Abstract
For the enzymatic production of chitosan oligosaccharides from chitosan, a chitosanase-producing bacterium, Bacillus sp. strain KCTC 0377BP, was isolated from soil. The bacterium constitutively produced chitosanase in a culture medium without chitosan as an inducer. The production of chitosanase was increased from 1.2 U/ml in a minimal chitosan medium to 100 U/ml by optimizing the culture conditions. The chitosanase was purified from a culture supernatant by using CM-Toyopearl column chromatography and a Superose 12HR column for fast-performance liquid chromatography and was characterized according to its enzyme properties. The molecular mass of the enzyme was estimated to be 45 kDa by means of sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The enzyme demonstrated bifunctional chitosanase-glucanase activities, although it showed very low glucanase activity, with less than 3% of the chitosanase activity. Activity of the enzyme increased with an increase of the degrees of deacetylation (DDA) of the chitosan substrate. However, the enzyme still retained 72% of its relative activity toward the 39% DDA of chitosan, compared with the activity of the 94% DDA of chitosan. The enzyme produced chitosan oligosaccharides from chitosan, ranging mainly from chitotriose to chitooctaose. By controlling the reaction time and by monitoring the reaction products with gel filtration high-performance liquid chromatography, chitosan oligosaccharides with a desired oligosaccharide content and composition were obtained. In addition, the enzyme was efficiently used for the production of low-molecular-weight chitosan and highly acetylated chitosan oligosaccharides. A gene (csn45) encoding chitosanase was cloned, sequenced, and compared with other functionally related genes. The deduced amino acid sequence of csn45 was dissimilar to those of the classical chitosanase belonging to glycoside hydrolase family 46 but was similar to glucanases classified with glycoside hydrolase family 8.
Chitosan is currently obtained by the deacetylation of chitin (poly-β-1,4-d-N-acetylglucosamine) that has been extracted from an abundant source of shrimp or crab shells. Deacetylated chitosans are produced by treating chitin in a concentrated alkaline solution (50%, wt/vol) and boiling it for several hours (34). Chitosan is also found in nature; it is found in the cell walls of fungi of the class Zygomycetes, in the chlorophycean algae Chlorella sp. (26), and in insect cuticles (2). These natural chitosans are synthesized by the tandem action of chitin synthetase and chitin deacetylase, as shown for Mucor rouxii and Colletotrichum lindemuthianum (9). Both chemical and enzymatic procedures for chitosan production result in the incomplete deacetylation of chitin, which yields chitosans in the intermediate range of the degree of deacetylation (DDA). Consequently, chitosan can be considered as a partly deacetylated derivative of chitin; it must have diverse structures containing hetero-linkages of N-acetylglucosamine (GlcNAc)-glucosamine (GlcN) and GlcN-GlcNAc, as well as homo-linkages of GlcNAc-GlcNAc and GlcN-GlcN.
In spite of their abundance in nature, the commercial utilization of chitin and chitosan has remained undeveloped for a long time (3). The insolubility of chitin in common solvents limits the utilization of chitin as a natural resource. Many derivatives have been synthesized from chitin to evaluate their usefulness (21, 46). Chitosan has advantages over chitin because of its high solubility in acidic solutions and its polycationic nature. In the last two decades, applications of chitosan have been developed in many industries; chitosan has emerged as a new biomaterial for food, pharmaceutical, textile, and other industries, as well as for wastewater treatment (3, 15, 19). Recently, chitosan oligosaccharides have received growing attention because they perform a variety of biological activities, such as inhibiting the growth of bacteria and fungi (13, 17, 44, 47), exerting antitumor activity (41, 45), acting as immunopotentiating effectors (42, 43), and eliciting pathogenesis-related proteins in higher plants (16). Chain length and DDA are considered the most important factors influencing the biological activities of chitosan oligosaccharides. Chitosan oligosaccharides of longer than a hexamer in chain length perform more potent antimicrobial, antitumor, and immunopotentiating activities than shorter oligosaccharides, even though shorter oligosaccharides also perform activities of significance (25). An increase in the DDA of chitosan oligosaccharides enhances antimicrobial and anticholesterolemic activities. Therefore, strict control of both the chain length and the DDA is a prerequisite for the production of valuable chitosan oligosaccharides.
Chemical and enzymatic processes have been used in various industries to produce chitosan oligosaccharides. Acid hydrolysis produces a large amount of short-chain oligosaccharides, including a monomeric unit which requires an additional process for the fractionation of longer oligosaccharides, resulting in low yields of oligosaccharides (6, 35). Alternatively, an enzymatic process to produce oligosaccharides by using chitosanases has been tried (14). Chitosanases have been found in abundance in a variety of microorganisms, including bacteria (24, 31, 32, 48) and fungi (1, 38). Because most of these chitosanases hydrolyze chitosan by an endo-type cleavage, these enzymes produce oligosaccharides ranging from dimer to octamer of GlcN in their compositions.
To obtain a novel chitosanase that can be used for large-scale production of chitosan oligosaccharides, many bacterial isolates with chitosanase activities have been screened. Among these bacteria, a bacterial strain with high chitosanase activity, Bacillus sp. strain KCTC 0377BP, was isolated. This bacterium constitutively produced chitosanase at the highest level in a culture medium. This paper describes the purification and characterization of a chitosanase from Bacillus sp. strain KCTC 0377BP and the applications of the enzyme for chitosan oligosaccharide production.
MATERIALS AND METHODS
Materials.
Chitin, chitosan, and glycol chitin were purchased from Sigma-Aldrich Co. (St. Louis, Mo.). Colloidal chitosan was prepared by using the methods of Roberts (34). Partially deacetylated chitosan (39 to 94% DDA) was prepared by using the methods of Yabuki et al. (48). A chitosan oligomer standard series, from chitobiose to chitoheptaose, and its monomer d-glucosamine-HCl were purchased from Wako (Osaka, Japan) and Sigma-Aldrich Co., respectively. Shodex standard P-82 (pullulan; molecular masses of 48.0, 23.7, 12.2, and 5.8 kDa), used for molecular weight measurements of the low-molecular-weight (LMW) chitosan, were purchased from Showa Denko (Tokyo, Japan). All other reagents were of the highest grade available.
Preparation of chitosan substrate.
Chitosan substrate (10 mg/ml) was prepared by suspending 10 g of powder chitosan in 400 ml of distilled water; it was dissolved while being stirred with an addition of 90 ml of 1 M acetic acid. The solution was made with up to 1 liter of water, and the pH was adjusted to 5.0 by using 1 M sodium acetate. This chitosan substrate contained a 100 mM concentration of sodium acetate and was used only for the chitosanase assay. The high amount of sodium salt contained by this chitosan substrate interfered with high-performance liquid chromatography (HPLC) and thin-layer chromatography (TLC). Therefore, the chitosan substrate for the production of chitosan oligosaccharides and LMW chitosan was prepared as follows: 10 g of chitosan powder was suspended in 400 ml of distilled water and dissolved while being stirred in 5 ml of concentrated HCl. This solution was made with up to 1 liter of water, and the pH was adjusted to 5.0 by using 1 N NaOH.
Screening of chitosanase-producing bacteria and culture conditions.
The soil samples were suspended in sterilized distilled water and spread onto minimal salt (MS)-chitosan agar plates containing 0.5% (wt/vol) colloidal chitosan with MS medium (0.5% yeast extract, 0.2% K2HPO4, 0.1% KH2PO4, 0.07% MgSO4 · 7H2O, 0.05% NaCl, 0.05% KCl, 0.01% CaCl2, 1.3% Bacto Agar [pH 6.8]) and incubated at 30°C for 7 days. After that, halo-forming strains on the MS-chitosan plates were selected as chitosanase producers, and these strains were cultivated in an MS-chitosan broth at 30°C for 5 days on a rotary shaker at 180 rpm. At 1-day intervals, aliquots were removed from the culture broths, and after centrifugation (6,000 × g, 10 min), the supernatants were used for the chitosanase assay and end product analysis of the chitosanase reactions. By means of this screening method, a bacterial strain, no. 44 (see Results), was selected for its high chitosanase activity and high production of long-chained chitosan oligosaccharides. The morphological characteristics of the isolate strain were determined by using Bergey's Manual of Systematic Bacteriology (40), and the physiological characteristics were examined by using the Vitek system and the API 50CHB system (Biomérieux, Inc., St. Louis, Mo.).
Optimization of the chitosanase production.
For the commercial production of chitosanase from Bacillus sp. strain KCTC 0377BP, the fermentation conditions in culture medium were optimized. The bacterium was cultivated in 500-ml Erlenmeyer flasks, each containing 100 ml of medium, at temperature ranges of 25 to 37°C and pH ranges of 4.0 to 7.0; afterwards, the levels of chitosanase produced by the bacterium were compared. After selecting the optimum temperature and pH, various carbon sources (glycerol, glucose, glucosamine, sucrose, soluble starch, and chitosan) and nitrogen sources (yeast extract, polypeptone, peptone, tryptone, NH4Cl, (NH4)2SO4, and corn steep liquor) were tested for maximum chitosanase production. A pilot-scale production of chitosanase was carried out with a 500-liter fermentor (KF-500 L; KoBiotech, Incheon, South Korea) with a working volume of 300 liters.
Purification of chitosanase.
Bacillus sp. strain KCTC 0377BP was cultivated in an MS-soluble starch broth containing 2% soluble starch as a carbon source at 30°C for 3 days on a rotary shaker at 180 rpm. After centrifugation of the culture broth (2 liters) at 6,000 × g for 10 min, the supernatant solution was placed into dialysis tubes. Polyethylene glycol (PEG; average molecular mass, 6,000 Da) powder was scattered around the dialysis tubes, and the PEG-treated dialysis tubes were maintained at 4°C until the tubes had shrunken to 1/10 of their original volumes. The enzyme solution concentrated by PEG was dialyzed against a 10 mM sodium acetate buffer (pH 5.0) and then loaded onto a CM-Toyopearl 650 M column (2.4 by 5 cm; Tosoh, Tokyo, Japan) equilibrated with the same buffer. After the column was washed with the equilibration buffer, the chitosanase was eluted with a 0 to 0.5 M linear gradient of NaCl. The chitosanase fractions were concentrated and diafiltrated by means of ultrafiltration (molecular cutoff, 10 kDa; Millipore, Bedford, Mass.) with a 100 mM sodium phosphate buffer containing 100 mM KCl (pH 5.0). After that, the enzyme was subjected to gel permeation fast-performance liquid chromatography (FPLC) by using a Superose 12HR column (1 by 30 cm; Pharmacia LKB, Uppsala, Sweden) equilibrated with the same diafiltration buffer.
Enzyme assay.
Chitosanase activity was determined by measuring the reducing sugars produced from chitosan. Three hundred microliters of the enzyme was mixed with 300 μl of chitosan substrate, and the reaction mixture was incubated for 10 min at 50°C. In order to stop the reaction and remove the undigested chitosan, 10 μl of 10 N NaOH was added to the reaction mixture, and the mixture was centrifuged at 12,000 × g for 5 min. The reducing sugars in the supernatant were measured by using a dinitrosalicylic acid method (27) with d-glucosamine as the calibration standard. One unit of chitosanase was defined as the amount of enzyme that liberated 1 μmol of d-glucosamine per min under the conditions described above.
SDS-PAGE and activity staining of chitosanase.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was carried out by using the method of Laemmli (22). After SDS-PAGE, proteins were stained with 0.05% Coomassie brilliant blue R-250. For activity staining, the removal of SDS from the gel and the renaturation of the chitosanase were carried out by incubating the gel in a 100 mM sodium acetate buffer (pH 5.0) containing 1% Triton X-100 for 24 h. After several washings with a 10 mM sodium acetate buffer (pH 5.0), 2.5% agarose gel containing 0.2 mg of chitosan/ml in a 10 mM sodium acetate buffer (pH 5.0) was laid onto the gel and incubated for 1 h at 50°C. The chitosanase band was visualized by staining the agarose gel with 0.1% Congo red.
Determination of protein and N-terminal amino acid sequence.
Protein concentration was determined by using the method of Bradford (7) with bovine serum albumin as the calibration standard. The purified chitosanase was separated by SDS-PAGE and transferred onto a polyvinylidene difluoride membrane (Schleicher & Schuell) by means of electroblotting. The blotted protein band was cut out from the membrane and subjected to N-terminal amino acid sequence analysis on an Applied Biosystems model 491 automatic protein sequencer.
Analysis of chitosan oligosaccharides.
Chitosan oligosaccharides were qualitatively analyzed by means of TLC by using a silica gel plate (Kieselgel 60 F254; Merck). After the chitosan oligosaccharides were developed with a solvent system of n-propanol-30% ammonia water (2:1, vol/vol), sugar spots on the plates were visualized by spraying 0.1% ninhydrin into n-butanol-saturated water and by baking them in an oven at 110°C for 10 min. For the quantitative analysis of chitosan oligosaccharides, HPLC analysis was carried out with an Amide NH2-80 column (0.46 by 25 cm; Showa Denko). The reaction products were eluted with a solvent of acetonitrile and 250 mM phosphoric acid water (60:40, vol/vol) at a flow rate of 0.8 ml/min and detected with a refractive index detector. By using authentic d-glucosamine and chitosan oligosaccharides from dimer to heptamer as standards, the amounts of chitosan oligosaccharides in the reaction products were calculated from peak areas.
The average molecular weight (Mav) of the chitosan hydrolysates was estimated by gel permeation HPLC with an Ultrahydrogel 120 column (0.78 by 30 cm; molecular weight, 100 to about 5,000; Waters) and an Ultrahydrogel 250 column (0.78 by 30 cm; molecular weight, 5,000 to about 50,000). Chitosan hydrolysates were eluted with a solvent of 1 M NaCl in 0.2% acetic acid at flow rate of 0.3 ml/min and then detected with a refractive index detector. The Mav of the chitosan hydrolysates was calculated by using pullulans as molecular weight standards.
Cloning of the chitosanase gene.
Chromosomal DNA of Bacillus sp. strain KCTC 0377BP was isolated according to the procedure of Sambrook and Russell (37), partially digested with Sau3AI, and separated on a 0.7% agarose gel. The DNA fragments in the range of 3 to 7 kb were excised from the gel and purified with a QIAGEN (Chatsworth, Calif.) kit. The DNA fragments were ligated with the dephosphorylated BamHI-digested pUC19, and the ligate was used for the transformation of Escherichia coli DH5α with electroporation. E. coli transformants were cultivated on MacConkey agar plates containing 50 μg of ampicillin/ml at 37°C. White colonies, carrying pUC19 with a DNA insert, were toothpicked onto Luria-Bertani agar plates containing 50 μg of ampicillin/ml and incubated for 2 days at 37°C. After that, the agar plates were overlaid with a chitosan-agarose solution containing 1 mg of chitosan/ml and 0.5% agarose in 50 mM sodium acetate (pH 5.0) and incubated for 1 more day. Chitosanase-producing transformants, showing clear halos around the colonies, were selected by staining with 0.1% Congo red.
Nucleotide sequence analysis.
Restriction fragments of the chitosanase gene were subcloned into pUC19 and used for a sequencing reaction with a BigDye sequencing kit (PE Applied Biosystems, Foster City, Calif.) according to the instructions of the manufacturer. The sequencing reactions were analyzed on an Applied Biosystems model 310 automated DNA sequencer. The nucleotide sequence data were analyzed with Lasergene software (DNASTAR, Inc., Madison, Wis.). Similarity searches in GenBank were carried out by using a BLAST program.
Nucleotide sequence accession number.
The nucleotide sequence data have been deposited in GenBank under accession number AF334682.
RESULTS
Isolation of chitosanase-producing bacterium.
More than 500 bacterial strains, which each formed a clear halo on MS-chitosan plates, were isolated as chitosanase producers. Among these bacteria, strain no. 44 was ultimately selected as a chitosanase producer because it showed the highest chitosanase activity and the extracellular chitosanase of this bacterium efficiently produced chitosan oligosaccharides ranging from trimer to octamer, which are known for having strong immunopotentiating activity. The taxonomic identification was based on the following criteria: the organism was gram positive; it was rod shaped (diameter, larger than 1.0 μm) without flagella; it showed spore formation; it was motile, aerobic, catalase positive, negative by Voges-Proskauer test, positive for reduction of nitrate, indole production negative, gelatine liquefaction negative, starch hydrolysis positive, and casein hydrolysis positive; it utilized d-glucose, N-acetylglucosamine, and trehalose, but it did not utilize sucrose, galactose, raffinose, mannitol, maltose, or inositol. It resembles Bacillus megaterium in morphological and biochemical characteristics, but it has some different characteristics from B. megaterium. The strain was tentatively designated as belonging to Bacillus sp., deposited in the Korean Culture Type Collection, and named Bacillus sp. strain KCTC 0377BP.
Production of chitosanase.
When Bacillus sp. strain KCTC 0377BP was cultivated in an MS-chitosan broth (0.5% chitosan) under various temperatures and pH conditions, it showed maximum enzyme production, with chitosanase production of 1.2 U/ml at a temperature of 30°C and a pH of 6.8. However, production of chitosanase increased to a maximum of 4.0 U/ml when chitosan was replaced with 0.5% of soluble starch as a carbon source. When various concentrations of soluble starch from 0.5 to 5% were used in the fermentation medium, the chitosanase activity increased to a maximum of 13.4 U/ml at a 2% concentration, but concentrations higher than 2% of soluble starch repressed chitosanase production. Among the nitrogen sources (0.5%) studied, polypeptone proved to be the best (14.5 U/ml). Chitosanase activity was further increased to a maximum of 45.8 U/ml by using 2.5% of polypeptone. In flask cultivations, the production of chitosanase was increased 38-fold by the optimization both of carbon and nitrogen sources and of culture conditions.
The fermentation conditions optimized in flask cultivation were applied to a pilot-scale production of chitosanase. When Bacillus sp. strain KCTC 0377BP was cultivated in a 500-liter fermentor at 100 rpm of agitation at an air supply rate of 1 vol/vol/min with 0.005% silicon oil, the chitosanase activity and cell growth reached approximately 100 U/ml and an absorbance at 600 nm of 100, respectively. Ultimately, the chitosanase activity was increased 83-fold by the optimization of the culture medium and the culture conditions.
Purification of chitosanase.
Because Bacillus sp. strain KCTC 0377BP produced chitosanase constitutively, the bacterium was cultivated in an MS broth containing 2% soluble starch as a carbon source. After cultivation, the chitosanase was concentrated with PEG from supernatant and purified with CM-Toyopearl 650 M column chromatography and a Superose 12HR column FPLC. The purification steps are summarized in Table 1. The purified enzyme had a specific activity of 1,700 U/mg and showed a single chitosanase band with a molecular mass of 45,000 Da (Fig. 1).
TABLE 1.
Summary of the purification steps of chitosanase from Bacillus sp. strain KCTC 0377BP
| Purification step | Total protein (mg) | Total activity (U) | Sp act (U/mg) | Purification factor | Yield (%) |
|---|---|---|---|---|---|
| Culture supernatant | 162 | 11,700 | 72 | 1 | 100 |
| PEG concentration | 82 | 10,500 | 130 | 2 | 90 |
| CM-Toyopearl 650 M | 7.5 | 8,500 | 1,100 | 15 | 73 |
| Superose 12HR FPLC | 3.1 | 5,400 | 1,700 | 24 | 46 |
FIG. 1.
SDS-PAGE of purified chitosanase of Bacillus sp. strain KCTC 0377BP. (a) Coomassie brilliant blue R-250 staining. (b) Activity staining. Lanes: S, standard marker proteins; 1, culture supernatant solution; 2, CM-Toyopearl 650 M column chromatography; 3, Superose 12HR column FPLC.
Commercial purification processes are somewhat different from the small-scale process described above. In brief, the culture supernatant was separated by continuous centrifugation and was concentrated by ultrafiltration (molecular cutoff, 10 kDa). The concentrated culture supernatant was further purified by CM-Sepharose CL-6B column chromatography and then powdered by means of lyophilization. The specific activity and recovery yield of the final lyophilized enzyme were 1,000 U/mg and 68.0%, respectively.
Substrate specificity of chitosanase.
The chitosanase showed the highest activity for soluble chitosan among the substrates tested, such as chitosan oligosaccharides, colloidal chitin, carboxymethyl cellulose (CMC), and peptidoglycan (Table 2). The enzyme could not hydrolyze chitobiose or chitotriose at all, but it hydrolyzed chitotetraose and longer chitosan oligosaccharides. This means that the chitosanase requires at least four glucosamine residues for catalytic activity. Even after a prolonged overnight reaction of the enzyme, it could not hydrolyze a hexamer of GlcNAc, crystalline chitin, Avicel PH-101 (Fluka, Buchs, Switzerland), and peptidoglycan, but it hydrolyzed colloidal chitin and CMC by 2.8 and 2.5%, respectively, of the activity for soluble chitosan. Resistance of (GlcNAc)6 to the enzyme action indicated that the enzyme has no chitinase activity which cleaves the β-1,4 linkage of GlcNAc-GlcNAc. The little activity of the colloidal chitin could be explained by the manner in which the enzyme cleaved β-1,4 linkages of GlcN-GlcN, GlcN-GlcNAc, and GlcNAc-GlcN, which occur in colloidal chitin, in general, with about a 10% DDA. Interestingly, although the relative activity was less than 3% of the activity for chitosan, the chitosanase showed carboxymethyl cellulase (CMCase) activity. This low CMCase activity of the chitosanase was also confirmed in the recombinant E. coli strain harboring the chitosanase gene (data not shown). Chitosanases with CMCase activity have been found in Myxobacter strain AL-1, B. megaterium P1, and Bacillus cereus S1 (20, 32, 33). In Myxobacter strain AL-1 and B. cereus S1, chitosanase and CMCase activities were of comparable rates (20% of the activity for chitosan). However, 43-kDa chitosanase of B. megaterium P1 possessed very low CMCase activity (0.4% of the activity for chitosan).
TABLE 2.
Substrate specificity of the chitosanase from Bacillus sp. strain KCTC 0377BP
| Substrate | Relative activity (%)a |
|---|---|
| Chitosan oligosaccharides | |
| Chitobiose | ND |
| Chitotriose | ND |
| Chitotetraose | 15.0 |
| Chitopentaose | 16.8 |
| Chitohexaose | 20.0 |
| Chitoheptaose | 23.8 |
| Soluble chitosan | 100.0 |
| Colloidal chitin | 2.8 |
| CMC | 2.5 |
| Peptidoglycan | ND |
| Soluble starch | ND |
A total of 0.5% (wt/vol) of substrates were used for chitosanase assay. Relative activity was expressed as rates relative to the activity of enzyme on soluble chitosan. ND, not detectable.
As described earlier, chitosans have chemical diversity in their DDA. Such diversity of chitosan can influence their susceptibility to chitosanase. When the relative activities of chitosanase for chitosans with various DDA were examined, chitosanase activities were found to have increased with the increase in DDA of chitosan (data not shown). However, the chitosanase still retained high activities against chitosans with a low DDA. When chitosan with a 39% DDA was used, the enzyme showed 72% of the activity for the chitosan (DDA, 94%). This result suggested that GlcNAc residue, occurring in GlcNAc-GlcN or GlcN-GlcNAc linkages in chitosan, does not greatly decrease the hydrolytic activity of the enzyme even though the enzyme has no activity in regard to GlcNAc-GlcNAc linkage.
Reaction pattern of the chitosanase.
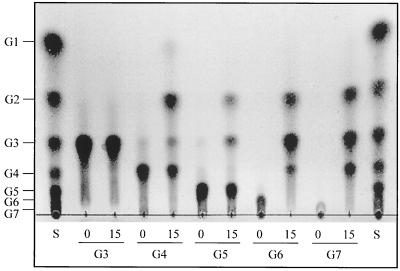
The reaction products of the chitosanase were examined by using soluble chitosan and several chitosan oligosaccharides as the substrates. A TLC analysis revealed that the chitosanase had liberated chitosan oligosaccharides from chitosan, mainly longer than (GlcN)2, by means of endo-type cleavage. The enzyme could not hydrolyze (GlcN)2 (data not shown) or (GlcN)3 at all. (GlcN)4 and (GlcN)5 were resistant to the enzyme action, as shown in Fig. 2. Even after 15 h of incubation of these oligosaccharides, only a fraction of the original (GlcN)4 and (GlcN)5 were hydrolyzed into mainly (GlcN)2 plus (GlcN)2 and (GlcN)3 plus (GlcN)2, respectively. However, chitosan oligosaccharides (GlcN)6 and (GlcN)7 were completely hydrolyzed after 15 h of incubation. These results suggest that for a fast reaction, chitosan substrates should have a chain length equal to or longer than that of (GlcN)6. Figure 2 indicates that (GlcN)6 was cleaved mainly into (GlcN)3 plus (GlcN)3 and to a lesser extent into (GlcN)2 plus (GlcN)4 [subsequently, (GlcN)4→(GlcN)2 + (GlcN)2]; it also indicates that (GlcN)7 was cleaved into (GlcN)3 plus (GlcN)4 [subsequently, (GlcN)4→(GlcN)2 + (GlcN)2]. From the cleavage patterns of the chitosan oligosaccharides, it was suggested that the substrate-binding cleft of the chitosanase accommodated at least six GlcN residues for the superior hydrolysis and that cleavage of the glycosidic linkage occurs preferably at the center of the bound hexameric unit, yielding (GlcN)3 as a main product. The fact that the chitosanase showed higher activity toward chitosan than that of the short-chained oligosaccharides implied that the extra bindings of chitosan, other than six GlcN residues bound to the cleft, can enhance the hydrolysis rate of the enzyme for chitosan.
FIG. 2.
Thin-layer chromatogram of the reaction products of Bacillus sp. strain KCTC 0377BP chitosanase on chitosan oligosaccharides. The reactions were conducted at 40°C for 15 h by using chitotriose (G3), chitotetraose, (G4), chitopentaose (G5), chitohexaose (G6), and chitoheptaose (G7) as the substrates. Lanes: S, standard mixture of chitosan oligosaccharides, ranging from glucosamine (G1) to chitoheptaose; 0, chitosan oligosaccharide not treated with chitosanase; 15, chitosan oligosaccharide treated with chitosanase for 15 h.
Kinetic characterization.
The double reciprocal plot of initial velocity versus the soluble chitosan concentration deviated from linearity at a substrate concentration higher than 2.5 mg of chitosan/ml. From the extrapolation of initial velocities at the concentrations of chitosan from 2.0 to 0.2 mg/ml, kcat and Km values were estimated to be 1,600 s−1 and 1.1 mg of chitosan/ml, respectively.
Effects of temperature and pH.
After incubation at 40 and 45°C for 24 h, the purified chitosanase retained almost its full activity. However, it became inactive at temperatures above 50°C. The half-lives of the chitosanase at 50, 55, and 60°C were 5 h, 10 min, and 5 min, respectively. When the enzyme activities were measured under standard assay conditions (pH 5.0, 10 min) except for temperature, the maximum activity was observed at 60°C. The enzyme was fairly stable in a pH range of 4.0 to 8.0, but it was completely inactive at pH 2.0 within 1 h. The optimum activity of the enzyme was found in a pH range of 4.0 to 6.0. The enzyme can be classified as an acidic enzyme with a moderate thermostability.
Effect of metal ions and other reagents on chitosanase activity.
Chitosanase activity was assayed after the addition of metal ion Mg2+, Cu2+, Ca2+, Zn2+, Mn2+, Ba2+, Fe2+, Hg2+, Co2+, or Ag+ to the reaction mixtures at a final concentration of 1 mM. The metal ions Hg2+ and Mn2+ inhibited chitosanase activity by 68 and 69%, respectively. However, the other metal ions did not significantly affect the enzyme activity. Chitosanase inhibition by Hg2+ is a general characteristic of the chitosanase group, but inhibition by Mn2+ was a peculiar characteristic, because other chitosanases that were reported on were not inhibited by this metal ion. Chitosanase activity dropped to around 50% with the addition of 0.4 M KCl or 0.4 M NaCl.
The N-terminal amino acid sequence.
The N-terminal 20-amino-acid sequence was determined to be A-A-K-E-M-K-P-F-P-Q-Q-V-N-Y-A-G-V-I-K-P. This sequence was 90% identical to the N-terminal sequence of an acidic endocellulase of Bacillus sp. strain KSM-330 (30). However, our chitosanase sequence had no similarity to the N-terminal sequences of other known chitosanases belonging to glycoside hydrolase family 46.
Production of chitosan oligosaccharides.
Chitosan hydrolysis with the enzyme was influenced by the DDA, the viscosity, the concentration of chitosan, the kinds of acid used for dissolving the chitosan, the amount of the enzyme, and other reaction conditions such as pH, temperature, and agitation. In order to obtain the best reaction conditions, these parameters were optimized at a fixed reaction time of 24 h. The optimum pH and temperature for the production of chitosan oligosaccharides were pH 5.0 and 40°C, respectively. With a chitosan concentration in the range of 2.5 to 80 mg/ml, the enzyme showed the highest initial velocity at a chitosan concentration of 5 mg/ml but dropped to 3% of maximum activity at 80 mg/ml. For the purpose of large-scale production of chitosan oligosaccharides, a chitosan concentration of 20 to 40 mg/ml was chosen because a chitosan concentration of 5 mg/ml required a much larger reactor volume and an additional concentration process. Because of the increase of viscosity and substrate inhibition, higher concentrations of chitosan required more enzymes per gram of chitosan. Generally, at 20 to 40 mg of chitosan/ml of solution, 2 to 8 U of enzyme/g of chitosan was needed for oligosaccharide production within 24 h at 40°C and pH 5.0.
Three types of chitosan solution which were solubilized with lactic acid, hydrochloric acid, and acetic acid showed the same productivity as that of chitosan oligosaccharides. The chitosan substrate with a low DDA yielded a lower content of oligosaccharide and also caused many unidentified peaks on an HPLC chromatogram. From an industry point of view, the most important things might be the content and the composition of oligosaccharides. These parameters were well traced by analyzing the Mav and the molecular weight distribution of chitosan hydrolysates by using gel permeation chromatography (GPC). Table 3 shows the changes of the Mav and oligosaccharide composition of the chitosan hydrolysates according to reaction time. This result indicated that the chitosan hydrolysates with desired oligosaccharide composition could be produced by a controlled hydrolysis of chitosan with the aid of GPC analysis. Even though the contents of long-chained oligosaccharides such as hexamer, heptamer, and octamer, which are known to have higher immunostimulating activity, varied according to the degree of hydrolysis of chitosan, our enzyme produced relatively high amounts of these oligosaccharides compared with other commercial oligosaccharides.
TABLE 3.
Change of the Mav, total oligosaccharide content, and oligosaccharide composition of chitosan hydrolysatesa
| Reaction time (h) | GPC analysis
|
Chitosan oligosaccharide composition (%) for chitosan hydrolysate:e
|
Total oligosaccharide (%) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| RTb | Mav | G2 | G3 | G4 | G5 | G6 | G7 | G8 | G9 | ||
| 30 | 13.70 | 2,000 (G10) | 1.7 | 4.5 | 4.6 | 7.2 | 7.8 | 6.7 | 5.0 | 3.7 | 41.2 |
| 36 | 13.90 | 1,590 (G8)c | 1.7 | 5.1 | 6.8 | 10.1 | 10.3 | 7.8 | 5.8 | 3.9 | 51.5 |
| 42 | 14.30 | 1,180 (G6) | 1.8 | 6.8 | 9.6 | 12.9 | 12.3 | 8.3 | 5.6 | 3.0 | 60.3 |
| 48 | 14.35 | 1,120 (G6) | 1.9 | 7.3 | 10.5 | 13.7 | 12.1 | 7.6 | 4.9 | 2.5 | 60.5 |
| 54 | 14.40 | 1,100 (G6) | 2.0 | 9.2 | 12.4 | 14.9 | 12.5 | 7.1 | 3.7 | 1.9 | 63.7 |
| 60 | 14.60 | 1,000 (G5) | 2.3 | 15.4 | 15.6 | 16.5 | 10.5 | 3.9 | 0.9 | NDd | 65.1 |
One hundred milliliters of 40 mg of soluble chitosan/ml was treated with 8 U of chitosanase at 40°C. At the time intervals indicated, samples were taken and average molecular weights were analyzed by GPC with Ultrahydrogel 120, and the oligosaccharide composition was determined by HPLC with an Amide NH2-80 column as described in Materials and Methods.
Retention time (min).
Approximate degree of polymerization of GlcN for each of the Mav values.
ND, not detectable.
G2 to G10, chitosan oligosaccharides from chitobiose (G2) to chitodecaose (G10).
Production of LMW chitosan.
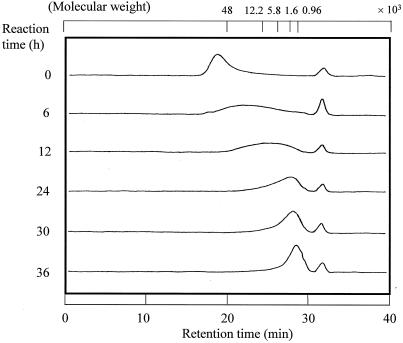
LMW chitosan has advantages over chitosan, such as high solubility in water, low viscosity, and specific biological activities including anticholesterolemic and antimicrobial effects. Our chitosanase could be used for the production of LMW chitosan by controlling the degree of hydrolysis of chitosan. When chitosan was treated with a small amount of chitosanase, the Mav of chitosan hydrolysates decreased with the reaction time (Fig. 3). The LMW chitosans produced by chitosanase had various molecular weight distributions: 21,500 to 1,500 [Mav = 4,800, ∼(GlcN)24], 9,700 to 1,100 [Mav = 2,200, ∼(GlcN)11] and 4,400 to 900 [Mav = 1,500, ∼(GlcN)8]. Solvent fractionation of these chitosan hydrolysates will further narrow down the molecular weight distribution of LMW chitosans.
FIG. 3.
GPC of chitosan hydrolysates. Twenty milligrams of chitosan solution/ml was hydrolyzed by chitosanase (2 U/g of chitosan) at 40°C for 36 h. During the reaction, aliquots were sampled and their Mavs were estimated by GPC (Ultrahydrogel 250; Waters). Molecular weight indicates pullulan standard P-82 (48,000, P-50; 12,200, P-10; 5,800, P-5) and predicted molecular weight (1,600 and 960) from a pullulan molecular weight standard curve.
Production of HA chitosan oligosaccharides.
Direct enzymatic production of chitin oligosaccharides has not yet been reported. The insolubility of chitin in water and the unavailability of a suitable enzyme limit the enzymatic production of chitin oligosaccharides. Because our chitosanase has a high activity toward highly acetylated (HA) chitosan, it could be used for the production of oligosaccharides from HA chitosans with 30 to 50% DDA. When chitosan (39% DDA) was hydrolyzed by the enzyme, oligosaccharides with a Mav of 920 [∼(GlcN)5] were obtained. Of these oligosaccharides, 72% were in the molecular weight range of 670 [∼(GlcN)3] to 1,300 [∼(GlcN)7]. We tentatively designated these oligosaccharides as HA chitosan oligosaccharides. Considering the fact that the chitin oligosaccharides have higher immunostimulating and anticancer activities than chitosan oligosaccharides (42, 45), the HA chitosan oligosaccharides were expected to have higher biological activities than chitosan oligosaccharides.
Cloning, sequencing, and analysis of chitosanase gene.
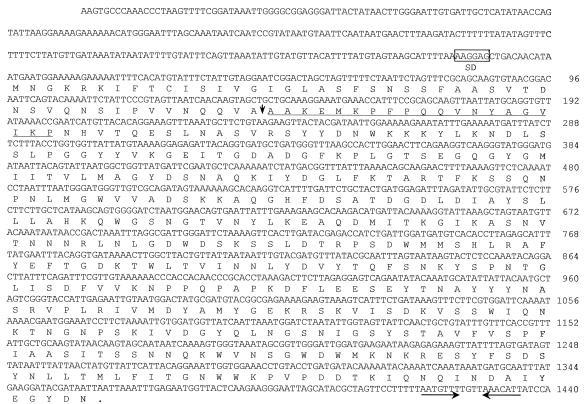
From about 20,000 transformants of E. coli, a transformant showing chitosanase activity on chitosan overlay agarose plates was selected. It contained a recombinant plasmid, pBC27, harboring a 2.7-kb insert DNA. Analysis of the nucleotide sequence of the chitosanase gene (csn45) revealed an open reading frame of 1,362 nucleotides that encodes a protein of 454 amino acids with a predicted molecular mass of 50,691 Da (Fig. 4). A potential ribosome-binding site, AAGGAG, was found upstream of the start codon and the 13-bp palindrome sequence, corresponding to a hairpin loop that was found downstream to the TAA termination codon. The deduced amino acid sequence at positions 48 to 68 is identical to the N-terminal 20-amino-acid sequence of the purified chitosanase. This suggested that the N-terminal 47 amino acids of the unprocessed chitosanase may function as a signal peptide, which is necessary for the extracellular secretion of the chitosanase. The molecular mass of processed chitosanase (407 amino acids) was predicted to be 45,808 Da, which is comparable to the molecular mass of chitosanase (45 kDa) estimated by SDS-PAGE.
FIG. 4.
Nucleotide sequence of the csn45 gene and deduced amino acid sequence of the gene product. The putative ribosome-binding site (AAGGAG) is boxed. The N-terminal amino acid sequencing of the purified chitosanase from Bacillus sp. strain KCTC 0377BP is underlined. The signal peptide cleavage site is shown with a vertical arrow. The putative inverted repeat sequence is indicated by facing arrows with a solid line.
The deduced protein sequence of the chitosanase was compared with entries in the GenBank and SWISS-PROT databases. The chitosanase contains a typical, conserved amino acid sequence, ATDGDLDIAYSLLLAHKQWGSNG, of glycoside hydrolase family 8. Furthermore, the enzyme showed a high sequence similarity with family 8 glycoside hydrolases such as Bacillus sp. strain KSM330 acidic cellulase (30) (76% similarity), a chitosanase-glucanase bifunctional enzyme from“Paenibacillus fukuinensis” (18) (74% similarity), B. circulans WL-12 endo-β-1,3-1,4-glucanase (8) (44% similarity), Clostridium cellulolyticum cellulase C (4) (29% similarity), Clostridium josui cellulase 2 (10) (29% similarity), Clostridium thermocellum endoglucanase A (5) (29% similarity), and Erwinia chrysanthemi cellulase Y (12) (25% similarity). The alignment of family 8 catalytic domains is shown in Fig. 5. The conserved Glu-122 (amino acid position of csn45), which was identified as a catalytic residue in cellulases, was expected to act as a proton donor in catalysis. The Asp-183, conserved in all members of family 8 glycosyl hydrolases, has the potential to participate as a nucleophile in the catalysis of family 8 hydrolases.
FIG. 5.
Alignment of putative catalytic domain of family 8 glycoside hydrolases. Numbers refer to amino acid residues at the start of the respective lines; all sequences are numbered from Met-1 of the peptide. The active sites, such as proton donor and nucleophile, are indicated by asterisks. Symbols (bacterial names followed by accession numbers): CSN_45, Bacillus sp. strain KCTC 0377BP chitosanase (AF334682); BAC_KSM, Bacillus sp. strain KSM330 endoglucanase (P29019); PAE_D2, P. fukuinensis D2 chitosanase (AB006819); BAC_CIR, B. circulans endoglucanase (P19254); CLO_CEL, C. cellulolyticum cellulase C (P37699); CLO_JOS, C. josui cellulase 2 (P37701); CLO_THE, C. thermocellum endoglucanase A (P04955).
The chitosanase of this study has no significant similarity to other known chitosanases of B. circulans MH-K1 (36), Streptomyces sp. strain N174 (11), or Nocardioides sp. strain N106 (24), which were classified as family 46 hydrolases. These family 46 chitosanases have a sequence similarity in the N-terminal regions containing catalytic residues of Glu-22 and Asp-40 (amino acid positions of Streptomyces sp. strain N174 chitosanase). This suggests that the chitosanase of this study has evolved from a different ancestral gene from that of family 46 chitosanases.
DISCUSSION
In order to develop a chitosanase suitable for the production of chitosan oligosaccharides, a bacterium with high chitosanase activity was isolated from soil, and it was designated Bacillus sp. strain KCTC 0377BP. In previous reports, most microorganisms needed chitosan as an inducer for the production of chitosanase, but some microorganisms (20, 23, 39) constitutively produced chitosanase without chitosan. Our strain also produced chitosanase without chitosan as an inducer, and this characteristic has advantages at the fermentation stage because chitosan increases the viscosity of the culture medium and makes production costs high. In an MS-chitosan medium, the bacterium produced chitosanase of approximately 1.2 U/ml, but its productivity increased to about 100 U/ml in the 500-liter fermentor when the culture conditions were optimized. The decreased production of chitosanase in the medium containing chitosan might be due to the inhibitory activity of chitosan on bacterial growth (29).
Because about 5 U of chitosanase was required for hydrolyzing 1 g of chitosan within 24 h at 40°C, 1 liter of culture supernatant (100 U/ml) could hydrolyze 20 kg of chitosan into chitosan oligosaccharides. This process is economically and environmentally sounder than acid hydrolysis, whereby the large-scale production of chitosan oligosaccharides of high yield and high purity is possible.
The molecular mass of the purified chitosanase (45 kDa) is the same or similar to those of other chitosanases from Bacillus species (20, 33, 50); however, the enzymatic characteristics of the purified chitosanase, such as substrate specificity and specific activity, were different from those of other chitosanases from Bacillus species. As shown in Table 4, the specific activities of other purified chitosanases are in the range of 5.84 to 334 U/mg, but the chitosanase of Bacillus sp. strain KCTC 0377BP has the highest specific activity, at 1,700 U/mg. Most of the enzymes show high activity only for the chitosans with a DDA of 70 to 100% (Table 4). However, Bacillus sp. strain KCTC 0377BP chitosanase showed a high relative activity against a broad range of deacetylated chitosan (40 to 100%). This broad range of specificity is advantageous for the production of partially deacetylated chitosan oligosaccharides and for the economical use of chitosan.
TABLE 4.
Comparison of properties of purified chitosanases produced by microorganismse
| Microorganism | Mol mass (kDa)a | Action modef | Optimum pH | Sp. act (U/mg) | Substrate specificity (% DDA)b | Inducibilityc | End productsd | Source or reference |
|---|---|---|---|---|---|---|---|---|
| Bacillus sp. strain KCTC 0377BP | 45 | Endo | 4.0-6.0 | 1,700 | Chitosan (40-100), CMC (2.5) | C | G3-G7 | This study |
| Aspergillus oryzae IAM2660 | 40 | Endo | 5.5 | 17.2 | Chitosan (70-100) | C | Oligomer | 51 |
| 135 | Exo | 5.5 | 38.8 | Chitosan (90-100) | I | G1 | ||
| Nocardia orientalis IFO 12806 | 97 | Exo | 5.5 | 35.8 | Chitosan (100) | I | G1 | 28 |
| Acinetobacter sp. strain CHB101 | 37 | Endo | 5-9 | 334 | Chitosan (70-90) | C | G2, G3 | 39 |
| 30 | Endo | 5-9 | 800 | Glycol chitosan | C | |||
| Enterobacter sp. strain G-1 | 50 | Exo | 7.0 | 5.84 | Chitosan (80) | I | G2 | 49 |
| Myxobacter strain AL-1 | 31 | Endo | 5.0, 6.8 | ND | CMC, chitosan | ND | ND | 32 |
| Matsuebacter chitosanotabidus 3001 | 34 | Endo | 4.0 | 250 | Chitosan (90) | I | G1-G3 | 31 |
| B. cereus S1 | 45 | Endo | 6.0 | 196 | Chitosan (ND), CMC (21.6) | C | G2-G4 | 20 |
| B. megaterium P1 chitosanase A | 43 | Endo | 4.5-6.5 | 154.8 | Chitosan (81), CMC (0.4) | I | Oligomer | 33 |
| Bacillus sp. strain KFB-C108 | 48 | Endo | 6.5 | 110 | Chitosan (ND) | I | G3-G5 | 50 |
Determined by SDS-PAGE analysis with purified enzyme.
Substrate specificity for chitosan and CMC. Data in parentheses indicate appropriate DDA of chitosan for the activity (more than 50% relative activity) and relative activity of CMC for chitosan.
C and I are abbreviations for constitutive and inducible, respectively.
End product of the enzymes using chitosan as the substrate.
ND, not determined.
Exo, exocleavage type; endo, endocleavage type.
The deduced amino acid sequence of csn45 showed a dissimilarity from the reported family 46 chitosanases. However, the active site and activity-related sequences of the enzyme have the same pattern as those of glucanases belonging to glycoside hydrolase family 8 (Fig. 5). Although family 8 glycoside hydrolases are a typical glucanase group, only one of the chitosanases of Paenibacillus fukuinensis was assigned to family 8 in a recent study (18). Therefore, the chitosanase of this study would be the second reported chitosanase to be assigned to glycoside hydrolase family 8. The chitosanase D2 of P. fukuinensis had bifunctional activities of chitosanase and glucanase, which are similar to the activities of the chitosanase of this study. Although our chitosanase and chitosanase D2 of P. fukuinensis both belong to glycoside hydrolase family 8 and both have similar active sites and catalytic modules, our chitosanase shows a higher similarity to family 8 and some different characteristics from chitosanase D2. The D2 enzyme has a unique discoidin domain at its C terminus (this domain is not typical of the family 8 group); however, our chitosanase lacks this domain. According to a truncation study of carboxyl-terminal residues of the D2 enzyme, its discoidin domain is not related to the activity of the enzyme because a D2 enzyme truncated in the discoidin domain showed comparable activity to that of an intact enzyme. A better understanding of the structure-function relationship of these bifunctional enzymes requires more research.
From a commercial standpoint, it is important to select an appropriate acid, such as acetic acid, lactic acid, or hydrochloric acid, for dissolving the chitosan. Chitosan should be dissolved with the appropriate acid for the particular application the chitosan oligosaccharides will be used for (e.g., acetic acid and hydrochloric acid for food and lactic acid for cosmetics industries). All chitosan solutions, which are prepared by using three types of acids, were effectively hydrolyzed by the chitosanase at similar rates, and this showed that the chitosanase under discussion could be used in many chitosan-related industries.
From a commercial standpoint, chitosanase of Bacillus sp. strain KCTC 0377BP has many beneficial characteristics, such as constitutive production, high productivity, high specific activity, simple purification, relatively strong enzyme stability, high activity against a broad range of deacetylated chitosan, and the possibility of using various acids for chitosan solubilization. Furthermore, this enzyme can be used for the production of chitosan oligosaccharides with desired oligosaccharide compositions, LMW chitosan, and HA chitosan oligosaccharides. Our intensive and cooperative study, together with Amicogen, Inc., proved that the chitosanase of Bacillus sp. strain KCTC 0377BP is suitable for the commercial production of chitosan oligosaccharides and other chitosan hydrolysates.
In conclusion, we have characterized the chitosanase of Bacillus sp. strain KCTC 0377BP with respect to its industrial applications, and our results demonstrate that this is a valuable enzyme for the commercial production of chitosan oligosaccharides and other chitosan hydrolysates.
REFERENCES
- 1.Alfonso, C., M. J. Martinez, and F. Reyes. 1992. Purification and properties of two endochitosanases from Mucor rouxii implicated in its cell wall degradation. FEMS Microbiol. Lett. 95:187-194. [Google Scholar]
- 2.Aruchami, M., N. Gowri, and G. Sundara-Rajulu. 1986. Chitin deacetylases in invertebrates, p. 263-265. In R. A. A. Muzzarelli, C. Jeuniaux, and G. W. Gooday (ed.), Chitin in nature and technology. Plenum Press, New York, N.Y.
- 3.Austin, P. R., C. J. Brine, J. E. Castle, and J. P. Zikakis. 1981. Chitin: new facets of research. Science 212:749-753. [DOI] [PubMed] [Google Scholar]
- 4.Bagnara-Tardif, C., C. Gaudin, A. Belaich, P. Hoest, T. Citard, and J. P. Belaich. 1992. Sequence analysis of a gene cluster encoding cellulases from Clostridium cellulolyticum. Gene 119:17-28. [DOI] [PubMed] [Google Scholar]
- 5.Beguin, P., P. Cornet, and J. P. Aubert. 1985. Sequence of a cellulose gene of the thermophilic bacterium Clostridium thermocellum. J. Bacteriol. 162:102-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bosso, C., J. Defaye, A. Domard, A. Gadelle, and C. Pedersen. 1986. The behavior of chitin towards anhydrous hydrogen fluoride. Preparation of β-(1→4)-linked 2-acetamido-2-deoxy-d-glucopyranosyl oligosaccharides. Carbohydr. Res. 156:57-68. [Google Scholar]
- 7.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 8.Bueno, A., C. R. Vazquez de Aldana, J. Correa, and F. del Rey. 1990. Nucleotide sequence of a 1,3-1,4-β-glucanase-encoding gene in Bacillus circulans WL-12. Nucleic Acids Res. 18:4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis, L. L., and S. Bartnicki-Garcia. 1984. The co-ordination of chitin and chitosan synthesis in Mucor rouxii. J. Gen. Microbiol. 130:2095-2102. [DOI] [PubMed] [Google Scholar]
- 10.Fujino, T., S. Karita, and K. Ohmiya. 1993. Nucleotide sequences of the celB gene encoding endo-1,4-β-glucanase 2, ORF1 and ORF2, forming a putative cellulose gene cluster of Clostridium josui. J. Ferment. Bioeng. 76:243-250. [Google Scholar]
- 11.Fukamizo, T., and R. Brzezinski. 1997. Chitosanase from Streptomyces sp. strain N-174: a comparative review of its structure and function. Biochem. Cell Biol. 75:687-696. [DOI] [PubMed] [Google Scholar]
- 12.Guiseppi, A., J. L. Aymeric, B. Cami, F. Barras, and N. Creuzet. 1991. Sequence analysis of the cellulose-encoding celY gene of Erwinia chrysanthemi: a possible case of interspecies gene transfer. Gene 106:109-114. [DOI] [PubMed] [Google Scholar]
- 13.Hadwiger, L., T. Ogawa, and H. Kuyama. 1994. Chitosan polymer sizes effective in inducing phytoalexin accumulation and fungal suppression are verified with synthesized oligomers. Mol. Plant Microbe Interact. 7:531-533. [DOI] [PubMed] [Google Scholar]
- 14.Hirano, S. 1988. Production and application of chitin and chitosan in Japan, p. 37-43. In G. Skjak-Braek, T. Anthonsen, and P. Sandford (ed.), Chitin and chitosan. Elsevier Applied Science, London, United Kingdom.
- 15.Hudson, S. M. 1997. Applications of chitin and chitosan as fiber and textile chemicals, p. 590-599. In A. Domard, G. A. F. Roberts, and K. M. Varum (ed.), Advances in chitin science. Jacques Andre Publisher, Lyon, France.
- 16.Kendra, D. F., D. Christian, and L. A. Hadwiger. 1989. Chitosan oligomers from Fusarium solani/pea interactions, chitinase/β-glucanase digestion of sporelings and from fungal wall chitin actively inhibit fungal growth and enhance disease resistance. Physiol. Mol. Plant Pathol. 35:215-230. [Google Scholar]
- 17.Kendra, D. F., and L. A. Hadwiger. 1984. Characterization of the smallest chitosan oligomer that is maximally antifungal to Fusarium solani and elicits pisatin formation in Pisum sativum. Exp. Mycol. 8:276-281. [Google Scholar]
- 18.Kimoto, H., H. Kusaoke, I. Yamamoto, Y. Fujii, T. Onodera, and A. Taketo. 2002. Biochemical and genetic properties of Paenibacillus glycosyl hydrolase having chitosanase activity and discoidin domain. J. Biol. Chem. 277:14695-14702. [DOI] [PubMed] [Google Scholar]
- 19.Knorr, D. 1991. Recovery and utilization of chitin and chitosan in food processing waste management. Food Technol. 45:114-122. [Google Scholar]
- 20.Kurakake, M., S. Yo-u, K. Nakagawa, M. Sugihara, and T. Komaki. 2000. Properties of chitosanase from Bacillus cereus S1. Curr. Microbiol. 40:6-9. [DOI] [PubMed] [Google Scholar]
- 21.Kurita, K. 1997. Preparation and evaluation of novel types of chitin derivatives, p. 320-327. In A. Domard, G. A. F. Roberts, and K. M. Varum (ed.), Advances in chitin science. Jacques Andre Publisher, Lyon, France.
- 22.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 227:680-685. [DOI] [PubMed] [Google Scholar]
- 23.Lee, H. W., J. W. Choi, D. P. Han, N. W. Lee, S. L. Park, and D. H. Yi. 1996. Identification and production of constitutive chitosanase from Bacillus sp. HW-002. J. Microbiol. Biotechnol. 6:12-18. [Google Scholar]
- 24.Masson, J. Y., I. Boucher, W. A. Neugebauer, D. Ramotar, and R. Brzezinski. 1995. A new chitosanase gene from a Nocardioides sp. is a third member of glycosyl hydrolase family 46. Microbiology 141:2629-2635. [DOI] [PubMed] [Google Scholar]
- 25.Matheson, D. S., B. J. Green, and S. J. Friedman. 1984. Effect of d-glucosamine on human natural killer activity in vitro. J. Biol. Response Modif. 3:445-453. [PubMed] [Google Scholar]
- 26.Mihara, S. 1961. Change in glucosamine content of Chlorella cells during the course of their life cycle. Plant Cell Physiol. 2:25-29. [Google Scholar]
- 27.Miller, G. L. 1959. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 31:426-428. [Google Scholar]
- 28.Nanjo, F., R. Katsumi, and K. Sakai. 1990. Purification and characterization of an exo-β-d-glucosaminidase, a novel type of enzyme, from Nocardia orientalis. J. Biol. Chem. 265:10088-10094. [PubMed] [Google Scholar]
- 29.No, H. K., N. Y. Park, S. H. Lee, and S. P. Meyers. 2002. Antibacterial activity of chitosans and chitosan oligomers with different molecular weights. Int. J. Food Microbiol. 74:65-72. [DOI] [PubMed] [Google Scholar]
- 30.Ozaki, K., N. Sumitomo, and O. Ito. 1991. Molecular cloning and nucleotide sequence of the gene encoding an endo-1,4-β-glucanase from Bacillus sp. KSM-330. J. Gen. Microbiol. 137:2299-2305. [DOI] [PubMed] [Google Scholar]
- 31.Park, J. K., K. Shimono, N. Ochiai, K. Shigeru, M. Kurita, Y. Ohta, K. Tanaka, H. Matsuda, and M. Kawamukai. 1999. Purification, characterization, and gene analysis of a chitosanase (ChoA) from Matsuebacter chitosanotabidus 3001. J. Bacteriol. 181:6642-6649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pedraza-Reyes, M., and F. Gutierrez-Corona. 1997. The bifunctional enzyme chitosanase-cellulase produced by the gram-negative microorganism Myxobacter sp. AL-1 is highly similar to Bacillus subtilis endoglucanases. Arch. Microbiol. 168:321-327. [DOI] [PubMed] [Google Scholar]
- 33.Pelletier, A., and J. Sygusch. 1992. Purification and characterization of three chitosanase activities from Bacillus megaterium P1. Appl. Environ. Microbiol. 56:844-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roberts, G. A. F. 1992. Chitin chemistry. Macmillan Press, Basingstoke, United Kingdom.
- 35.Rupley, J. A. 1964. The hydrolysis of chitin by concentrated hydrochloric acid, and the preparation of low-molecular-weight substrates for lysozyme. Biochim. Biophys. Acta 83:245-255. [DOI] [PubMed] [Google Scholar]
- 36.Saito, J., A. Kita, Y. Higuchi, Y. Nagata, A. Ando, and K. Miki. 1999. Crystal structure of chitosanase from Bacillus circulans MH-K1 at 1.6-Å resolution and its substrate recognition mechanism. J. Biol. Chem. 274:30818-30825. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 38.Shimosaka, M., M. Nogawa, Y. Ohno, and M. Okazaki. 1993. Chitosanase from the plant pathogenic fungus, Fusarium solani f. sp. phaseoli—purification and some properties. Biosci. Biotechnol. Biochem. 57:231-235. [DOI] [PubMed] [Google Scholar]
- 39.Shimosaka, M., M. Nogawa, X. Y. Wang, M. Kumehara, and M. Okazaki. 1995. Production of two chitosanases from a chitosan-assimilating bacterium, Acinetobacter sp. strain CHB101. Appl. Environ. Microbiol. 61:438-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sneath, P. H. A. 1986. Endospore-forming Gram-positive rods and cocci, p. 1104-1139. In P. H. A. Sneath, N. S. Mair, M. E. Sharpe, and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 2. Williams & Wilkins, Baltimore, Md. [Google Scholar]
- 41.Suzuki, K., T. Mikami, Y. Okawa, A. Tokoro, S. Suzuki, and M. Suzuki. 1986. Antitumor effect of hexa-N-acetylchitohexaose and chitohexaose. Carbohydr. Res. 151:403-408. [DOI] [PubMed] [Google Scholar]
- 42.Suzuki, K., A. Tokoro, Y. Okawa, S. Suzuki, and M. Suzuki. 1985. Enhancing effect of N-acetyl-chitooligosaccharides on the active oxygen-generating and microbicidal activities of peritoneal exudate cells in mice. Chem. Pharm. Bull. 36:886-888. [DOI] [PubMed] [Google Scholar]
- 43.Suzuki, S., Y. Okawa, Y. Okura, K. Hashimoto, and M. Suzuki. 1982. Immunoadjuvant effect of chitin and chitosan, p. 210-212. In S. Hirano and S. Tokura (ed.), Chitin and chitosan. Japanese Society of Chitin and Chitosan, Tottori University, Tottori, Japan.
- 44.Tokoro, A., M. Kobayashi, N. Tatewaki, K. Suzuki, Y. Okawa, T. Mikami, S. Suzuki, and M. Suzuki. 1989. Protective effect of N-acetyl chitohexaose on Listeria monocytogenes infection in mice. Microbiol. Immunol. 33:357-367. [DOI] [PubMed] [Google Scholar]
- 45.Tokoro, A., N. Tatewaki, K. Suzuki, T. Mikami, S. Suzuki, and M. Suzuki. 1988. Growth-inhibitory effect of hexa-N-acetylchitohexaose and chitohexaose against Meth-A solid tumor. Chem. Pharm. Bull. 36:784-790. [DOI] [PubMed] [Google Scholar]
- 46.Tokura, K. 1988. Structure and chemical modification of chitin and chitosan, p. 45-50. In G. Skjak-Braek, T. Anthonsen, and P. Sandford (ed.), Chitin and chitosan. Elsevier Applied Science, London, United Kingdom.
- 47.Ueno, K., T. Yamaguchi, N. Sakairi, N. Nishi, and S. Tokura. 1997. Antimicrobial activity by fractionated chitosan oligomers, p. 156-161. In A. Domard, G. A. F. Roberts, and K. M. Varum (ed.), Advances in chitin science. Jacques Andre Publisher, Lyon, France.
- 48.Yabuki, M., A. Uchiyama, K. Suzuki, A. Ando, and T. Fujii. 1988. Purification and properties of chitosanase from Bacillus circulans MH-K1. J. Gen. Appl. Microbiol. 34:255-270. [Google Scholar]
- 49.Yamasaki, Y., I. Hayashi, Y. Ohta, T. Nakagawa, M. Kawamukai, and H. Matsuda. 1993. Purification and mode of action of chitosanolytic enzyme from Enterobacter sp. G-1. Biosci. Biotechnol. Biochem. 57:444-449. [DOI] [PubMed] [Google Scholar]
- 50.Yoon, H. G., S. C. Ha, Y. H. Lim, and H. Y. Cho. 1998. New thermostable chitosanase from Bacillus sp.: purification and characterization. J. Microbiol. Biotechnol. 86:449-454. [Google Scholar]
- 51.Zhang, X. Y., A. L. Dae, X. K. Zhang, and K. Kuroiwa. 2000. Purification and characterization of chitosanase and exo-β-d-glucosaminidase from Koji Mold, Aspergillus oryzae IAM2660. Biosci. Biotechnol. Biochem. 64:1896-1902. [DOI] [PubMed] [Google Scholar]