Abstract
This study was designed to isolate different strains of the genus Bifidobacterium from the fecal material of neonates and to assess their ability to produce the cis-9, trans-11 conjugated linoleic acid (CLA) isomer from free linoleic acid. Fecal material was collected from 24 neonates aged between 3 days and 2 months in a neonatal unit (Erinville Hospital, Cork, Ireland). A total of 46 isolates from six neonates were confirmed to be Bifidobacterium species based on a combination of the fructose-6-phosphate phosphoketolase assay, RAPD [random(ly) amplified polymorphic DNA] PCR, pulsed-field gel electrophoresis (PFGE), and partial 16S ribosomal DNA sequencing. Interestingly, only 1 of the 11 neonates that had received antibiotic treatment produced bifidobacteria. PFGE after genomic digestion with the restriction enzyme XbaI demonstrated that the bifidobacteria population displayed considerable genomic diversity among the neonates, with each containing between one and five dominant strains, whereas 11 different macro restriction patterns were obtained. In only one case did a single strain appear in two neonates. All genetically distinct strains were then screened for CLA production after 72 h of incubation with 0.5 mg of free linoleic acid ml−1 by using gas-liquid chromatography. The most efficient producers belonged to the species Bifidobacterium breve, of which two different strains converted 29 and 27% of the free linoleic acid to the cis-9, trans-11 isomer per microgram of dry cells, respectively. In addition, a strain of Bifidobacterium bifidum showed a conversion rate of 18%/μg dry cells. The ability of some Bifidobacterium strains to produce CLA could be another human health-promoting property linked to members of the genus, given that this metabolite has demonstrated anticarcinogenic activity in vitro and in vivo.
Immediately after birth, the human gastrointestinal tract (GIT) is sterile but rapidly becomes colonized by bacteria. Full-term vaginally delivered neonates are inoculated during birth by the vaginal and fecal flora of the mother, such that Escherichia coli and enterococci can be detected in the stools of infants within 24 h after birth (3, 11). The composition of the gut microbiota is profoundly influenced by the feeding regime of the neonate, with the result that in breast-fed infants, bifidobacteria are among the dominant microorganisms, whereas formula-fed infants are colonized by a more diverse microbiota, including bifidobacteria, bacteroides, clostridia, and streptococci (11, 22). In addition, the bacterial flora of the gut of preterm infants differs from term infants, in part due to the extensive use of broad-spectrum antibiotics, different nursing and hygiene practices, and the relatively aseptic neonatal intensive care environment, which can lead to outgrowth and resulting high counts of facultatively anaerobic bacteria, whereas colonization by bifidobacteria can be delayed (12, 18, 36). A similar situation is observed after caesarean section, where colonization with microorganisms from the hospital environment rather than from the birth canal of the mother occurs, with the result that the presence of anaerobic bacteria is delayed (3).
A number of studies have reported beneficial effects associated with ingestion of Bifidobacterium species in a viable form (see reference 1 for a review), including tumor inhibitory effects (7, 26, 27, 30), activation or regulation of the immune system (38), and improvement of gastrointestinal disturbances (8, 28). At present, the genus Bifidobacterium includes 28 species and 5 subspecies (5, 33), many of which have been isolated from fecal sources. Typical species in breast-fed or formula-fed infants are Bifidobacterium breve and B. longum (B. infantis) (20).
We recently reported that some human-derived bifidobacteria were capable of producing the fatty acid metabolite conjugated linoleic acid (CLA) (10). It is well known that CLA is produced by some rumen bacteria as an intermediate in the biohydrogenation of linoleic acid to stearic acid and, consequently, the main dietary sources of CLA are milk and ruminant fats. Many potentially beneficial health effects have been ascribed to CLA when it is consumed as a mixture of the two isomers, cis-9, trans-11 and trans-10, cis-12 CLA, most notably anticarcinogenic, immune modulation, antiatherosclerotic, and antiobesity activities (see reference 2 for a review). Microbial CLA production has only been reported for a few species in addition to rumen microflora, including propionibacteria used as dairy starter cultures (13), strains of the intestinal flora of rats (9), and some lactic cultures (19). We have previously shown that some strains of bifidobacteria can produce CLA, mainly the cis-9, trans-11 isomer (10). The neocolonization of the human infant intestine with bifidobacteria provides us with the earliest opportunity to determine whether or not CLA production is a feature of the initial flora. Such metabolic activity may have an important influence on the evolving and interactive intestinal mucosal environment, thus influencing long-term health. The present study was undertaken to isolate and genomically characterize strains of the genus Bifidobacterium (using pulsed-field gel electrophoresis [PFGE]) from infant feces, so that genetically distinct strains could be assessed for their ability to produce the beneficial fatty acid CLA from free linoleic acid.
MATERIALS AND METHODS
Subjects.
Fecal material was sampled from 24 neonates (12 premature and 12 full-term) aged from 3 days to 2 months. All samples were collected at the neonatal unit of the Erinville Hospital, Cork, Ireland. Fully informed consent was obtained from all parents. Ethical approval was not necessary, since the collection of body excreta does not require Ethics Committee approval. In order to obtain as much diversity as possible, isolates were obtained from neonates with diverse case histories and included 10 neonates who were breast fed, eight who were formula fed, and 6 who were both breast and formula fed. In addition, 11 of these neonates had been on antibiotic treatment prior to sampling.
Fecal sampling.
In most cases, fecal swab samples were taken from the neonates, which did not allow accurate enumeration of bacteria but did allow the isolation of strains from the samples. Samples were stored at 5°C and delivered to the laboratory for processing within 5 h of sampling. The swabs were vortex mixed in maximum recovery diluent (Oxoid, Ltd., Hampshire, United Kingdom) and serially diluted to enumerate the CFU/swab. Alternatively, the swabs were streaked directly onto agar plates.
Growth conditions and selection of colonies.
Serial dilutions of fecal samples were pour-plated onto modified MRS agar (mMRS) supplemented with 0.05% (wt/vol) l-cysteine hydrochloride (98% pure; Sigma Chemical Co., St. Louis, Mo.) and 1.5% (wt/vol) agar (Oxoid). To preselect for bifidobacteria, 100 μg of Mupirocin (Oxoid)/ml was added to the medium as antimicrobial susceptibility disks by using the disk diffusion method (25). Agar plates were incubated anaerobically (anaerobic jars with Anaerocult A gas packs; Merck, Darmstadt, Germany) at 37°C for 72 h. Twelve colonies were randomly selected from each sample and subcultured in mMRS broth for 24 to 48 h.
F-6-PPK activity.
The fructose-6-phosphate phosphoketolase (F-6-PPK) assay enables detection of the enzyme F-6-PPK, an enzyme characteristic of bifidobacterial carbohydrate metabolism (5). F-6-PPK activity can be detected in the intracellular extracts of bacterial cultures by using Triton X-100 detergent-mediated cell lysis based on protocols previously described (6, 24). B. breve NCFB 2258 and Propionibacterium freudenreichii subsp. shermanii 9093 were used as positive and negative controls, respectively, in these assays.
RAPD PCR.
Genomic DNA was extracted from the isolates, and a RAPD [random(ly) amplified polymorphic DNA] PCR was performed with the random primer R1 (5′-ATGTAACGCC-3′) synthesized by Sigma-Genosys, Ltd., Pampisford, United Kingdom. Details of procedures for genomic DNA isolation and RAPD PCR conditions were as previously described (31).
PFGE.
High-molecular-weight DNA was isolated from stationary-phase cultures by using previously described procedures (32). The restriction enzymes XbaI or SpeI (New England Biolabs) were used, and the DNA fragments were resolved with a contour-clamped homogeneous electric field CHEF-DR III pulsed-field system (Bio-Rad Laboratories) at 6 V/cm for 18 h with a 1- to 15-s linear ramp pulse time and with 0.5× Tris base-borate-EDTA running buffer maintained at 14°C. Gels were stained in distilled water containing 0.5 μg of ethidium bromide/ml for 30 min and then destained for 60 min. Pulsed-field gel electrophoresis (PFGE) gels were visualized by UV transillumination, and the sizes of the PFGE fragments were estimated by comparison with a low-range PFGE marker ranging from 2.03 to 194 kb (no. N0350S; New England Biolabs).
16S rDNA sequencing.
Two 16S ribosomal DNA (rDNA) primers—CO1 for the 5′ end (5′-AGTTTGATCCTGGCTCAG-3′) and CO2 for the 3′ end (5′-TACCTTGTTACGACT-3′)—were used to generate an approximate 1.5-kb 16S rDNA product under PCR conditions described previously (32); this product was partially sequenced by using the primer CO1 by MWG sequencing service (Ebersberg, Germany). Comparison of the 16S rDNA sequences obtained by using the BLAST program allowed the assignment of a strain to a particular species. In general, when 16S rDNA similarity values exceed 97%, the strains are considered to belong to the same species (34).
Microbial production of CLA.
The Bifidobacterium isolates were tested for bioconversion of free linoleic acid (0.5 mg ml−1) to cis-9, trans-11 CLA in mMRS medium after incubation for 72 h, and the fatty acid profile of the supernatant was assessed by gas-liquid chromatography (GLC) as previously described (10, 35). cis-9, trans-11 and trans-9, trans-11 CLA standards were from Matreya, Inc., and linoleic acid was from Sigma.
RESULTS
Since the purpose of the present study was to screen neonatally derived bifidobacteria for CLA production, initial experiments were designed to isolate genetically distinct strains of the genus that predominate in the neonatal GIT. These strains were obtained from stool samples or swabs taken from neonates with a range of different case histories, i.e., neonates that were either breast or formula fed, that were either full-term or preterm, and that did or did not receive antibiotics (Table 1).
TABLE 1.
Samples, description, and selection of bifidobacteria by F-6-PPK detection
| Sample no. | Full-term or preterm | Age | Feedinga | Antibiotic treatment (duration [days])b | Count on mMRS + MUP (CFU/swab)c | No. of colonies assessed | No. of F-6-PPK-positive colonies (%) |
|---|---|---|---|---|---|---|---|
| 1 | Full-term | 4 days | F | None | 0 | 0 | |
| 2 | Preterm | 3 wk | B | AMP + GEN (5) | Swab streak | 13 | 1 (7.7) |
| 3 | Preterm | 5 days | B | None | Swab streak | 10 | 0 (0) |
| 4 | Full-term | 4 days | F | None | 0 | 0 | |
| 5 | Full-term | 3 wk | B | None | Swab streak | 12 | 0 (0) |
| 6 | Full-term | 4 days | F | None | 0 | 0 | |
| 7 | Full-term | 4 days | F | AMP + GEN (2) | 0 | 0 | |
| 8 | Full-term | 3 days | F | None | Swab streak | 11 | 11 (100) |
| 9 | Preterm | 39 days | B + F | AMP + GEN (5), CEF + TEI (5) | 0 | 0 | |
| 10 | Preterm | 12 days | B + F | AMP | <10 | 0 | |
| GEN (5) | |||||||
| 11 | Preterm | 5 days | F | AMP + GEN (2) | 0 | 0 | |
| 12 | Preterm | 4 days | B + F | AMP + GEN (2) | One colony | 0 | |
| 13 | Preterm | 8 days | B | AMP + GEN (2) | 0 | 0 | |
| 14 | Preterm | 2 wk | B | AMP + GEN (2) | 6 × 107 | 12 | 0 (0) |
| 15 | Full-term | 4 days | B | AMP + GEN (2) | 7 × 103 | 1 | 0 (0) |
| 16 | Full-term | 4 days | B | None | 4 × 105 | 11 | 9 (81.8) |
| 17 | Full-term | 4 days | F | None | 1 × 104 | 12 | 0 (0) |
| 18 | Full-term | 10 days | B + F | None | 1 × 104 | 2 | 0 (0) |
| 19 | Preterm | 1 wk | B | AMP + GEN (2) | 5 × 101 | 12 | 0 (0) |
| 20 | Full-term | 3 days | F | None | 7 × 106 | 12 | 12 (100) |
| 21 | Preterm | 4 days | B | AMP + GEN (2) | 0 | 0 | |
| 22 | Preterm | 6 wk | B + F | None | 4 × 105 | 7 | 1 (14.3) |
| 23 | Full-term | 5 days | B | None | 5 × 106 | 12 | 12 (100) |
| 24 | Preterm | 2 mo | B + F | None | 4 × 107 | 2 | 0 (0) |
B, breast-feeding; F, formula.
AMP, ampicillin; GEN, gentamicin; CEF, cefotaxime; TEI, teicoplanin.
Swab streak, swab that was streaked directly onto an agar plate. MUP, mupirocin.
Isolation of neonatally derived bifidobacteria.
In 8 of the 24 fecal samples, no isolates were obtained on the mupirocin-containing media (Table 1). For the remaining 16 fecal samples, 46 colonies from 6 samples were found to be F-6-PPK positive (6 of 24 samples), which confirmed their Bifidobacterium status. Interestingly, Bifidobacterium isolation was rare from infants which had received antibiotic treatment. Indeed, of the samples obtained from neonates subjected to antibiotic treatment, only one generated a Bifidobacterium colony (sample 2). Most of the samples containing bifidobacteria were produced by full-term infants (four of six of bifidobacteria containing samples). Only two premature neonates yielded bifidobacteria (sample 2 [3 weeks old] and sample 22 [6 weeks old]); however, it is notable that the neonates that produced the highest numbers of CFU/swab on mupirocin-containing media (107 [samples 14 and 24]) were not colonized by any bifidobacteria.
Genetic fingerprinting of neonatal isolates.
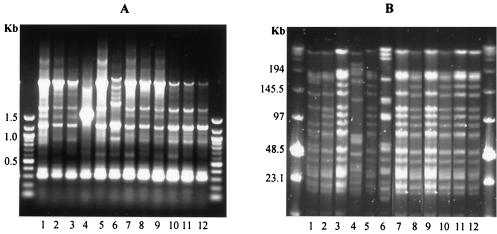
In an initial attempt to group all of the isolates, RAPD PCR was performed with the single random primer R1. Under the conditions described, primer R1 generally generated about 7 to 10 DNA fragments which ranged in size from 100 bp to about 3 kb per isolate (Fig. 1A). Figure 1A shows a RAPD PCR gel of all isolates from fecal sample 23. The method allowed the grouping of most of the isolates but did not discriminate between some of the very closely related strains (see below). Interestingly, RAPD PCR had grouped these isolates into only three genotypes as indicated in lanes 1, 4, and 6 (Fig. 1A). Consequently, PFGE was performed on the same isolates from fecal sample 23, after genomic digestion with the restriction enzyme XbaI (Fig. 1B). It is evident that some of these strains shared a very close genetic relationship, since their PFGE DNA fingerprints were very similar, making them indistinguishable by RAPD PCR. For example, isolates 2, 7, and 9 and isolates 3, 5, 8, 10, and 12 (Fig. 1B) differed from each other by only one DNA fragment (a DNA fragment of ∼150 kb present in isolates 3, 5, 8, 10, and 12 appeared to be absent in isolates 2, 7 and 9). Isolate 1 differed from isolates 2, 7 and 9 by 3 DNA fragments. Fragments of ∼55 and ∼160 kb were present in isolate 1 but not in isolates 2, 7, and 9, whereas isolates 2, 7, and 9 harbored a fragment of ∼100 kb, which appeared to be absent in isolate 1 (Fig. 1B).
FIG. 1.
Genetic fingerprinting by RAPD PCR of all 12 isolates from fecal sample 23 (A) and their corresponding PFGE genomic patterns when restricted with XbaI (B). Molecular weight markers are shown in outside lanes of both gels.
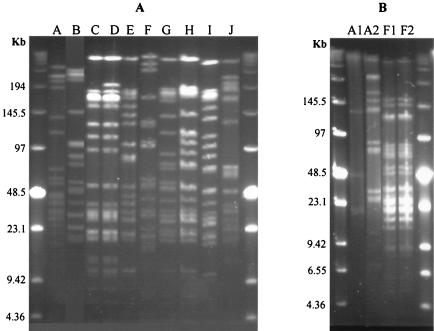
In order to make a final grouping and characterization of all 46 different isolates positive for the F-6-PPK test, further PFGE analyses were performed. XbaI restriction of the 46 isolates from six different neonates resulted in 10 different PFGE macrorestriction patterns, termed A through J (Table 2). In all, one to four DNA fragments differed between PFGE patterns G, H, and I (Fig. 2A), and 16S rDNA partial sequencing (see below) demonstrated that the different isolates belong to the same species (B. longum). PFGE profiles from all 46 isolates demonstrated that some neonates consisted of one dominant strain type (sample 8 and 20), whereas others harbored multiple strains (Table 2); for example, sample 23 harbored at least five different strains (see Fig. 1B). In only two cases were isolates with the same XbaI pattern obtained from two different individuals (Fig. 2A). In one of these cases, further analysis with SpeI allowed differentiation between two of the isolates (Fig. 2B). Thus, the PFGE data demonstrated that there were 11 genetically distinct Bifidobacterium strains among the 46 isolates.
TABLE 2.
Distribution of PFGE patterns among fecal samples and numbers of isolates sharing the same pattern
| Sample no. | PFGE pattern | No. of Bifidobacterium isolates |
|---|---|---|
| 2 | A | 1 |
| 8 | B | 11 |
| 16 | C | 7 |
| D | 1 | |
| E | 1 | |
| 20 | A | 12 |
| 22 | F | 1 |
| 23 | G | 1 |
| H | 3 | |
| I | 6 | |
| J | 1 | |
| F | 1 |
FIG. 2.
(A) Genomic digests with XbaI of the 10 distinct patterns (A to J) obtained by PFGE from 46 isolates. PFGE pattern A shared by samples 2 and 20. Pattern F shared by sample 22 and one of the strains in sample 23. (B) SpeI restriction of samples 2, 20 (PFGE patterns A1 and A2), 22, and 23 (PFGE patterns F1 and F2). Molecular weight markers are shown in outside lanes of both gels.
16S rDNA sequencing.
Using this approach, all 11 different strains exhibiting different PFGE patterns showed homology values of ≥97% with different species within the genus Bifidobacterium (Table 3). The most commonly found strain in the fecal samples was B. breve, which was found in three out of six of the Bifidobacterium containing samples. Moreover, B. longum was present in two out of the six samples containing F-6-PPK positive isolates and represents 6 of 11 of the different PFGE profiles. In contrast, Bifidobacterium bifidum was found in two different fecal samples in the present study, while Bifidobacterium adolescentis was present in only in sample 23. Interestingly, the B. bifidum strains found in two different neonates (sample 2 and 20) gave identical PFGE patterns after restriction with XbaI, but when restricted with SpeI the strains appeared different (Fig. 2B). The same occurred when strains from two other neonates were compared; samples 22 and 23 had a strain in common that had indistinguishable PFGE patterns (B. breve) after restriction with the same enzyme. In the latter case, they were still indistinguishable after restriction with SpeI (Fig. 2B).
TABLE 3.
16S rDNA partial sequencing of genetically distinct strains (PFGE patterns A to J)
| PFGE pattern | 16S rDNA partial sequencing
|
|
|---|---|---|
| % Homologya | Strain | |
| A | 99 | B. bifidum |
| B | 97 | B. breve |
| C | 99-100 | B. longum |
| D | 97 | B. longum |
| E | 99 | B. longum |
| F | 97 | B. breve |
| G | 100 | B. longum |
| H | 98 | B. longum |
| I | 100 | B. longum |
| J | 97 | B. adolescentis |
That is, the percent homology with Bifidobacterium species.
Microbial production of CLA.
After incubation for 72 h with 0.5 mg of the linoleic acid ml−1, the cis-9, trans-11 CLA isomer was the main isomer produced, a finding indicative of linoleic acid isomerase activity. Previous studies have demonstrated 0.5 mg of linoleic acid ml−1 to be the optimum concentration for most Bifidobacterium cultures (10); however, since the ability to grow in a medium containing this concentration varied among the isolates, the results are expressed as the percent conversion per microgram of dry cells. B. breve NCFB 2258 was used as a positive control since the strain was previously shown to be an efficient producer of CLA (10), in this study converting 36.7% of linoleic acid to cis-9, trans-11 CLA per microgram of dry cells (Table 4). The ability to generate CLA from free linoleic acid differed considerably among the strains, ranging from a nondetectable level to a conversion level of 29% per microgram of dry cells. The most efficient producers were shown to belong to the species B. breve, exhibiting between 7.7 and 29% conversion of free linoleic acid to cis-9, trans-11 CLA per microgram of dry cells (Table 4), with some of the B. breve strains also producing small amounts of the trans-9, trans-11 CLA isomer (Fig. 3). In addition, a strain of B. bifidum showed CLA-producing ability with a conversion rate of 17.9% per microgram of dry cells, with smaller amounts produced by a B. longum strain. The two different strains sharing patterns A and F, respectively, differed in their CLA production ability. PFGE pattern A (B. bifidum) strain from sample 2 converted 17.9%, whereas the strain from sample 20 did not produce any detectable levels of CLA (Table 4). This supports the fact that they could be discriminated after restriction with the additional restriction enzyme SpeI. PFGE pattern F (B. breve) strain from sample 22 converted only 7.7%, whereas the strain from sample 23 converted 27.4%, although their PFGE profiles were indistinguishable after genomic digestion with XbaI and SpeI (Fig. 2B).
TABLE 4.
CLA production by different PFGE pattern strains and B. breve NCFB 2258
| PFGE profile | Mean % conversion ± SDa |
|---|---|
| B. breve NCFB 2258 | 36.7 ± 12.2 |
| A1 | 17.9 ± 4.7 |
| A2 | 0 |
| B | 29.0 ± 6.4 |
| C | 0 |
| D | 0 |
| E | 3.2 ± 1.8 |
| F1 | 7.7 ± 2.3 |
| F2 | 27.4 ± 6.9 |
| G | 0 |
| H | 0 |
| I | 0 |
| J | 0 |
That is, the percent conversion of linoleic acid to cis-9, trans-11 CLA/microgram of dry cells. Analyses were done at least in triplicate.
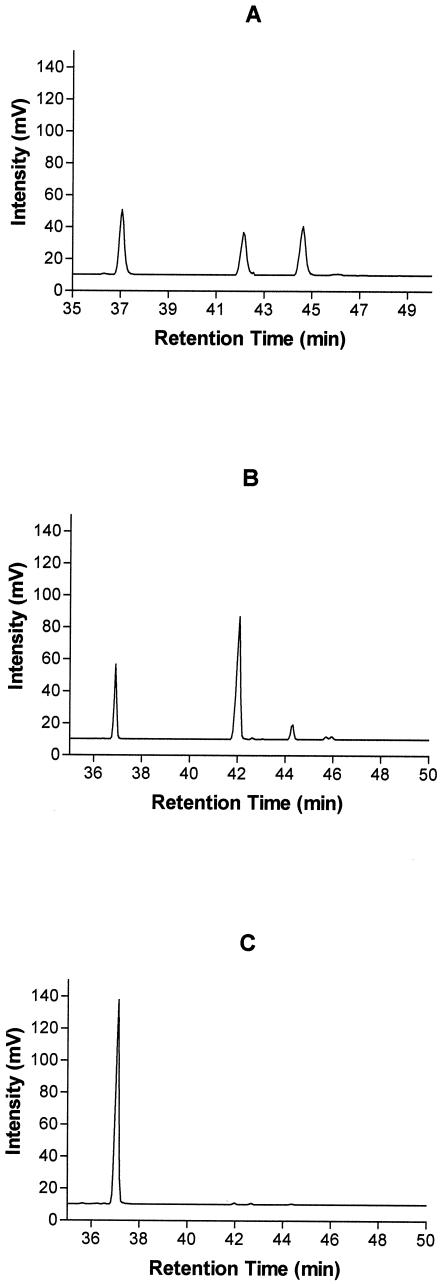
FIG. 3.
Gas-liquid chromatogram of linoleic acid, cis-9, trans-11 isomer of CLA and trans-9, trans-11 CLA fatty acid standards (A), fatty acids in mMRS medium containing linoleic acid (0.5 mg ml−1) after incubation with B. breve PFGE pattern B (B), and B. longum PFGE pattern C (C). Peak 1, linoleic acid; peak 2, cis-9, trans-11 CLA; peak 3, trans-9, trans-11 CLA.
DISCUSSION
The infant GIT is initially sterile but becomes colonized by different microorganisms, with Bifidobacterium being among the dominant microorganisms soon after birth (22). The surprisingly low isolation rate of bifidobacteria from the fecal samples in the present study (25% of total samples) is possibly due to the incidence of antibiotic treatment prior to sampling and/or premature birth. Indeed, previous studies have demonstrated an isolation rate of only ca. 10% of anaerobic bacteria from infants during antibiotic treatment, after which a slow regrowth of bifidobacteria was reported (3). In the present study, we isolated bifidobacteria from only 1 of the 11 neonates that had received antibiotic treatment. In this case, the neonate had not received the treatment recently prior to sampling. In our study, the only two premature neonates that yielded bifidobacteria were 3 and 6 weeks old and therefore old enough to have developed a normal intestinal microflora. In extremely low birthweight infants (<1,000 g), it has been reported that Lactobacillus and Bifidobacterium spp. were identified in stool samples of only 1 of 29 tested infants at day 30 after birth, whereas predominant species were Enterococcus faecalis, E. coli, Staphylococcus epidermidis, Enterobacter cloacae, Klebsiella pneumoniae, and Staphylococcus haemolyticus (12). In such cases, the relatively aseptic neonatal intensive care environment, coupled with extensive use of broad spectrum antibiotics and nursing and hygiene practices, probably result in a colonization delay for many species with an outgrowth by a limited number of other species (12). Indeed, in another study on 29 preterm infants that were hospitalized in a neonatal intensive care unit, E. coli, Enterococcus sp., and K. pneumoniae were the most commonly found bacteria in the stool samples of all preterm infants (29).
Considerable genomic diversity was evident among the neonatal Bifidobacterium population isolated in the present study, with two neonates having one dominant strain, one having three strains, and another having as many as five different strains, based on PFGE macrorestriction patterns. An earlier study reported that a mixture of many different species in the same neonatal sample was not a common trait (4). A combination of two Bifidobacterium species was found in 28.1% of 128 breast-fed infants tested, a combination of three in 7.8%, and a combination of four in only 2.3% of the same group. Moreover, 14.1% were completely lacking the genus Bifidobacterium. In our study, the most commonly found strain in the fecal samples was B. breve, which was found in three of six of the Bifidobacterium-containing samples, with B. longum also found to be prevalent, representing 6 of 11 of the different PFGE profiles. Other species isolated in our study included B. bifidum and B. adolescentis. These bifidobacterial species have been frequently found in the feces of infants. In a study by Matsuki et al. (20) on 27 healthy, vaginally delivered, and breast-fed infants (22 to 46 days old), the most commonly found Bifidobacterium species were B. breve (isolated from 70% of the infants), B. infantis (isolated from 41% of the infants), and B. longum (isolated from 37% of the infants). A combination of two species was found in 19% of the infant fecal samples, a combination of three species was found in 22% of the samples, a combination of four species was found in 7.4% of the samples, a combination of five species was found in 3.7% of the samples, and a combination of six species was found in none of the infants. No bifidobacteria were found in 11% of the infants tested (20). The observed diversity reported in these studies may be due to different approaches used for identification, different ages of the neonates, and different delivery methods. An interesting finding of our study was the identification of B. breve strains, which had almost identical PFGE patterns, differing in only one or two bands. Indeed, a similar observation was recently reported in Bifidobacterium strains isolated from a porcine cecum (32). In these cases, it is tempting to suggest that these strains may be undergoing an increased evolutionary rate imposed on them by the local environment of the intestine.
Since the main purpose of the present study was to isolate neonatally derived bifidobacteria to investigate their ability to produce CLA isomers, all genetically distinct strains were tested for their CLA biosynthetic ability. The data show that strains of B. breve were the most efficient CLA producers, while a strain of B. bifidum also exhibited considerable activity. Indeed, the CLA production rates by the novel B. breve strains isolated in the present study were nearly as efficient as B. breve NCFB 2258, previously found to be the most efficient producer of cis-9, trans-11 CLA from linoleic acid in a screening study involving a range of strains of bifidobacteria, lactobacilli, lactococci, and pediococci (10). Other Bifidobacterium species reported with CLA biosynthetic capabilities include B. dentium (10) and B. pseudocatenulatum (23). Surprisingly, the two B. breve strains sharing PFGE pattern F (identical PFGE patterns after digestion with XbaI and SpeI) did not show comparable rates of CLA production. The strain from sample 22 (F1) produced CLA at a rate of 7.7%/μg of dry cells, whereas the strain from sample 23 (F2) produced 27.4%/μg of dry cells. The explanation for this is unknown, but it may be due to point mutations or subtle differences in the genetic elements responsible for CLA production between the two strains. The reason why some bacteria convert linoleic acid to CLA is unclear. Jiang et al. (13) observed a positive correlation between CLA production and ability to tolerate free linoleic acid, which indicates that the conversion could function as a detoxification process in the bacterial cell. The presence of conjugated dienes in the cell membranes changes the fluidity and thereby improves cellular resistance to stress (14, 37). The “bent” structure of cis-9, cis-12 linoleic acid could increase the fluidity and permeability when interfering with the membranes and impair vital cell functions. Isomerization of the cis-12 bond of linoleic acid to trans-11 during conversion to cis-9, trans-11 CLA straightens the fatty acid structure, thereby resulting in decreased fluidity. cis-9, trans-11 CLA is produced by the action of linoleic acid isomerase, an enzyme present in the anaerobic rumen bacteria Butyrivibrio fibrisolvens (15, 16). This CLA isomer has received considerable attention in recent years because of its remarkable health benefits including anticarcinogenic, anti-inflammatory, and immunomodulating properties (2). The potential for such benefits may well extend to preterm infants for whom necrotizing enterocolitis is the most common intestinal emergency in the neonatal period (21). This condition carries a significant morbidity and mortality and recent studies have suggested benefits from probiotic therapy in these preterm infants (17). Indeed, it is tempting to suggest that the ability of bifidobacteria to produce the health promoting bioactive metabolite CLA in vivo may be associated with some of their suggested therapeutic effects.
The present study demonstrates that bifidobacteria populations with ability to produce CLA develop in the neonate shortly after birth and that there is considerable genomic diversity among the neonatal Bifidobacterium population. These bacteria may offer potential in the functional food industry aiming to develop new probiotic food products. The ability of particular strains to efficiently produce CLA perhaps should influence the choice of bifidobacteria for supplementation in preterm infants at risk of necrotizing enterocolitis.
Acknowledgments
This study was funded by the Irish government under National Development Plan 2000-2006, by EU project QLK1-2002-02362, and in part by SFI funds.
REFERENCES
- 1.Arunachalam, K. D. 1999. Role of bifidobacteria in nutrition, medicine, and technology. Nutr. Res. 19:1559-1597. [Google Scholar]
- 2.Belury, M. A. 2002. Dietary conjugated linoleic acid in health: physiological effects and mechanisms of action. Annu. Rev. Nutr. 22:505-531. [DOI] [PubMed] [Google Scholar]
- 3.Bennet, R., and C. E. Nord. 1987. Development of the fecal anaerobic microflora after cesarean section and treatment with antibiotics in newborn infants. Infection 15:332-336. [DOI] [PubMed] [Google Scholar]
- 4.Biavati, B., P. Castagnoli, F. Crociani, and L. D. Trovatelli. 1984. Species of the Bifidobacterium in the feces of infants. Microbiologica 7:341-345. [PubMed] [Google Scholar]
- 5.Biavati, B., M. Vescovo, S. Torriani, and V. Bottazzi. 2000. Bifidobacteria: history, ecology, physiology, and applications. Ann. Microbiol. 50:117-131. [Google Scholar]
- 6.Bibiloni, R., P. F. Pérez, and G. L. De Antoni. 2000. An enzymatic-colorimetric assay for the quantification of Bifidobacterium. J. Food Prot. 63:322-326. [DOI] [PubMed] [Google Scholar]
- 7.Biffi, A., D. Corandini, R. Larsen, L. Riva, and G. Di Fronzo. 1997. Antiproliferative effect of fermented milk on the growth of a human cancer cell line. Nutr. Cancer 28:93-99. [DOI] [PubMed] [Google Scholar]
- 8.Black, F., K. Einarsson, A. Lidbeck, K. Orrhage, and C. E. Nord. 1991. Effect of lactic acid producing bacteria on the human intestinal microflora during ampicillin treatment. Scand. J. Infect. Dis. 23:247-254. [DOI] [PubMed] [Google Scholar]
- 9.Chin, S. F., J. M. Storksson, W. Liu, K. J. Albright, and M. W. Pariza. 1994. Conjugated linoleic acid (9,11- and 10,12-octa-decadienoic acid) is produced in conventional but not germ-free rats fed linoleic acid. J. Nutr. 124:694-701. [DOI] [PubMed] [Google Scholar]
- 10.Coakley, M., R. P. Ross, M. Nordgren, G. Fitzgerald, D. Devery, and C. Stanton. 2003. Conjugated linoleic acid biosynthesis by human-derived Bifidobacterium species. J. Appl. Microbiol. 94:138-145. [DOI] [PubMed] [Google Scholar]
- 11.Favier, C. F., E. E. Vaughan, W. M. De Vos, and A. D. L. Akkermans. 2002. Molecular monitoring of succession of bacterial communities in human neonates. Appl. Environ. Microbiol. 68:219-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gewolb, I. H., R. S. Schwalbe, V. L. Taciak, T. S. Harrison, and P. Panigrahi. 1999. Stool microflora in extremely low birthweight infants. Arch. Dis. Child Fetal Neonatal 80:167-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang, J., L. Björck, and R. Fonden. 1998. Production of conjugated linoleic acid by dairy starter cultures. J. Appl. Microbiol. 85:95-102. [DOI] [PubMed] [Google Scholar]
- 14.Junker, F., and J. L. Ramos. 1999. Involvement of the cis/trans isomerase Cti in solvent resistance of Pseudomonas putida DOT-T1E. J. Bacteriol. 181:5693-5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kepler, C. R., K. P. Hirons, J. J. McNeill, and S. B. Tove. 1966. Intermediates and products of the biohydrogenation of linoleic acid by Butyrivibrio fibrisolvens. J. Biol. Chem. 241:1350-1354. [PubMed] [Google Scholar]
- 16.Kepler, C. R., and S. B. Tove. 1967. Biohydrogenation of unsaturated fatty acids. J. Biol. Chem. 242:5686-5692. [PubMed] [Google Scholar]
- 17.Kitajima, H., Y. Sumida, R. Tanaka, N. Yuki, H. Takayama, and M. Fujimura. 1997. Early administration of Bifidobacterium breve to preterm infants: randomized controlled trial. Arch. Dis. Child Fetal Neonatal 76:101-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lawrence, G., J. Bates, and A. Gaul. 1982. Pathogenesis of necrotising enterocolitis. Lancet i:137-139. [DOI] [PubMed] [Google Scholar]
- 19.Lin, T. Y., C. W. Lin, and C. H. Lee. 1999. Conjugated linoleic acid concentration as affected by lactic cultures and added linoleic acid. Food Chem. 67:1-5. [Google Scholar]
- 20.Matsuki, T., K. Watanabe, R. Tanaka, M. Fukuda, and H. Oyaizu. 1999. Distribution of bifidobacterial species in human intestinal microflora examined with 16S rRNA-gene-targeted species-specific primers. Appl. Environ. Microbiol. 65:4506-4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Millar, M., M. Wilks, and K. Costeloe. 2003. Probiotics for preterm infants? Arch. Dis. Child Fetal Neonatal 88:F354-F358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mitsuoka, T. 1996. Intestinal flora and human health. Asia Pacific, J. Clin. Nutr. 5:2-9. [PubMed] [Google Scholar]
- 23.Oh, D. K., G. H. Hong, Y. Lee, S. Min, H. S. Sin, and S. K. Cho. 2003. Production of conjugated linoleic acid by isolated Bifidobacterium strains. World J. Microbiol. Biotechnol. 19:907-912. [Google Scholar]
- 24.Orban, J. J., and J. A. Patterson. 2000. Modification of the phosphoketolase assay for rapid identification of bifidobacteria. J. Microbiol. Methods 40:221-224. [DOI] [PubMed] [Google Scholar]
- 25.Rada, V. 1997. Detection of Bifidobacterium species by enzymatic methods and antimicrobial susceptibility testing. Biotechnol. Tech. 11:909-912. [Google Scholar]
- 26.Reddy, B. S., and A. Rivenson. 1993. Inhibitory effect of Bifidobacterium longum on colon, mammary, and liver carcinogenesis induced by 2-amino-3-methylimidazo[4,5-f]quinoline, a food mutagen. Cancer Res. 53:3914-3918. [PubMed] [Google Scholar]
- 27.Rowland, I. R., C. J. Rumney, J. T. Coutts, and L. C. Lievense. 1998. Effect of Bifidobacterium longum and inulin on gut bacterial metabolism and carcinogen-induced aberrant crypt foci in rats. Carcinogenesis 19:281-285. [DOI] [PubMed] [Google Scholar]
- 28.Saavedra, J. M., N. A. Bauman, I. Oung, J. A. Perma, and R. H. Yolken. 1994. Feeding of Bifidobacterium bifidum and Streptococcus thermophilus to infants in hospital for prevention of diarrhoea and shedding of rotavirus. Lancet 344:1046-1049. [DOI] [PubMed] [Google Scholar]
- 29.Schwiertz, A., B. Gruhl, M. Löbnitz, P. Michel, M. Radke, and M. Blaut. 2003. Development of the intestinal bacterial composition in hospitalized preterm infants in comparison with breast-fed, full-term infants. Pediatr. Res. 54:393-399. [DOI] [PubMed] [Google Scholar]
- 30.Sekine, K., T. Toida, M. Saito, M. Kuboyama, T. Kawashima, and Y. Hashimoto. 1985. A new morphologically characterized cell wall preparation (whole peptidoglycan) from Bifidobacterium infantis with a higher efficacy on the regression of an established tumor in mice. Cancer Res. 45:1300-1307. [PubMed] [Google Scholar]
- 31.Simpson, P. J., C. Stanton, G. F. Fitzgerald, and R. P. Ross. 2002. Genomic diversity within the genus Pediococcus as revealed by randomly amplified polymorphic DNA PCR and pulsed-field gel electrophoresis. Appl. Environ. Microbiol. 68:765-771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Simpson, P. J., C. Stanton, G. F. Fitzgerald, and R. P. Ross. 2003. Genomic diversity and relatedness of bifidobacteria isolated from a porcine cecum. J. Bacteriol. 185:2571-2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simpson, P. J., R. P. Ross, G. F. Fitzgerald, and C. Stanton. 2004. Bifidobacterium psychroaerophilum sp. nov. and Bifidobacterium aerophilum sp. nov., isolated from a porcine cecum. Int. J. Syst. Evol. Microbiol. 54:401-406. [DOI] [PubMed] [Google Scholar]
- 34.Stackebrandt, E., and B. M. Goebel. 1994. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44:846-849. [Google Scholar]
- 35.Stanton, C., F. Lawless, G. Kjellmer, D. Harrington, R. Devery, J. F. Connolly, and J. J. Murphy. 1997. Dietary influences on bovine milk cis-9, trans-11-conjugated linoleic acid content. J. Food Sci. 62:1083-1086. [Google Scholar]
- 36.Stark, P. L., and A. Lee. 1982. The bacterial colonization of the large bowel of preterm low-birth-weight neonates. J. Hyg. 89:59-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weber, F. J., S. Isken, and J. A. M. de Bont. 1994. cis/trans isomerisation of fatty acids as a defense mechanism of Pseudomonas putida strains to toxic concentrations of toluene. Microbiology 140:2013-2017. [DOI] [PubMed] [Google Scholar]
- 38.Yasui, H., and M. Ohwaki. 1991. Enhancement of immune response in Peyer's patch cells cultured with Bifidobacterium breve. J. Dairy Sci. 7:1187-1195. [DOI] [PubMed] [Google Scholar]