Abstract
Metformin is a widely used first-line antidiabetic drug that has been shown to protect against a variety of specific diseases in addition to diabetes, including cardiovascular disorders, polycystic ovary syndrome, and cancer. However, the precise mechanisms underlying the diverse therapeutic effects of metformin remain elusive. Here, we report that transforming growth factor-β1 (TGF-β1), which is involved in the pathogenesis of numerous diseases, is a novel target of metformin. Using a surface plasmon resonance-based assay, we identified the direct binding of metformin to TGF-β1 and found that metformin inhibits [125I]-TGF-β1 binding to its receptor. Furthermore, based on molecular docking and molecular dynamics simulations, metformin was predicted to interact with TGF-β1 at its receptor-binding domain. Single-molecule force spectroscopy revealed that metformin reduces the binding probability but not the binding force of TGF-β1 to its type II receptor. Consequently, metformin suppresses type II TGF-β1 receptor dimerization upon exposure to TGF-β1, which is essential for downstream signal transduction. Thus, our results indicate that metformin is a novel TGF-β suppressor with therapeutic potential for numerous diseases in which TGF-β1 hyperfunction is indicated.
Metformin was originally derived from the French lilac Galega officinalis, and it is currently a widely prescribed biguanide used as a first-line antidiabetic drug. Metformin is safe and effective in the treatment of diabetes and does not induce hypoglycemia. Beyond its known blood glucose lowering effects, metformin has been shown to elicit beneficial effects on cardiovascular diseases1, polycystic ovary syndrome2, diabetic nephropathy3, and cancer4. However, the mechanisms underlying the pleiotropic effects of metformin remain elusive. Transforming growth factor-β1 (TGF-β1) is involved in the progression of many diseases, including cancer and fibrotic, cardiovascular, and immunological diseases. It is associated with fibrosis, epithelial-to-mesenchymal transition (EMT) and inflammation which are important pathological changes in the aforementioned diseases. Our previous study revealed that metformin inhibits cardiac fibrosis by inhibiting the TGF-β1-Smad3 signaling pathway5, which raises an intriguing possibility that metformin may prevent organ fibrosis via targeting TGF-β1 signaling. To date, the molecular and pharmacological mechanisms underlying metformin’s ability to inhibit the TGF-β1 signaling pathway are unknown, and identifying these mechanisms represents an important challenge that may elucidate new molecular targets of metformin and promote target-based novel strategies against many diseases beyond diabetes in which TGF-β1 signaling malfunctions are indicated.
The TGF-β1 signaling pathway requires two cell-surface serine/threonine kinase receptors, type II (TβRII) and type I (TβRI) TGF-β receptors. In general, TGF-β1 binds to TβRII, which recruits TβRI to form a heteromeric complex of TβRI-TβRII. The formation of this complex leads to the phosphorylation of TβRI and subsequent phosphorylation of receptor-regulated Smads (R-Smads, i.e., Smad2, Smad3, etc), which bind a co-Smad (Smad4). The R-Smad/co-Smad complexes translocate and accumulate in the nucleus and subsequently regulate target genes6. In our previous study, individual GFP-tagged TβRII molecules were imaged on the cell membrane using total internal reflection fluorescence microscopy (TIRFM) to study receptor activation. We demonstrated that TβRII occurs as a monomer in the resting state, and dimerizes upon TGF-β1 stimulation7, which supports the idea that the receptor dimerization is essential for receptor activation.
Here, we identified a direct interaction between metformin and TGF-β1 using a surface plasmon resonance (SPR)-based assay, and found that metformin inhibits [125I]-TGF-β1 binding to its receptor. We modeled the interaction mode between metformin and TGF-β1 using molecular docking and molecular dynamics simulations. We further investigated the binding force between TGF-β1 and TβRII using atomic force microscopy (AFM) and the dimerization of TβRII upon TGF-β1 stimulation using single molecule fluorescence imaging. This study revealed a novel mechanism underlying the inhibition of the TGF-β1 signaling pathway by metformin that explains the beneficial effects of metformin against organ fibrosis and cancer progression.
Results
Metformin directly interacts with TGF-β1, inhibits binding to its receptor, and attenuates downstream signaling
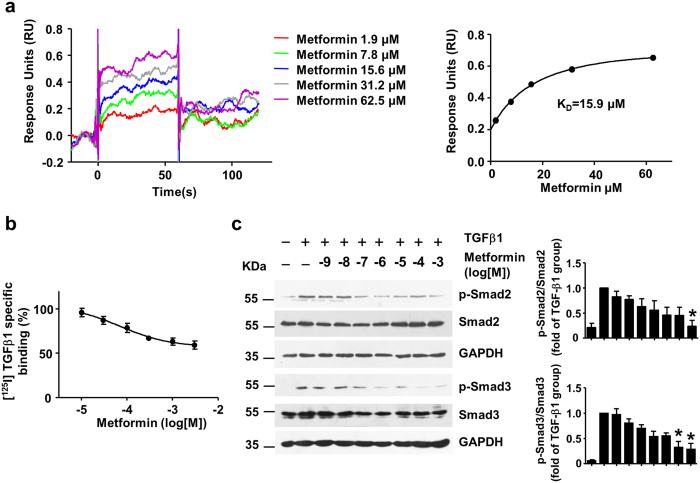
The SPR-based assay suggested that the binding of metformin to TGF-β1 occurred with a KD value of 15.9 μM (Fig. 1a). Next, we measured the effect of metformin on the specific binding of [125I]-TGF-β1 to 3T3 fibroblasts. As shown in Fig. 1b, metformin inhibited [125I]-TGF-β1 binding to its receptor in 3T3 fibroblasts in a dose-dependent manner (log[IC50] = −4.16 ± 0.53). In contrast, HCT 116 cells that harbor a loss of function mutation in TβRII displayed negligible binding of [125I]-TGF-β1. And metformin has little effect on the [125I]-TGF-β1 binding to HCT 116 cells, suggesting that metformin inhibits the binding of [125I]-TGF-β1 to TβRII (see Supplementary Fig. S1). Consistent with these results, western blotting analyses indicated that metformin decreased TGF-β1-induced phosphorylation of Smad2 and Smad3 (pSmad2 and pSmad3) in 3T3 fibroblasts (Fig. 1c, Kruskal–Wallis ANOVA combined with post-hoc Dunn’s multiple comparison test, TGF-β1+ Metformin 0.1 mM vs. TGF-β1, pSmad3, P* = 0.016; TGF-β1+ Metformin 1 mM vs. TGF-β1, pSmad2, P* = 0.041, pSmad3, P* = 0.017) and H9C2 cells (see Supplementary Fig. S2). Notably, metformin didn’t inhibit TGF-β1 induced Smad2/3 phosphorylation until 30 minutes after treatment (see Supplementary Fig. S3).
Figure 1.
Metformin interacts with TGF-β1 to block its binding with receptors and attenuates downstream signaling (a) Sensorgrams for the binding of metformin and TGF-β1. TGF-β1 was covalently coupled to a CM5 chip, and metformin was injected in a two-fold dilution concentration series ranging from 62.5 μM to 1.9 μM. The steady-state values were calculated from the sensorgrams and plotted against the concentration. The data were fit to a single site binding model to calculate the KD. (b) Metformin inhibited [125I]-TGF-β1 receptor binding to 3T3 mouse fibroblasts. The results are expressed as the percentage of specific binding in the absence of metformin (n = 4). (c) Metformin and TGF-β1 (0.5 ng/mL) were premixed for 2 h and then 3T3 cells were treated with the mixture for 30 minutes. Western blot analysis and quantification of phosphorylated-Smad2 (p-Smad2), Smad2, p-Smad3, Smad3 and GAPDH were performed. Data are mean ± SEM from 4 independent experiments. Kruskal–Wallis ANOVA combined with post-hoc Dunn’s multiple comparison test (two tailed) was performed. *P < 0.05 vs. TGF-β1 group.
Metformin attenuates TGF-β1 downstream signaling through AMPK independent pathway
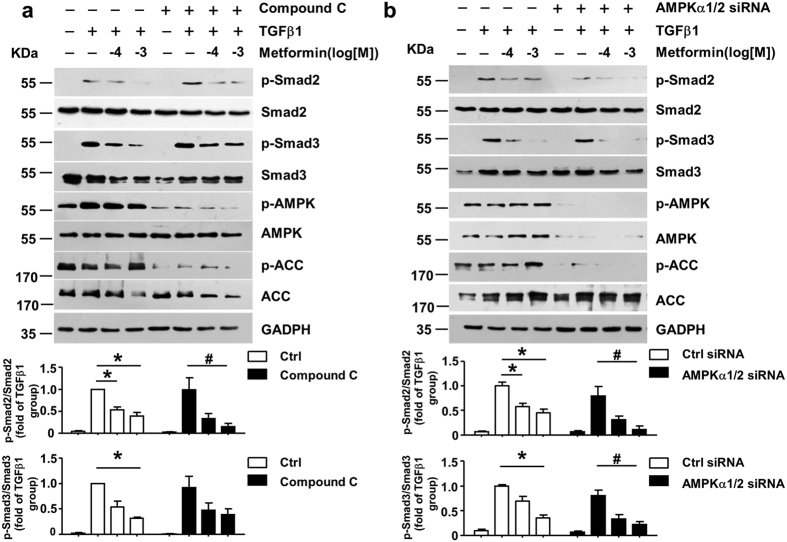
Metformin is a well-known AMP-activated protein kinase (AMPK) activator. Phosphorylation of AMPK and Acetyl-CoA carboxylase (ACC), an AMPK target, were both decreased in AMPK inhibitor Compound C pretreatment group (Fig. 2a) and AMPKα1/2 siRNA transfected group (Fig. 2b), demonstrating the loss of AMPK activity. Notably, metformin still decreased p-Smad2/3 induced by TGF-β1, to a degree similar with that in the vehicle treatment group (Fig. 2a,b). This result indicated that metformin attenuates TGF-β1-induced p-Smad2/3 independent of AMPK pathway.
Figure 2.
Metformin attenuates TGF-β1 downstream signaling through AMPK independent pathway (a) AMPK inhibitor, Compound C (5 μM) were pretreated in 3T3 cells for 4 h. n = 5. (b) Negative control siRNA and AMPKα1/2 siRNA (80 nM) were transfected in 3T3 cells for 48 h. n = 6. Next, metformin and TGF-β1 (0.5 ng/mL) were premixed for 2 h and then the 3T3 cells were treated with the mixture for 30 minutes. Western blot analysis and quantification of phosphorylated-Smad2 (p-Smad2), Smad2, p-Smad3, Smad3, p-AMPK, AMPK, p-ACC, ACC and GAPDH were performed. Data are mean ± SEM. Kruskal–Wallis ANOVA combined with post-hoc Dunn’s multiple comparison test (two tailed) was performed. *P < 0.05 vs. TGF-β1 group, #P < 0.05 vs. TGF-β1 plus compound C or TGF-β1 plus AMPKα1/2 siRNA group.
Metformin antagonizes TGF-β1 signaling via a direct interaction with TGF-β1
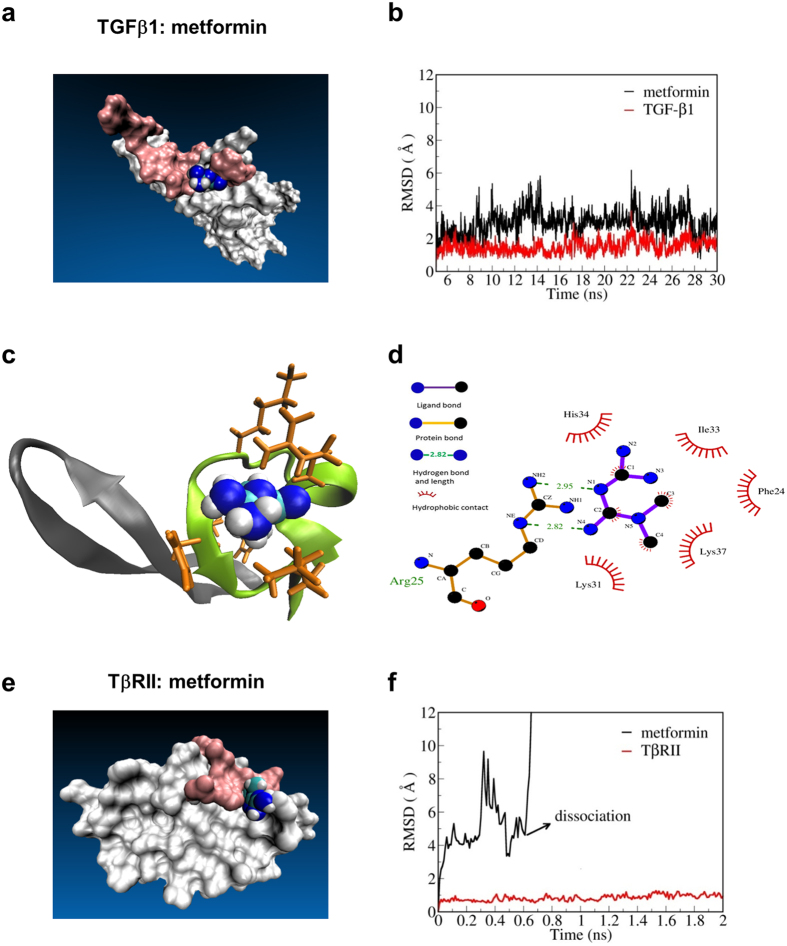
To further assess the potential binding of metformin to TGF-β1 and its receptor, we performed molecular docking and molecular dynamics (MD) simulation. The binding of metformin to TGF-β1 was stable, determined by the root mean square deviation (RMSD) of metformin relative to TGF-β1 (Fig. 3a,b). Metformin tended to bind in a cave-like structure consisting of the β-strand1 and β-strand2 of TGF-β1 (Fig. 3c). Importantly, this site was partially overlaid with the binding interface of TβRII. The residues in direct contact with TGF-β1 are depicted in Fig. 3d. The binding of metformin was largely attributed to the shape complementarity and hydrogen bond interaction between the guanidine group and Arg25. In addition, the nonpolar components (methyl groups) of metformin were nestled in the hydrophobic bottom of the cave. Thus, the binding stability of metformin to TGF-β1 was further evaluated according to the binding free energy using Molecular Mechanics/Poisson Boltzmann Surface Area methods. The estimated value of the binding free energy (ΔGbind) was −68.50 kJ/mol, which was considered to be sufficiently strong for such a small compound. However, metformin could not stably bind to the putative binding site of TβRII(extracellular domain) (Fig. 3e). The molecule quickly diffused away from the initial binding site during the molecular simulation (Fig. 3f).
Figure 3.
Molecular docking and molecular dynamics simulations for TβRII :metformin and TGF-β1:metformin (a) Structure of the TGF-β1:metformin complex after relaxation. The surface region of TGF-β1 recognized by type II TGF-β receptors (TβRII) is shown in pink. (b) Root-mean-square deviation (RMSD) of TGF-β1 and metformin relative to TGF-β1 during the last 25-ns trajectory. (c) Binding site of metformin consists of the β-sheet1 and β-sheet2 of TGF-β1. (d) Residues in direct contact with metformin (depicted using the LIGPLOT program with a cutoff of 3.9 Å). (e) Structure of the TβRII: metformin complex after relaxation. The surface region of TβRII recognized by TGF-β1 is shown in pink. (f) RMSD of TβRII and metformin relative to TβRII.
Metformin inhibits the binding probability of TGF-β1 and TβRII but has no effect on the binding force of TGF-β1 and TβRII
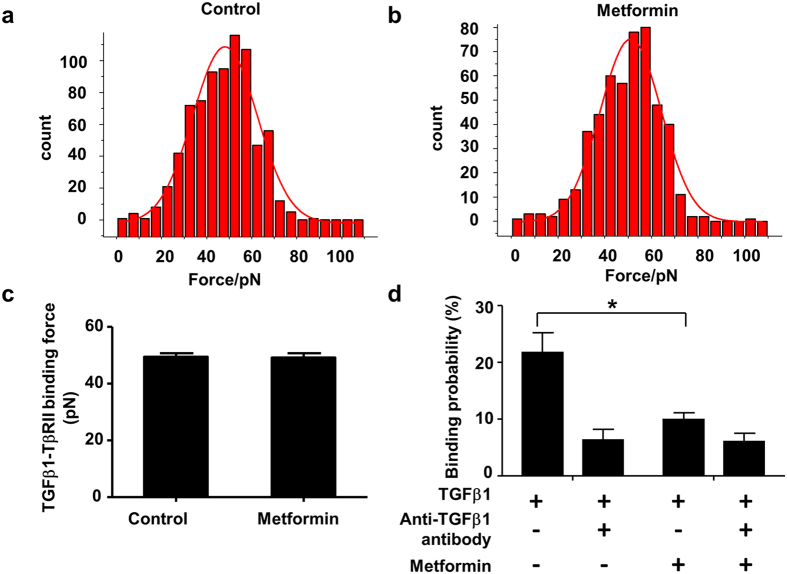
To determine how metformin inhibits the binding of TGF-β1 to its receptor, we next performed TGF-β1–TβRII binding force measurements on live cells using AFM-based single-molecule force spectroscopy. Blocking experiments were performed by the addition of free TGF-β1 monoclonal antibodies. As shown in Fig. 4a,b, the force distribution histogram displayed a single maximum by a Gaussian fit and the binding probability was less than 30%, indicating that the single molecule forces were measured. In the cells treated with metformin (50 μM), similar binding forces (measured as the averaged histogram peak value) were observed for TGF-β1 with TβRII on the cell surface as the control (Mann-Whitney U test with exact method, control vs. metformin: 49.5 ± 1.3 vs.49.3 ± 1.4 pN, P > 0.05, Fig. 4c). However, metformin decreased the binding probabilities from 21.7 ± 3.5% to 9.9 ± 1.2%, which was similar to the result of the TGF-β1 antibody treatment (6.4 ± 1.9%, Fig. 4d).
Figure 4. Metformin inhibits the binding probability but not binding force of TGF-β1 and TβRII.
TGF-β1–TβRII binding force measurements were obtained with TGF-β1-modified atomic force microscopy (AFM) tips on HeLa cells, which express TβRII. Blocking experiments were performed by the addition of free TGF-β1 monoclonal antibodies into the solution. (a) Histograms of binding forces of TGF-β1/TβRII in the untreated cells and (b) the cells treated with 50 μM metformin. (c) Binding forces of TGF-β1/TβRII in cells treated with/without 50 μM metformin. (d) Binding probability of TGF-β1and TβRII when the cells were treated with metformin and/or the blocking reagent (anti-TGF-β1 antibody). Data were expressed as the mean ± SEM from 3 independent experiments. Mann-Whitney U test (one tailed) with exact method was used. *P < 0.05.
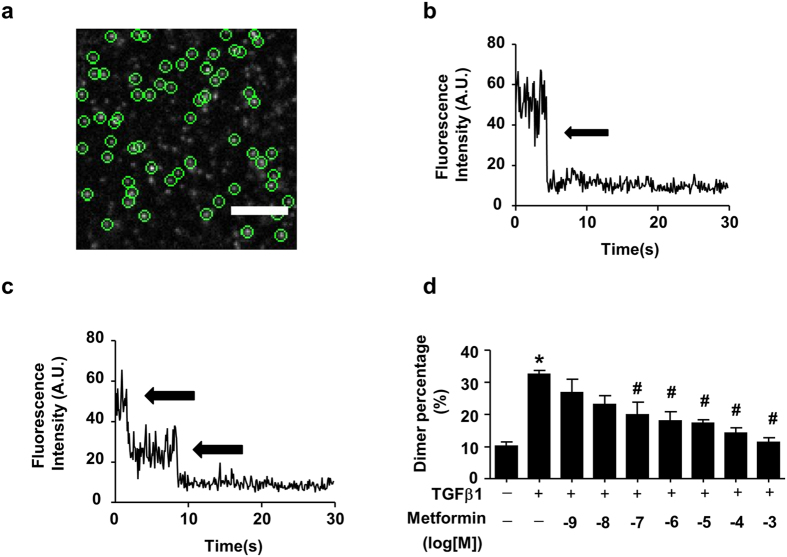
Metformin inhibits TGF-β1-induced TβRII dimerization
Because TGF-β1-induced TβRII dimerization is a consequence of TGF-β ligand–receptor interaction and essential for receptor activation7, we next determined the effect of metformin on the formation of ligand-induced TβRII dimers. By analyzing the photobleaching traces (Fig. 5a), we found that 88.8% (778 out of 876 spots from 14 fixed cells) of individual TβRII-GFP molecules were monomers because they bleached in one step (Fig. 5b), 10.7% (94 of 876 spots) were dimers because they bleached in two steps (Fig. 5c), and that 0.5% (4 of 876) bleached in three steps. Following the TGF-β1 stimulation, 67.7% (529 of 781 spots) bleached in one step as monomers, 31.6% (247 of 781spots from sixteen fixed cells) bleached in two steps as dimers and 0.6% (5 of 781) bleached in three steps. As shown in Fig. 5d, metformin inhibited the percentage of dimers induced by TGF-β1 in a dose-dependent manner.
Figure 5. Metformin inhibits TGF-β1-induced TβRII dimerization.
(a) Typical single-molecule image of TβRII-GFP on the HeLa cell membrane. After transfection with TβRII-GFP for 4 h, HeLa cells were imaged using total internal reflection fluorescence microscopy (TIRFM). The diffraction-limited spots (5 × 5 pixel regions) enclosed with green circles represent the signals from individual TβRII-GFP molecules, and they were chosen for the intensity analysis. Scale bar, 5 μm. (b,c) Two representative time course graphs of GFP emissions after background correction demonstrating one- and two-step bleaching, respectively. The arrows indicate the bleaching steps. The individual TβRII-GFP molecules were monomers when they were bleached in one step (b) and dimers when they were bleached in two steps (c). (d) Metformin inhibited TGF-β1-induced TβRII dimerization as shown by single-molecule imaging. Fraction of two-step bleaching events for TβRII-GFP molecules (counted spots were set as 100%) was represented as the dimer percentage. Prior to single-molecule fluorescence imaging, metformin and TGF-β1 (10 ng/mL) were premixed for 2 h, and HeLa cells were then treated with the mixture for 15 min at 37 °C. The data were presented as the mean ± SEM (n = 5–16). ANOVA combined with Tukey’s post-hoc test (two tailed) was used. *P < 0.05 vs. control group, #P < 0.05 vs. TGF-β1 group.
Discussion
In the present study, we demonstrated that metformin antagonized TGF-β1 signaling by interacting with the TGF-β1 ligand, thereby blocking the binding of TGF-β1 to TβRII and resulting in decreased downstream signaling. This finding provides new indications for metformin, including numerous TGF-β1 hyperfunction-associated diseases. Based on the interaction between metformin and TGF-β1, new compounds with similar properties could be further developed.
It is generally accepted that metformin acts via the activation of AMPK and the inhibition of mitochondrial respiratory-chain complex 18. Our previous study has shown that metformin inhibits TGF-β1 induced collagen synthesis independent of AMPK activation5. Here, we further discovered that metformin antagonizes TGF-β1 signaling by directly binding to TGF-β1 which is independent of AMPK activation. Consistent with the well-established role of TGF-β1 in the exacerbation of fibrosis9, our previous study and other studies have shown that metformin treatment attenuates cardiac fibrosis5, liver fibrosis10 and renal fibrosis11. In addition, metformin has been shown to inhibit TGF-β1-induced EMT which plays a key role in carcinoma progression and organ fibrosis12,13. Moreover, clinical trials have suggested that metformin is associated with decreased cancer risk and improved prognosis in cancer patients14,15. These findings support the idea that metformin exerts a protective effect against organ fibrosis and malignant tumor progression by blocking TGF-β1.
In addition to fibrosis and tumors, TGF-β is involved in numerous other diseases. Thus, targeting the TGF-β signaling pathway has become attractive for drug development. Currently, therapeutic strategies against the TGF-β family include three approaches: 1) inhibition at the translational level using antisense oligonucleotides, 2) inhibition of the ligand-receptor interaction using ligand traps and anti-receptor monoclonal antibodies, and 3) inhibition of the receptor-mediated signaling cascade using inhibitors and aptamers of TGF-β receptor kinases9,16. However, these approaches have specific challenges that limit their application, such as the limited ability of an antisense oligonucleotides and monoclonal antibodies to reach the targeted tissue17,18. In contrast, metformin is a small molecule compound that can easily reach the targeted tissue. Inhibitors of TGF-β receptor kinases have side effects that occur through the potential cross-inhibition of other kinases9. Conversely, metformin has been shown to be safe and have fewer side effects over decades of use. In addition, metformin has beneficial effects beyond targeting TGF-β1 and based on the interaction mode between metformin and TGF-β1, additional compounds can be developed to target TGF-β with higher specificity and potency.
In summary, our study identified metformin as a novel TGF-β1 suppressor, and this action underlies the pleiotropic effects of the drug. This finding strongly supports the clinical use of metformin as a treatment for numerous diseases beyond diabetes where TGF-β1 signaling malfunctions are indicated. In addition, our study provides insights that can be used in the development of new compounds targeting TGF-β1.
Methods
Reagents
Metformin was purchased from Sigma-Aldrich (St. Louis, MO, USA). Compound C was purchased from EMD Millipore Corp. (Billerica, MA, USA). Recombinant human TGF-β1 was purchased from PeproTech (Rocky Hill, NJ, USA). [125I]-TGF-β1 was purchased from Perkin Elmer Inc. (Waltham, MA, USA).
Equipment and settings
For single molecule fluorescence imaging, a custom-built TIRFM system was used as previously described7. The experiment was performed on TIRFM with 100X/1.45NA Plan Apochromat TIR objective (Olympus, Japan) and a 14-bit back-illuminated electron-multiplying charge-coupled device camera (Andor iXon DU-897 BV). Imaging was performed at room temperature. GFP was excited at 488-nm by an argon laser (Melles Griot,Carlsbad, CA, USA) with the power of 1 mW measured after the laser passing through the objective. Movies of 200–300 frames were acquired for each sample at a frame rate of 10 Hz. Sequences of images were stored directly to a computer hard drive for subsequent analysis by IQ live cell imaging software (Andor Technology, BT, UK).
SPR spectroscopy
Experiments were performed at 25 °C using a Biacore T200 and the data were analyzed using Biacore T200 evaluation software 2.0 (GE Healthcare, Stockholm, Sweden). TGF-β1 was covalently coupled to a CM5 chip (GE Healthcare) and metformin was injected in a two-fold dilution concentration series ranging from 62.5 μM to 1.9 μM. The steady-state values were calculated from the sensorgrams and plotted against the concentrations. The data were fit to a single site binding model to calculate the KD.
TGF-β1 binding assay
The binding experiments were performed as previously described19. Briefly, 3T3 cells (American Type Culture Collection, ATCC® CRL-2752™) or HCT 116 cells (American Type Culture Collection, ATCC® CRL-247™) were seeded onto a 24-well-plate and cultured in DMEM supplemented with 10% Fetal Bovine Serum and antibiotics (100 U/mL penicillin-streptomycin). When cells were at a near-confluent stage, 50 pM [125I]-TGF-β1 with or without different concentrations of metformin were added. After 4 h at 4 °C, the medium was removed and cells were washed five times with ice-cold binding buffer (50 mM HEPES, 128 mM NaCl, 5 mM KCl, 5 mM MgSO4, and 1.2 mM CaCl2). The cells were then solubilized using binding buffer containing 1%Triton X-100 and the radioactivity was measured. Non-specific binding was determined in the presence of unlabeled TGF-β1 (10 nM).
Western blotting analysis
Western blotting analyses were performed using specific antibodies as previously described5. Total proteins were extracted from 3T3 cells or H9C2 cells (American Type Culture Collection, ATCC® CRL-1446™) by use of RIPA buffer (6.5 mM Tris, pH 7.4, 15 mM NaCl, 1 mM EDTA, 0.1% SDS, 0.25% sodium deoxycholate, 1% NP-40). Bicinchoninic acid reagents were used to measure the protein concentration. Equal amounts of proteins were separated by SDS-PAGE and transferred to nitrocellulose filter membranes. Phospho-Smad2 Ser465/467 (3108S), Smad2 (5339S), phospho-Smad3 Ser423/425 (9520S), Smad3 (9513S), phospho-AMPKα Thr172 (2531S), AMPKα (2532S), phosphor-ACC Ser79 (3661S), and ACC (3662S) antibodies were purchased from Cell Signaling Technology (Danvers, MA, USA). GAPDH antibodies were purchased from Santa Cruz Biotechnology (sc-47724, Santa Cruz, CA, USA). The blots were immunoreacted with primary antibodies (4 °C, 16 h) and secondary antibodies conjugated with horseradish peroxidase (room temperature, 1 h). Protein bands were visualized by enhanced chemiluminescence detection and the intensity was quantified by use of Image-J software.
siRNA knockdown
3T3 fibroblasts at 30% confluence were transfected with AMPKα1/2 siRNA (sc-45313, Santa Cruz Biotechnology, Santa Cruz, CA, USA), or negative control siRNA (1027281, QIAGEN, Valencia, CA, USA) at 80 nM with HiPerFect transfection reagent (301705, QIAGEN, Valencia, CA, USA). Experiments were performed with these cells at 48 h post-transfection.
Theoretical study: molecular docking and molecular dynamics simulation
The geometry structure of metformin was optimized with Hartree-Forck methods at 6–31 + G* level of theory. The crystal structures of TGF-β1 and the extracellular domain of TβRII, were retrieved from the PDB archives (3KFD)20. Autodock4.2 suite21 was first applied to predict the preferential binding poses of ligand (metformin) in both TGF-β1 and TβRII. Then the structure of both TGF-β1 and TβRII bound with metformin were obtained for further evaluation by MD simulation. Amber99SB-ILDN forcefield22 for protein and General Amber force field23 for ligand was used. The charge parameters of ligand were taken from restrained electrostatic potential calculation24. The protein-ligand complex was solvated with TIP3P water. Sodium and Chloride ions were added to neutralize the system. All simulations were carried out with the GROMACS4.6.1 packages25 and were run in NPT ensemble. The temperature (T = 300k) and pressure (p = 1atm) was kept constant using velocity scaling methods and Berendsen barostat methods, respectively. Based on the results of simulation, Molecular Mechanics/Poisson Boltzmann Surface Area methods26 was used to estimate the binding free energy of metformin on protein.
Single molecule fluorescence imaging
Hela cells (American Type Culture Collection, ATCC® CCL-2™) were transfected with TβRII-GFP plasmid for 4 h. Prior to the single-molecule fluorescence imaging, the cells were treated with TGF-β1 (10 ng/ml for 15 min) and different concentrations of metformin and then washed twice and fixed. The single-molecule fluorescence intensity and photobleaching steps were also analyzed as previously reported7.
AFM tips preparation and AFM force measurements
TGF-β1-modified AFM tips (type: NP-10, Bruker, Santa Barbara, CA, USA) were prepared as previously reported27. Hela cells were transfected with the TβRII-GFP plasmid for 24 h, and the force measurements were performed on a PicoSPM II system with a PicoScan 3000 controller and a large scanner (Agilent, Santa Clara, CA, USA). The AFM scanner was mounted on an inverted fluorescence microscope (Olympus IX71, Japan). The fluorescent protein-labeled cells were used to guide the AFM tips on the cell expressing TβRII. All of the force curves were measured with the contact mode at room temperature using a soft cantilever (0.06 N m−1). The loading rate of the force measurements was 1.0 × 104 pN/s. The force curves were recorded using PicoScan 5 software (Molecular Imaging, Tempe, AZ) and analyzed using a user-defined program in MATLAB (MathWorks Corp., Beijing, China).
Statistical Analysis
Data were expressed as the means ± SEM from at least 3 independent experiments. In parametric data, Student’s t test or ANOVA combined with Tukey’s post-hoc test was used to analyze the differences among groups if data were determined to be normal distribution by K-S test. For non-parametric data, Mann-Whitney U test with exact method was used to analyze the differences between two groups. A Kruskal–Wallis ANOVA combined with post-hoc Dunn’s multiple comparison test was performed when more than 2 groups were evaluated. A p-value < 0.05 was considered statistically significant.
Additional Information
How to cite this article: Xiao, H. et al. Metformin is a novel suppressor for transforming growth factor (TGF)-β1. Sci. Rep. 6, 28597; doi: 10.1038/srep28597 (2016).
Supplementary Material
Acknowledgments
This work was supported by the National Key Basic Research Program of People’s Republic of China (2013CB933701) and the Natural Science Foundation of China (NSFC, 81300067, 81530009). We thank Prof. Yue Feng from Emory University School of Medicine for the manuscript editing.
Footnotes
Author Contributions H.X. designed the experiment, collected and analyzed data, and wrote the manuscript. Z.X. and M.Z. collected and analyzed data and edited the manuscript. J.Z., Y.F., J.L., R.C. and J.S. collected and analyzed data. J.W. and Z.L. edited the manuscript. X.F. and J.L. designed the experiment and edited the manuscript. All authors reviewed the manuscript. Y.Z. is the guarantor of this work and takes responsibility for the contents of the article.
References
- El Messaoudi S., Rongen G. A. & Riksen N. P. Metformin therapy in diabetes: the role of cardioprotection. Curr Atheroscler Rep. 15, 314 (2013). [DOI] [PubMed] [Google Scholar]
- Tang T. & Balen A. H. Use of metformin for women with polycystic ovary syndrome. Hum Reprod Update. 19, 1 (2013). [DOI] [PubMed] [Google Scholar]
- Nasri H. et al. Bright renoprotective properties of metformin: beyond blood glucose regulatory effects. Iran J Kidney Dis. 7, 423–428 (2013). [PubMed] [Google Scholar]
- Leone A., Di Gennaro E., Bruzzese F., Avallone A. & Budillon A. New perspective for an old antidiabetic drug: metformin as anticancer agent. Cancer Treat Res. 159, 355–376 (2014). [DOI] [PubMed] [Google Scholar]
- Xiao H. et al. Metformin attenuates cardiac fibrosis by inhibiting the TGFbeta1-Smad3 signalling pathway. Cardiovasc Res. 87, 504–513 (2010). [DOI] [PubMed] [Google Scholar]
- Moustakas A. & Heldin C. H. The regulation of TGFbeta signal transduction. Development. 136, 3699–3714 (2009). [DOI] [PubMed] [Google Scholar]
- Zhang W. et al. Single-molecule imaging reveals transforming growth factor-beta-induced type II receptor dimerization. Proc Natl Acad Sci USA 106, 15679–15683 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foretz M., Guigas B., Bertrand L., Pollak M. & Viollet B. Metformin: from mechanisms of action to therapies. Cell Metab. 20, 953–966 (2014). [DOI] [PubMed] [Google Scholar]
- Akhurst R. J. & Hata A. Targeting the TGFbeta signalling pathway in disease. Nat Rev Drug Discov. 11, 790–811 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvnjak M. et al. Therapy of nonalcoholic fatty liver disease: current status. J Physiol Pharmacol. 60 Suppl 7, 57–66 (2009). [PubMed] [Google Scholar]
- Cavaglieri R. C., Day R. T., Feliers D. & Abboud H. E. Metformin prevents renal interstitial fibrosis in mice with unilateral ureteral obstruction. Mol Cell Endocrinol. 412, 116–122 (2015). [DOI] [PubMed] [Google Scholar]
- Cufi S. et al. Metformin against TGFbeta-induced epithelial-to-mesenchymal transition (EMT): from cancer stem cells to aging-associated fibrosis. Cell Cycle. 9, 4461–4468 (2010). [DOI] [PubMed] [Google Scholar]
- Thiery J. P., Acloque H., Huang R. Y. & Nieto M. A. Epithelial-mesenchymal transitions in development and disease. Cell. 139, 871–890 (2009). [DOI] [PubMed] [Google Scholar]
- Soranna D. et al. Cancer risk associated with use of metformin and sulfonylurea in type 2 diabetes: a meta-analysis. Oncologist. 17, 813–822 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizos C. V. & Elisaf M. S. Metformin and cancer. Eur J Pharmacol. 705, 96–108 (2013). [DOI] [PubMed] [Google Scholar]
- Sheen Y. Y., Kim M. J., Park S. A., Park S. Y. & Nam J. S. Targeting the transforming growth factor-beta signaling in cancer therapy. Biomol Ther (Seoul). 21, 323–331 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaraj N. S. & Datta P. K. Targeting the transforming growth factor-beta signaling pathway in human cancer. Expert Opin Investig Drugs. 19, 77–91 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chames P., Van Regenmortel M., Weiss E. & Baty D. Therapeutic antibodies: successes, limitations and hopes for the future. Br J Pharmacol. 157, 220–233 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami S. et al. Ursolic acid, an antagonist for transforming growth factor (TGF)-beta1. FEBS Lett. 566, 55–59 (2004). [DOI] [PubMed] [Google Scholar]
- Radaev S. et al. Ternary complex of transforming growth factor-beta1 reveals isoform-specific ligand recognition and receptor recruitment in the superfamily. J Biol Chem. 285, 14806–14814 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris G. M. et al. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J Comput Chem. 30, 2785–2791 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindorff-Larsen K. et al. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins. 78, 1950–1958 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Wolf R. M., Caldwell J. W., Kollman P. A. & Case D. A. Development and testing of a general amber force field. J Comput Chem. 25, 1157–1174 (2004). [DOI] [PubMed] [Google Scholar]
- Bayly C. I., Cieplak P., Cornell W. D. & Kollman P. A. A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges - the resp model. J. Phys Chem. 97, 10269–10280 (1993). [Google Scholar]
- Hess B., Kutzner C., van der Spoel D. & Lindahl E. GROMACS 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput. 4, 435–447 (2008). [DOI] [PubMed] [Google Scholar]
- Srinivasan J., Cheatham T. E., Cieplak P., Kollman P. A. & Case D. A. Continuum solvent studies of the stability of DNA, RNA, and phosphoramidate - DNA helices. J Am Chem Soc. 120, 9401–9409 (1998). [Google Scholar]
- Yu J. et al. Single-molecule force spectroscopy study of interaction between transforming growth factor beta1 and its receptor in living cells. J Phys Chem B. 111, 13619–13625 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.