixed thoroughly. After the addition of 1/5 volume of chloroform and mixing, samples were spun for 15 min at 10,000 rpm in a microcentrifuge. The RNA-containing water phase was collected and further purified by phenol extraction. RNA was precipitated with ethanol and dissolved in H2O. The RNA concentration was determined spectroscopically at 260 nm. Reverse transcriptase PCR (RT-PCR) was performed using RT-PCR beads (Amersham-Pharmacia, Freiburg, Germany) and a Perkin-Elmer thermocycler. Primers were used that generated internal fragments of PcDIP5 (0.35 kb) or PcGAP1 (0.5 kb) or the internal standard actA (0.8 kb), which served as a control to monitor the total amount of RNA used as a template. Samples were incubated at 42°C for 30 min for reverse transcription, followed by a standard PCR under the following conditions: 5 min at 95°C, followed by 22 cycles of 30 s at 95°C, 30 s at 55°C, and 40 s at 72°C. PCR products were analyzed by gel electrophoresis.
Materials.
Uniformly labeled dl-[3H]α-aminoadipate, with a specific activity of 38 Ci/mmol, was custom synthesized by Amersham Pharmacia Biotech.
Nucleotide sequence accession numbers.
The 2,580- and 1,997-kb sequences containing the P. chrysogenum PcDIP5 and PcGAP1 genes have been deposited in GenBank under accession numbers AY456273 and AY456274, respectively.
RESULTS
Uptake of α-AAA by P. chrysogenum Wisconsin 54-1255.
Since the general amino acid permease of P. chrysogenum transports most amino acids including glutamate and aspartate, and because α-AAA inhibits glutamate uptake mediated by the acidic amino acid permease (21), both of these permeases have been implicated in the uptake of α-AAA. However, this assumption has not been directly tested in experiments in which the uptake of α-AAA is measured. Nor have the individual contributions of the two permeases and other possible systems to α-AAA uptake in P. chrysogenum cells grown at penicillin-producing conditions been determined. For that purpose, transport studies were performed with dl-[3H]α-aminoadipate. The amino acid was used at a concentration of 25 μM. l-Leucine is a good substrate for the general amino acid permease and is not transported by the acidic amino acid permease (4, 21, 22). Therefore, l-leucine was used for competition studies to distinguish between the two transporters. α-AAA was readily accumulated by the cells. The inhibition profile of α-AAA uptake by P. chrysogenum in the presence of increasing concentrations of l-leucine showed two distinguishable inhibition effects (Fig. 1A). At low l-leucine concentrations, up to 100 μM, the uptake of α-AAA was readily reduced by 45 to 50%, whereas higher concentrations of l-leucine up to 2.5 mM only marginally inhibited the remaining α-AAA uptake. These data suggest that a major fraction of the α-AAA uptake is mediated by the general amino acid permease, but they also show that there is a residual uptake which is minimally inhibited by higher concentrations of l-leucine.
Cloning of the acidic amino acid permease gene PcDIP5.
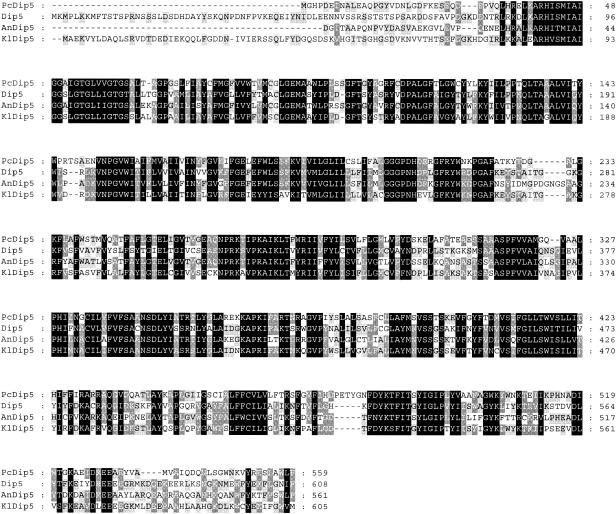
To define the exact identity of the transporters involved in α-AAA, it is necessary to clone the genes and analyze their activities separately. Since the residual α-AAA uptake in the presence of excess l-leucine could be inhibited by unlabeled glutamate (data not shown), we decided, in addition to the general amino acid permease, to clone the acidic amino acid permease. In S. cerevisiae, acidic amino acids (glutamate and aspartate) can be taken up by the general amino acid permease Gap1, as well as by the acidic amino acid permease Dip5 (33). A mutant strain that lacks both the Gap1 and Dip5 genes, M4276, has been described. This mutant is deficient in acidic amino acid transport and therefore cannot grow with glutamate or aspartate as the sole nitrogen source (33). In contrast to the parental strain, S. cerevisiae M4276 is also unable to accumulate α-AAA, which indicates that there are no other routes for uptake of this molecule in S. cerevisiae (data not shown). Strain M4276 was used for a functional complementation cloning strategy to identify transporters that mediate dicarboxylic amino acid uptake. A cDNA library of P. chrysogenum was expressed in S. cerevisiae M4276, and cells were selected on plates and liquid media containing l-glutamate as the sole nitrogen source. Cells that grow on these media must have acquired a transport system that is capable of glutamate uptake. This procedure yielded one clone that could grow with glutamate as the sole nitrogen source. The cDNA of the clone contained a 1,677-bp open reading frame encoding a 559-amino-acid protein that is 47% identical to Dip5 (Fig. 2) and 30 to 34% identical to Gap1 and other amino acid permeases of S. cerevisiae. The gene was designated PcDIP5.
FIG. 2.
Amino acid sequence alignment of the acidic amino acid permeases of P. chrysogenum (PcDip5) and S. cerevisiae (Dip5) (33) and putative acidic amino acid permeases of A. nidulans (AnDip5) and Kluyveromyces lactis (KlDip5). The A. nidulans sequence, 57% identical to that of PcDip5 and 50% identical to that of Dip5, was derived from the A. nidulans database of the Whitehead Institute after a BLAST search (2). This sequence is incomplete at the N terminus. The K. lactis sequence was obtained after a BLAST search at the National Center for Biotechnology Information; it shares 61% amino acid identity with Dip5 and 50% identity with PcDip5. Conserved and similar amino acids are highlighted in black and grey, respectively.
Cloning of the general amino acid permease gene PcGAP1.
Despite several attempts, selection for complementation of glutamate uptake in S. cerevisiae with the partial cDNA library did not yield a general amino acid permease. Therefore, an alternative PCR approach using degenerate primers based on the alignment of five known general amino acid permeases was employed. With these primers, a 424-bp PCR fragment was obtained that, upon translation into amino acids, showed high homology with S. cerevisiae Gap1. The fragment was used to probe a genomic library from which the full-length gene was isolated. The entire gene consists of a 1,751-bp open reading frame interrupted by three introns of 63, 55, and 35 bp and encodes a protein of 581 amino acids. After a BLAST search (2), the highest homology was found with known general amino acid permeases of S. cerevisiae (Gap1; 50% identity), Candida albicans (Gap1; 47%), and N. crassa (Naap1; 48%) (Fig. 3). The gene was therefore designated PcGAP1.
FIG. 3.
Sequence alignment of the general amino acid permeases of P. chrysogenum (PcGap1), S. cerevisiae (Gap1) (24), C. albicans (CaGap1) (6), Aspergillus fumigatus (AfGap1), and N. crassa (NcGap1; encoded by the pmg locus) (28). AfGap1 is a putative general amino acid permease, based on its homology with PcGap1 (78% amino acid identity) and Gap1 (55% identity), and was obtained from the A. fumigatus Genome Database at The Institute for Genomic Research after a BLAST search (2). NcGap1 was originally deposited with GenBank as NAAP1. Conserved and similar amino acids are highlighted in black and grey, respectively.
Overexpression of PcDIP5 and PcGAP1 in S. cerevisiae M4276 and transport characteristics.
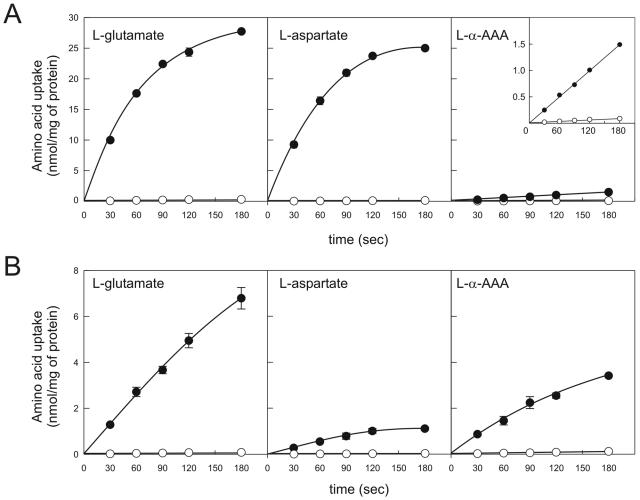
The plasmids isolated from the cDNA library, yEX-PcDIP5 and yEX-PcGAP1, were used for functional characterization of the transporters in S. cerevisiae M4276. As a negative control, M4276 was transformed with the empty vector yEX-C. Cells were incubated with CuSO4 for induction of expression and were analyzed for the uptake of a range of amino acids (42). Cells expressing PcDIP5 readily accumulated l-glutamate and l-aspartate, while α-AAA was taken up at a lower rate (Fig. 4A). l-Asparagine was accumulated slowly, whereas for l-glutamine and l-serine no significant difference was observed between the PcDip5 and control cells (data not shown). The affinity (Km) of PcDip5 for the uptake of l-glutamate and l-aspartate was about 35 μM, while the Km for α-AAA uptake appeared rather high, i.e., 800 μM. Since the 3H-labeled α-AAA represents a racemic mixture of the d and l configurations, further competition experiments were performed with the unlabeled stereoisomers. A 100-fold excess of unlabeled d-α-AAA did not inhibit uptake of labeled dl-α-aminoadipate, whereas an excess of l-α-AAA decreased uptake by more than 90%. This shows that PcDip5 is specific for the l configuration (data not shown). Taken together, these results demonstrate that the cloned permease, PcDip5, corresponds to the acidic amino acid permease of P. chrysogenum.
FIG. 4.
Time-dependent uptake of acidic amino acids by S. cerevisiae M4276 expressing PcDIP5 (A) or PcGAP1 (B). Cells containing the empty vector were used as controls (open symbols). The inset in panel A shows the uptake of α-AAA with an adjusted scale. Uptake assays were performed with cells grown to exponential phase in MP medium, and expression of PcDIP5 was induced with 0.2 mM CuSO4 5 h before harvesting.
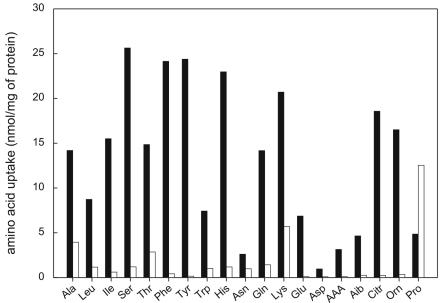
For cells expressing PcGAP1, the uptake of 19 amino acids, including the nonprotein amino acids l-citrulline, dl-α-aminoadipate, l-α-aminoisobutyric acid, and l-ornithine, was measured (Fig. 5). For all amino acids except l-proline, a significant increase of at least twofold in uptake was observed upon expression of PcGAP1. These data suggest either that l-proline is not a substrate for PcGap1 or that in cells overexpressing PcGAP1, the background uptake via endogenous proline permease(s) is reduced due to the expression of PcGAP1. The acidic amino acids glutamate and aspartate were accumulated more slowly by PcGAP1-expressing cells than by PcDIP5-expressing cells (Fig. 4B). However, since the total amount of PcGap1 or PcDip5 protein in these cells is unknown, the exact Vmax values cannot be compared. Strikingly, α-AAA was readily accumulated, with a Km of 230 μM. This affinity is more than 3 times higher than that observed for PcDip5. The general amino acid permeases of S. cerevisiae and N. crassa have been reported also to transport some d amino acids (20), but like PcDip5, PcGap1 was found to transport α-aminoadipate only in the l configuration (data not shown). These data demonstrate that the cloned gene PcGAP1 encodes the general amino acid permease of P. chrysogenum.
FIG. 5.
Uptake of amino acids by S. cerevisiae M4276 expressing PcGAP1. Cells were incubated for 3 min with the indicated radioactively labeled amino acids at a final concentration of 25 μM. Solid or open bars, levels of uptake by cells expressing PcGAP1 (yEX-PcGAP1) or cells with the empty vector (yEX-C), respectively.
Inhibition of PcDip5- or PcGap1-mediated α-AAA uptake by leucine.
The biphasic inhibition by l-leucine of α-AAA uptake by the P. chrysogenum mycelium (Fig. 1A) was explained by the presence of at least two different uptake routes for α-AAA: uptake via the general amino acid permease, which is strongly inhibited by l-leucine, and uptake via a specific amino acid permease that is nearly insensitive to l-leucine. To validate this hypothesis, we reinvestigated the inhibition by leucine of α-AAA uptake in S. cerevisiae M4276 expressing PcDIP5 and, separately, in M4276 expressing PcGAP1 (Fig. 1B and C). Indeed, PcGap1-mediated α-AAA uptake was strongly inhibited by l-leucine (Fig. 1C) at exactly the concentration range that was inhibitory in P. chrysogenum cells. In contrast, PcDip5-mediated uptake was inhibited by l-leucine only at high concentrations (Fig. 1B). The Ki for leucine inhibition of PcGap1-mediated uptake was 10 μM, and for PcDip5-mediated uptake the Ki was around 6.3 mM. This high Ki is not in perfect agreement with the inhibition pattern shown in Fig. 1B, which should have a flatter slope. Therefore, a nonspecific effect by leucine seems to inhibit α-AAA uptake via PcDip5.
The various Km and Ki values were used for computer modeling of α-AAA uptake by using the equation given in Materials and Methods (see “Amino acid transport assays with S. cerevisiae” above), which describes the relative contributions of the two transporters according to Michaelis-Menten kinetics. The data were fitted by regression to the data shown in Fig. 1A. The best-fitting curve (Fig. 1A) showed an imperfect fit with the data measured at higher leucine concentrations, indicating that the uptake process is more complex than that described by the equation. From the regression analysis, the relative contributions (maximal velocities) of PcDip5 and PcGap1 to α-AAA uptake in the absence of leucine could be calculated as corresponding to 51 and 49%, respectively. This is in good agreement with a more phenomenological analysis in which it is considered that nearly 95% of the α-AAA uptake activity of PcGap1 is already inhibited at 250 μM l-leucine, while the inhibition of PcDip5 at this concentration is, at most, 5% (Fig. 1B and C). This would also yield an estimated 50% contribution of each permease (Fig. 1A). Thus, PcDip5 and PcGap1 seem to make equal contributions to the uptake of α-AAA when each is present at 25 μM. Taking the difference in Km into account, a greater fraction of the α-AAA flux will be mediated by PcDip5 with increasing α-AAA concentrations. Taken together, these data strongly suggest that the uptake of α-AAA by P. chrysogenum involves two transporters, PcGap1 and PcDip5.
Expression of PcDIP5 and PcGAP1 in response to nitrogen source and carbon source.
In fungi, the activity of amino acid permeases is regulated both at the transcriptional and the posttranslational level (20, 37, 38, 40, 41). To analyze the expression of PcDIP5 and PcGAP1 in P. chrysogenum, an RT-PCR method was used. The mycelium was grown in liquid culture with different nitrogen and carbon sources and in penicillin production medium (with lactose as the carbon source and with urea and glutamate as nitrogen sources). After preculturing of P. chrysogenum Wisconsin 54-1255 on YPG medium, germinating conidia were transferred to minimal media containing NH4+ (a rich nitrogen source), urea (a poor nitrogen source), glutamate (a substrate of PcDip5 and PcGap1), serine, or lysine (substrates of PcGap1) as the sole nitrogen source and glucose as the carbon source. After 24 h of incubation, mRNA was isolated and used as a template for semiquantitative RT-PCR (Fig. 6A). Expression of PcDIP5 and PcGAP1 was induced when the mycelium was grown with glutamate, whereas in the presence of NH4+, expression was very low. With urea, PcDIP5 was not expressed, while PcGAP1 showed only low expression. With serine, both genes were expressed at moderate levels. In contrast to the findings for PcDIP5, lysine was as effective as glutamate at inducing the expression of PcGAP1. However, lysine appears not to be a very good nitrogen source, since the cultures grew slowly as mycelial pellets. This morphological behavior is indicative of stress.
FIG. 6.
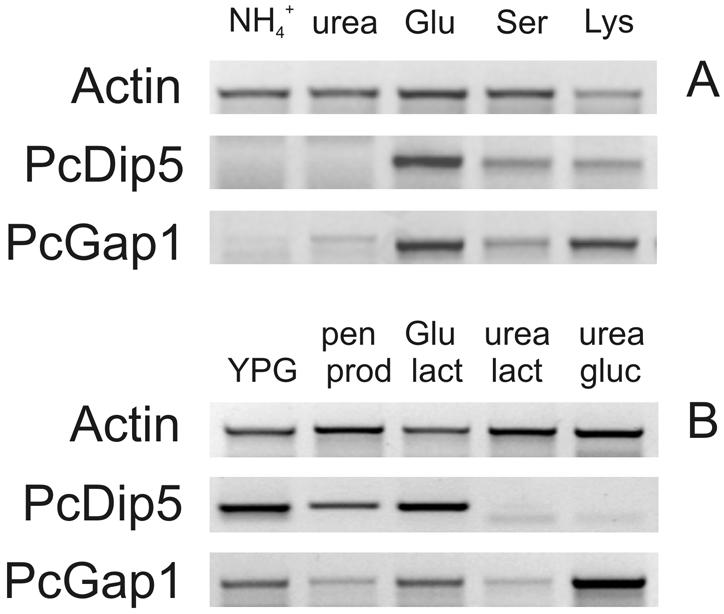
RT-PCR expression analysis of PcDIP5 and PcGAP1 in P. chrysogenum grown under different conditions. Expression of PcDIP5 and PcGAP1 was determined after 24 h of growth on minimal medium containing glucose as the carbon source and different nitrogen sources (A) or on various media as indicated (B). Penicillin production medium (pen prod) contains urea and glutamate as nitrogen and carbon sources, respectively. The other three media are minimal media with lactose (lact) or glucose (gluc) as the sole carbon source and glutamate or urea as the sole nitrogen source. Isolated mRNA was used as a template for semiquantitative RT-PCR. The expression analysis provides relative information for one gene only; analyses for different genes should not be compared.
To test whether PcDIP5 and PcGAP1 are expressed under conditions of penicillin production and/or are subject to carbon catabolite repression, P. chrysogenum Wisconsin 54-1255 was grown for 48 h on either YPG, penicillin production medium containing lactose as the carbon source, or minimal medium with glutamate or urea as the sole nitrogen source and lactose or glucose as the carbon source (Fig. 4B). Under penicillin-producing conditions, PcDIP5 and PcGAP1 were moderately expressed in comparison with the other tested growth conditions. Nevertheless, substantial rates of α-AAA uptake were observed via PcGap1 and PcDip5 under these conditions (see Fig. 1A). The pattern of expression did not change significantly after 72 h of growth (data not shown). Carbon catabolite repression was not observed in any of the media tested. Expression of PcGAP1 was much lower when cells were growing on lactose and urea than on glucose and urea (Fig. 6B, last two lanes). In addition, growth on YPG with glucose as the carbon source did not repress PcGAP1 or PcDIP5 expression. Although nitrogen catabolite repression was observed in the presence of NH4+, PcDIP5 and PcGAP1 seem not to be under the control of carbon catabolite repression. Both transporters are expressed when cells are grown with glutamate or serine as the sole nitrogen source, whereas PcGAP1 is also expressed when urea is used as the sole nitrogen source.
DISCUSSION
When α-AAA is fed to the penicillin-producing mycelium of P. chrysogenum, a higher intracellular α-AAA concentration and a higher penicillin synthesis rate can be observed (13, 19). The exact mechanism of uptake of α-AAA has not been studied before. Here we demonstrate that both the acidic and general amino acid permeases (PcDip5 and PcGap1) contribute to α-AAA uptake in P. chrysogenum when cells are grown under conditions that result in penicillin production. Although the studies with the cloned genes in S. cerevisiae indicate that PcGap1 has a higher affinity for α-AAA, it appears that each system contributes equally to α-AAA uptake by P. chrysogenum when α-AAA present at 25 μM. Taking the kinetic parameters into account, one can predict that the contribution of PcDip5 will increase with the α-AAA concentration and will reach a maximum at about 70 to 77% of total uptake. The transport assays were performed at pH 6.0, but since PcGap1 has a preference for the uncharged form of glutamate (21), the contribution of PcGap1 to α-AAA uptake may increase at lower pHs. Although we cannot exclude the possibility that in addition to PcGap1 and PcDip5, other permeases might be involved in α-AAA uptake, the studies with the S. cerevisiae mutant indicate that at least in yeast, Gap1 and Dip5 are the major permeases for uptake of this molecule. In other fungi also, no other transporters have been identified that could participate in the uptake of negatively charged amino acids.
Feeding of penicillin fermentations with external α-AAA is not a cost-effective process. Therefore, at first sight it appears that the transporters identified would be of little benefit to the fermentation. However, transporters not only function in the uptake of compounds but also prevent the loss of critical nutrients by cells. In this regard, the permeases identified may be important for the retention of intracellular α-AAA. In A. nidulans, two ammonium transporters have been identified, MeaA and MepA, that are required for the retention of intracellular ammonium (30). Surprisingly, MeaA, which exhibits a lower affinity for ammonium (Km, 3 mM), appears more important for retention than the high-affinity ammonium permease MepA (Km, 44 μM). It is not known to what extent losses of intracellular α-AAA in P. chrysogenum to the medium occur during industrial fermentation. However, processes such as fragmentation of the hyphae by agitation or leakage across the membrane could potentially lead to significant losses. PcGap1 and PcDip5 might be involved in reuptake. Our results suggest that PcDIP5 and PcGAP1 are both moderately expressed in shaking flask cultures when penicillin-producing conditions prevail, i.e., when lactose is used as the carbon source and urea and glutamate are nitrogen sources. However, these conditions are not identical to those in industrial fermentation (25, 31), where glucose is fed to the cells in a fed-batch fermentation. Carbon catabolite repression, however, was not observed for PcGAP1 or PcDIP5. On the contrary, the mRNA level of PcGAP1 was much higher when cells were grown in minimal medium with urea as the sole nitrogen source and glucose as the carbon source than in a medium with lactose as the sole carbon source. Also in YPG medium with glucose, significant levels of PcGAP1 and PcDIP5 expression were observed. It should be stressed that the final activities of these permeases also depend on posttranslational regulatory phenomena. For instance, for Gap1 of S. cerevisiae growing with glutamate as the sole nitrogen source, a high mRNA level is observed but the transport activity is low. This has been attributed to the sorting of Gap1 to the vacuole instead of the plasma membrane (11, 38).
In conclusion, externally added α-AAA can be taken up by P. chrysogenum via the acidic amino acid permease PcDip5 and the general amino acid permease PcGap1. Even though transport occurs with relatively low affinity, a significant flux of α-AAA uptake can be detected in mycelia grown under penicillin-producing conditions. These transporters may contribute to the maintenance of a high α-AAA concentration within the cell to allow a high β-lactam production capacity.
Acknowledgments
S. cerevisiae strain M4276 was kindly provided by M. C. Kielland-Brandt (Department of Physiology, Carlsberg Laboratory, Copenhagen, Denmark). We thank Wil Konings for his interest in this work.
This work was supported by the Eurofung, European Community (grant QLK3-CT-1999-00729).
REFERENCES
- 1.Aharonowitz, Y., G. Cohen, and J. F. Martin. 1992. Penicillin and cephalosporin biosynthetic genes: structure, organization, regulation, and evolution. Annu. Rev. Microbiol. 46:461-495. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Andre, B. 1995. An overview of membrane transport proteins in Saccharomyces cerevisiae. Yeast 11:1575-1611. [DOI] [PubMed] [Google Scholar]
- 4.Benko, P. V., T. C. Wood, and I. H. Segel. 1969. Multiplicity and regulation of amino acid transport in Penicillium chrysogenum. Arch. Biochem. Biophys. 129:498-508. [DOI] [PubMed] [Google Scholar]
- 5.Bhattacharjee, J. K. 1985. α-Aminoadipate pathway for the biosynthesis of lysine in lower eukaryotes. Crit. Rev. Microbiol. 12:131-151. [DOI] [PubMed] [Google Scholar]
- 6.Biswas, S., M. Roy, and A. Datta. 2003. N-Acetylglucosamine-inducible CaGAP1 encodes a general amino acid permease which co-ordinates external nitrogen source response and morphogenesis in Candida albicans. Microbiology 149:2597-2608. [DOI] [PubMed] [Google Scholar]
- 7.Brakhage, A. A. 1998. Molecular regulation of β-lactam biosynthesis in filamentous fungi. Microbiol. Mol. Biol. Rev. 62:547-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Breitling, R., O. Sharif, M. L. Hartman, and S. K. Krisans. 2002. Loss of compartmentalization causes misregulation of lysine biosynthesis in peroxisome-deficient yeast cells. Eukaryot. Cell 1:978-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Busch, S., H. B. Bode, A. A. Brakhage, and G. H. Braus. 2003. Impact of the cross-pathway control on the regulation of lysine and penicillin biosynthesis in Aspergillus nidulans. Curr. Genet. 42:209-219. [DOI] [PubMed] [Google Scholar]
- 10.Casqueiro, J., S. Gutierrez, O. Banuelos, M. J. Hijarrubia, and J. F. Martin. 1999. Gene targeting in Penicillium chrysogenum: disruption of the lys2 gene leads to penicillin overproduction. J. Bacteriol. 181:1181-1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, E. J., and C. A. Kaiser. 2002. Amino acids regulate the intracellular trafficking of the general amino acid permease of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 99:14837-14842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friedrich, C. G., and A. L. Demain. 1977. Homocitrate synthase as the crucial site of the lysine effect on penicillin biosynthesis. J. Antibiot. (Tokyo) 30:760-761. [DOI] [PubMed] [Google Scholar]
- 13.Friedrich, C. G., and A. L. Demain. 1978. Uptake and metabolism of α-aminoadipic acid by Penicillium chrysogenum Wis 54-1255. Arch. Microbiol. 119:43-47. [DOI] [PubMed] [Google Scholar]
- 14.Gietz, D., A. St Jean, R. A. Woods, and R. H. Schiestl. 1992. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 20:1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grauslund, M., T. Didion, M. C. Kielland-Brandt, and H. A. Andersen. 1995. BAP2, a gene encoding a permease for branched-chain amino acids in Saccharomyces cerevisiae. Biochim. Biophys. Acta 1269:275-280. [DOI] [PubMed] [Google Scholar]
- 16.Henriksen, C. M., J. Nielsen, and J. Villadsen. 1998. Cyclization of α-aminoadipic acid into the δ-lactam 6-oxo-piperidine-2-carboxylic acid by Penicillium chrysogenum. J. Antibiot. (Tokyo) 51:99-106. [DOI] [PubMed] [Google Scholar]
- 17.Hill, J. E., A. M. Myers, T. J. Koerner, and A. Tzagoloff. 1986. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast 2:163-167. [DOI] [PubMed] [Google Scholar]
- 18.Hillenga, D. J., H. J. Versantvoort, A. J. Driessen, and W. N. Konings. 1996. Basic amino acid transport in plasma membrane vesicles of Penicillium chrysogenum. J. Bacteriol. 178:3991-3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Honlinger, C., and C. P. Kubicek. 1989. Regulation of δ-(l-α-aminoadipyl)-l-cysteinyl-d-valine and isopenicillin N biosynthesis in Penicillium chrysogenum by the α-aminoadipate pool size. FEMS Microbiol. Lett. 53:71-75. [DOI] [PubMed] [Google Scholar]
- 20.Horak, J. 1986. Amino acid transport in eucaryotic microorganisms. Biochim. Biophys. Acta 864:223-256. [DOI] [PubMed] [Google Scholar]
- 21.Hunter, D. R., and I. H. Segel. 1971. Acidic and basic amino acid transport systems of Penicillium chrysogenum. Arch. Biochem. Biophys. 144:168-183. [DOI] [PubMed] [Google Scholar]
- 22.Hunter, D. R., and I. H. Segel. 1973. Control of the general amino acid permease of Penicillium chrysogenum by transinhibition and turnover. Arch. Biochem. Biophys. 154:387-399. [DOI] [PubMed] [Google Scholar]
- 23.Jaklitsch, W. M., W. Hampel, M. Rohr, C. P. Kubicek, and G. Gamerith. 1986. α-Aminoadipate pool concentration and penicillin biosynthesis in strains of Penicillium chrysogenum. Can. J. Microbiol. 32:473-480. [DOI] [PubMed] [Google Scholar]
- 24.Jauniaux, J. C., and M. Grenson. 1990. GAP1, the general amino acid permease gene of Saccharomyces cerevisiae. Nucleotide sequence, protein similarity with the other bakers yeast amino acid permeases, and nitrogen catabolite repression. Eur. J. Biochem. 190:39-44. [DOI] [PubMed] [Google Scholar]
- 25.Jorgensen, H., J. Nielsen, J. Villadsen, and H. Mollgaard. 1995. Analysis of penicillin V biosynthesis during fed-batch cultivations with a high-yielding strain of Penicillium chrysogenum. Appl. Microbiol. Biotechnol. 43:123-130. [DOI] [PubMed] [Google Scholar]
- 26.Koo, K., and W. D. Stuart. 1991. Sequence and structure of mtr, an amino acid transport gene of Neurospora crassa. Genome 34:644-651. [DOI] [PubMed] [Google Scholar]
- 27.Lara, F., M. R. del Carmen, G. Vazquez, and S. Sanchez. 1982. Induction of penicillin biosynthesis by l-glutamate in Penicillium chrysogenum. Biochem. Biophys. Res. Commun. 105:172-178. [DOI] [PubMed] [Google Scholar]
- 28.Margolis-Clark, E., I. Hunt, S. Espinosa, and B. J. Bowman. 2001. Identification of the gene at the pmg locus, encoding system II, the general amino acid transporter in Neurospora crassa. Fungal Genet. Biol. 33:127-135. [DOI] [PubMed] [Google Scholar]
- 29.Martin, J. F. 1998. New aspects of genes and enzymes for β-lactam antibiotic biosynthesis. Appl. Microbiol. Biotechnol. 50:1-15. [DOI] [PubMed] [Google Scholar]
- 30.Monahan, B. J., J. A. Fraser, M. J. Hynes, and M. A. Davis. 2002. Isolation and characterization of two ammonium permease genes, meaA and mepA, from Aspergillus nidulans. Eukaryot. Cell 1:85-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nielsen, J. 1994. Physiological engineering aspects of Penicillium chrysogenum. Ph.D. thesis. Center for Process Biotechnology, Lyngby, Denmark.
- 32.Regenberg, B., L. During-Olsen, M. C. Kielland-Brandt, and S. Holmberg. 1999. Substrate specificity and gene expression of the amino-acid permeases in Saccharomyces cerevisiae. Curr. Genet. 36:317-328. [DOI] [PubMed] [Google Scholar]
- 33.Regenberg, B., S. Holmberg, L. D. Olsen, and M. C. Kielland-Brandt. 1998. Dip5p mediates high-affinity and high-capacity transport of l-glutamate and l-aspartate in Saccharomyces cerevisiae. Curr. Genet. 33:171-177. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 35.Seaston, A., C. Inkson, and A. A. Eddy. 1973. The absorption of protons with specific amino acids and carbohydrates by yeast. Biochem. J. 134:1031-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sewell, A. K., F. Yokoya, W. Yu, T. Miyagawa, T. Murayama, and D. R. Winge. 1995. Mutated yeast heat shock transcription factor exhibits elevated basal transcriptional activation and confers metal resistance. J. Biol. Chem. 270:25079-25086. [DOI] [PubMed] [Google Scholar]
- 37.Sophianopoulou, V., and G. Diallinas. 1995. Amino acid transporters of lower eukaryotes: regulation, structure and topogenesis. FEMS Microbiol. Rev. 16:53-75. [DOI] [PubMed] [Google Scholar]
- 38.Stanbrough, M., and B. Magasanik. 1995. Transcriptional and posttranslational regulation of the general amino acid permease of Saccharomyces cerevisiae. J. Bacteriol. 177:94-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stuart, W. D., K. Koo, and S. J. Vollmer. 1988. Cloning of mtr, an amino acid transport gene of Neurospora crassa. Genome 30:198-203. [DOI] [PubMed] [Google Scholar]
- 40.Tazebay, U. H., V. Sophianopoulou, B. Cubero, C. Scazzocchio, and G. Diallinas. 1995. Post-transcriptional control and kinetic characterization of proline transport in germinating conidiospores of Aspergillus nidulans. FEMS Microbiol. Lett. 132:27-37. [DOI] [PubMed] [Google Scholar]
- 41.Tazebay, U. H., V. Sophianopoulou, C. Scazzocchio, and G. Diallinas. 1997. The gene encoding the major proline transporter of Aspergillus nidulans is upregulated during conidiospore germination and in response to proline induction and amino acid starvation. Mol. Microbiol. 24:105-117. [DOI] [PubMed] [Google Scholar]
- 42.Trip, H., M. E. Evers, W. N. Konings, and A. J. Driessen. 2002. Cloning and characterization of an aromatic amino acid and leucine permease of Penicillium chrysogenum. Biochim. Biophys. Acta 1565:73-80. [DOI] [PubMed] [Google Scholar]
- 43.van de Kamp, M., A. J. Driessen, and W. N. Konings. 1999. Compartmentalization and transport in β-lactam antibiotic biosynthesis by filamentous fungi. Antonie Leeuwenhoek 75:41-78. [DOI] [PubMed] [Google Scholar]
- 44.van der Lende, T. R., M. van de Kamp, M. Berg, K. Sjollema, R. A. Bovenberg, M. Veenhuis, W. N. Konings, and A. J. Driessen. 2002. δ-(l-α-Aminoadipyl)-l-cysteinyl-d-valine synthetase, that mediates the first committed step in penicillin biosynthesis, is a cytosolic enzyme. Fungal Genet. Biol. 37:49-55. [DOI] [PubMed] [Google Scholar]
- 45.Zaret, K. S., and F. Sherman. 1982. DNA sequence required for efficient transcription termination in yeast. Cell 28:563-573. [DOI] [PubMed] [Google Scholar]