Abstract
Pre-harvest contamination of forage maize by mycotoxin producing Fusarium species was investigated in the UK in 2011 and 2012. A total of 15 Fusarium species were identified from a collection of 1,761 Fusarium isolates recovered from maize stalks and kernels. This study characterized the diversity of Fusarium species present in forage maize in the UK. The predominant species detected were F. graminearum (32.9%) and F. culmorum (34.1%). Along with those species; F. avenacem, F. cerealis, F. equiseti, F. langsethiae, F. napiforme, F. oxysporum, F. poae, F. proliferatum, F. scripi, F. solani, F. subglutinans, F. tricinctum and, F. verticillioides were occasionally isolated. The trichothecene genotypes for F. graminearum were determined to be 84.9% deoxynivalenol (DON) and 15.0% nivalenol (NIV) while F. culmorum isolates were determined to have 24.9% DON and 75.1% NIV genotypes. A Bayesian model-based clustering method with nine variable number of tandem repeat markers was used to evaluate the population genetic structure of 277 F. graminearum isolates from the maize and wheat in the UK. There were three genetic clusters detected which were DON in maize, NIV in maize and DON in wheat. There were high admixture probabilities for 14.1% of the isolates in the populations. In conclusion, increased maize production in the UK and the high admixture rates in a significant portion of F. graminearum populations in maize and wheat will contribute to a new pathogen population which will further complicate breeding strategies for tolerance or resistance to this pathogen in both crops.
Keywords: Trichothecene, Fusarium graminearum, Wheat, Genetic relationships, Maize, F. verticillioides, F. culmorum
Introduction
Maize is one of the most important crops worldwide, with the cropping area of the 27 member states of the European Union (EU) reaching 9.7 million ha for grain maize and 5.9 million ha for forage maize in 2013. The largest EU maize producers are France, Romania, Germany, Hungary and Italy, which account for 6.1 million ha (FAOSTAT, 2013). With climate change and the establishment of maize varieties tolerant to cooler climates, regions like the UK have seen an increase in maize production. Presently, UK production is at 183 thousand ha (DEFRA, 2014) and is predominantly grown as forage maize on livestock enterprises as a supplement for livestock rations over the winter, with emerging production for bioenergy and grain. The main markets for maize in the EU are for human consumption, animal feed and bioenergy.
Animal pests, weeds and pathogens impact yield and quality of maize (OERKE, 2006) with ear, stalk and roots rots from Fusarium species considered to be the most economically significant in the EU (Meissle et al., 2010). The predominant Fusarium species associated with ear and stalk rots in the EU are F. graminearum followed by F. verticillioides, F. proliferatum and F. culmorum (Meissle et al., 2010). These Fusarium species are also capable of producing mycotoxins which contribute to pre-harvest contamination of human food and animal feed impacting health (Desjardins, 2006).
While maize is an emerging crop in the UK, the predominant arable crop is wheat, which accounts for 1.9 million ha (DEFRA, 2014). One of the most devastating diseases of wheat, and small grain cereals, is Fusarium head blight (FHB). The most commonly recovered Fusarium species in small grain cereals are F. graminearum and F. culmorum (Osborne & Stein, 2007). Control of the disease is focused on agricultural practices, fungicides and cultivar resistance. Fusarium surveys in cereals in Belgium (Waalwijk et al., 2003) and France (Assemat et al., 1996; Ioos, Belhadj & Menez, 2004) have found that F. graminearum is replacing F. culmorum as the predominant Fusarium species for FHB. Surveys of Fusarium species in maize is commonly done in European maize communities with recent studies in Belgium (Scauflaire et al., 2011), Germany (Goertz et al., 2010), Spain (Arino et al., 2007) and Switzerland (Eckard et al., 2011). However, there are presently no surveys being undertaken to determine the Fusarium species present in UK maize.
The major mycotoxin type of F. graminearum is the trichothecene (TCT) type-B mycotoxin class of fungi capable of producing deoxynivalenol (DON) and its derivatives (3AcDON, 15AcDON) or nivalenol (NIV) (Desjardins, 2006). The NIV-producing isolates of F. graminearum have been found to be more aggressive in maize than the DON-producing isolates (Carter et al., 2002). The emergence of NIV-producing populations has been found in regions with maize and wheat production (Pasquali et al., 2010; Sampietro et al., 2011). This has been attributed to maize commonly being rotated with wheat as a part of integrated pest management practices. Typically, maize residues remaining on the soil surface and are present for the following crop. These maize residues may be infested with F. graminearum from which ascospores are released to initiate infection of wheat under favorable conditions (Meissle et al., 2010). Research on the biology of F. graminearum is needed to understand the infection process and pathogenesis in the life cycle of this pathogen to develop control methods to reduce infection in wheat and other cereals.
Understanding that the natural boundaries and climates of agricultural regions are contributing factors to pathogen potential is an important area to focus research with Fusarium species. The UK has a temperate climate with high humidity year round and is separated from Europe by the English Channel and the North Sea. These natural climates and boundaries may select for pathogen profiles that are potentially unique compared to the agricultural regions found in continental Europe. Even with the widely understood effects that Fusarium species have on maize production in terms of yield and quality, the knowledge available on the occurrence of these species in UK maize is absent. Additionally, little is known about the population subdivision of F. graminearum in the UK based on differences of host species, mycotoxin profiles and pathogen dynamics in wheat and maize.
Therefore, the aim of this study was to determine the Fusarium species and the mycotoxin profiles of those species that are naturally occurring in forage maize in geographically diverse fields in the UK. Sampling strategies of maize would focus on identifying predominant mycotoxin Fusarium species and emerging Fusarium species in UK maize over a two year period. To investigate host specialization and pathogen dynamics of F. graminearum, polymorphic allele data was obtained by nine variable tandem repeat markers (VNTR) from 277 isolates of F. graminearum recovered from UK maize and wheat, genotyped for TCT-type and analyzed by a Bayesian model-based clustering method and population genetic software.
Materials and Methods
Maize sample collection
Sampling of maize plants was undertaken in 2011 and 2012 in the UK to assess the diversity of Fusarium species at harvest in five major production regions (North, North Central, South East, South West and West Central) at fifteen fields. The fifteen forage maize fields sampled were trail grounds for maize variety testing which were managed by Limagrain and the British Society of Plant Breeders (Table 1). Sixty-six plants were sampled at each field from the surrounding maize around each trail ground which allowed for the identification of Fusarium species present at greater than 5%. Data on the mean daily temperature and precipitation was acquired from the closest weather station to each sampling site from the British Atmospheric Data Centre (National Centre for Atmospheric Science, 2012).
Table 1. Sample location of maize samples taken in the UK in 2011 and 2012.
Sixty-six plants were harvested at each site when the maize plants had a dry matter content between 28 and 32%.
| UK maize region | Sample site | County | Coordinates | Maize cultivar | Year |
|---|---|---|---|---|---|
| North | 1 | Yorkshireb | 54°19″40′N, 1°27″76′W | NK Smile | 2011/2012c |
| North Central | 2 | Nottinghamshirea | 53°13″48′N, 0°55″80′W | Artist | 2011 |
| 3 | Lincolnshirea | 53°0″79′N, 0°45″02′W | Artist | 2011 | |
| 4 | Shropshireb | 52°43″06′N, 02°23″46′W | ES Picker | 2011/2012d | |
| South East | 5 | Sussexb | 50°50″55′N, 0°11″33′E | Kokon | 2011 |
| 6 | Kentb | 51°15″39′N, 1°05″05′E | Kougar | 2012 | |
| South West | 7 | Somerseta | 51°5″33′N, 2°59″17′W | ES Regain | 2011 |
| 8 | Devonb | 50°39″61′N, 3°20″00′W | ES Marco | 2011/2012e | |
| West Central | 9 | Oxfordshirea | 51°56″86′N, 1°12″79′W | MAS17E | 2011 |
| 10 | Gloucestershireb | 51°40″35′N, 2°37″47′W | Bonapart | 2011 | |
| 11 | Herefordshireb | 52°4.74′N, 2°56.31′W | Claxxon | 2011/2012c |
Notes.
Limagrain maize trials.
BSPB forage maize list trials.
150 m North East of 2011 field.
100 m North West of 2011 field.
100 m East of 2011 field.
The maize was sampled when dry matter (DM) content was between 28 and 32% as determined by five randomly sampled plants at each field. DM of 28–32% occurs when the maize plant has entered into the reproductive stages of plant development and the kernel begins to turn yellow on the outside while the inner fluid becomes milky white due to accumulating starch. The harvested maize plants were commercial hybrids naturally infected with Fusarium. The maize was precision sown in rows with 0.75–0.80 m spacing between rows with an expected density of 100,000 plants per hectare. Sampled plants were selected in a diagonal manner beginning at an arbitrary point at the edge of the field and working towards the center. Each plant was harvested at the base of the stalk, 2–3 cm above the ground, and retained in separate collection bags to avoid cross contamination of samples. Plant samples were stored at 4 °C until they were analyzed for Fusarium colonization.
Isolation and identification of Fusarium species in maize
Fusarium isolates from maize were recovered from kernels, and stalk tissues utilizing a standardized isolation technique. In a laminar flow bench, maize stalk sections were taken in 5 mm sections from either the base of the plant, below the primary ear or above the primary ear for the selected plant. Stalk samples were longitudinally split and tissues of the vascular bundle were removed aseptically from the innermost leaf of the stalk. Three pieces of stalk tissue from the same plant were transferred to 90-mm Petri plates with Spezieller Nährstoffarmer Agar (SNA). Under the same aseptic conditions as the stalk samples, maize kernels were manually removed from the ear and ten kernels were randomly selected for analysis. The maize kernels were surface sterilized in sodium hypochlorite (1% available chlorine) for 2 min and rinsed twice with sterile distilled H2O and dried on sterilized paper. After drying completely the maize kernels were transferred to 90-mm Petri plates with SNA at 5 kernels per plate. All samples were incubated at 20 °C under 12 h near-UV and 12 h dark cycle for 3–5 days. Hyphal tips of potential Fusarium colonies were transferred aseptically to new 90-mm Petri plates of SNA and incubated under the same conditions for 7–10 days. A conidia and mycelium solution from each sample was preserved in 1.5 ml of 25% glycerol-water and stored at −80 °C. All potential Fusarium isolates from the maize samples were retained and identified to species.
Fusarium species were initially identified to species based on the morphological characteristics (Leslie & Summerell, 2006) of the macroconidia, microconidia and general mycelium presentation from a single spore isolate grown for 7–10 days on SNA with an Olympus BH-2 light microscope. When macroconidia, microconidia and mycelial characteristics from SNA were insufficient for identifying the species further examination of the samples were done on different agar media. Potato Dextrose Agar (PDA) was used to identify colony pigment characteristics of aerial mycelium on the agar (Leslie & Summerell, 2006). Carnation Leaf Agar (CLA) was used to identify macroconidia, chlamydospores and the presentation of aerial mycelium.
DNA was amplified in multiplex PCR by Goertz et al., (2010) to confirm the identity of ten Fusarium species. The ten Fusarium species identified in the multiplex PCR are Fusarium avenaceum, F. culmorum, F. cerealis, F. equiseti, F. graminearum, F. oxysporum, F. poae, F. proliferatum, F. subglutinans and F. verticillioides. Single spore isolates were grown on PDA plates for 3 days and genomic DNA was extracted from harvested aerial mycelia with a cetyltrimethylammonium bromide (CTAB) method (Leslie & Summerell, 2006). For each amplification reaction 2 µl of template DNA (40 ng/µl) was mixed with 18 µl PCR master mix containing 12 µM of each primer, 6.0 µl of 10× PCR buffer plus 20 mM MgCl2 (Roche, Diagnostics, Mannheim, Germany), 400 µM dNTPs, 0.16 µl of Fast start Taq DNA polymerase (Roche Diagnostics, Mannheim, Germany). The PCR consisted of one cycle of 5 min at 94 °C; 35 cycles at 30 s at 94 °C (denaturing), 30 s at 60 °C (annealing), 1 min at 72 °C (extension); and one cycle of 10 min at 72 °C. After the PCR, 5 µl of the amplification product was used to reveal the presence/absence under UV light on 1.5% agarose gel (Sigma Aldrich, Darmstadt, Germany) containing ethidium bromide (0.5 µg/ml) and scored relative to a 100-bp HyperLadder (Bioline, London, UK).
Trichothecene genotyping for F. graminearum and F. culmorum
Multiplex PCR assays were used to characterize the trichothecene genotypes NIV, 3AcDON and 15AcDON based on the polymorphisms in the Tri5 gene cluster of the trichothecene biosynthetic pathway. Targeting more than one tri gene was undertaken to reduce misidentification due to gene cluster recombination (Pasquali et al., 2011). The first multiplex assay (Quarta et al., 2006) used primer pairs based on the Tri3 and Tri7 genes. The primers for the Quarta et al. multiplex assay are Tri3F971, Tri3F1325, Tri3R1679, Tri7F340, TriR965 with three DNA fragments for 15AcDON (708 bp), 3AcDON (354 bp) and NIV (625 bp). The second multiplex assay (Ward et al., 2008) used primer pairs based on the Tri12 gene. The primers for the Ward et al. multiplex assay are 12CON, 12NF, 12-15F, 12-3F with three DNA fragments for 15AcDON (670 bp), 3AcDON (410 bp) and NIV (840 bp). Both multiplex PCR reactions for determining the trichothecene chemotype were done in 20 µl reaction volumes with 30 ng DNA as described for Fusarium species-specific assays (Goertz et al., 2010). The amplification protocol consisted of one cycle of 5 min at 95 °C; 35 cycles at 30 s at 95 °C (denaturing), 30 s at 55 °C (annealing), 1 min at 72 °C (extension); and one cycle of 10 min at 72 °C. After the PCR, the amplification product was revealed as described for Fusarium species-specific assays (Goertz et al., 2010).
Statistical analysis
The incidence of Fusarium species was determined from the predicted mean at each field from a restricted maximum likelihood (REML) analysis to assess the significance of regional locations, chemotypes, and differences between years using Genstat (GenStat, 2011). Multiple strains of the same species recovered from the same plant were assessed qualitatively as their independence could not be critically assessed.
Sampling and isolation of F. graminearum in wheat
For the evaluation of the population structure of F. graminearum in maize and wheat the isolates from wheat were recovered from wheat kernel samples which were harvested in 2012 from wheat producers in the UK. A total of thirty-two isolates were recovered from thirty-two fields with one isolate representing each field. The locations of the UK fields are identified in an EU survey of Fusarium from 2001 to 2013 (Pasquali et al., 2016). The wheat kernels were screened for F. graminearum by surface sterilizing the kernels in sodium hypochlorite (0.37% available chlorine) with 0.1% Tween20 for 5 min and then rinsing twice with sterile distilled water and dried on sterilized paper. 20 wheat kernels per sample were then transferred to 9 cm Petri plates with SNA. Wheat kernel samples were then incubated under the same conditions as maize samples with F. graminearum isolates identified as described above.
Population genetics analysis with variable number tandem repeat markers
Nine VNTR markers (Suga, Gale & Kistler, 2004) were used for the analyses of population genetic structure in the F. graminearum populations which were; HK630, HK913, HK917, HK957, HK967, HK977, HK1043, HK1059 and HK1073. There were 245 F. graminearum isolates from the maize collection and 32 isolates from wheat collection used for the population genetics analysis. VNTR amplifications were performed with 20 ng of DNA per reaction in 10 µl of master mix as described in the Fusarium species-specific assays. PCR was performed as described by Ward et al. (2008) which consisted of 2 min at 95°C; 25 cycles of 1 min at 95 °C, 1 min at 59 °C and 1 min at 72 °C with a final extension stage of 10 min at 72 °C. Reaction products were scored relative to GS500 ROX (Applied Biosystems) internal size standard using an ABI 3730xl Genetic analyzer with Genemapper 4.0 software. Irresolvable alleles and missing data were treated as missing data.
Genetic diversity and FST based genetic distances (Slatkin, 1995) were estimated with ARELQUIN 3.5 (Excoffier, Laval & Schneider, 2005). Pairwise FST distance estimates were determined based on the number of different alleles and the significance of pairwise FST estimates using 1,000 permutations. The analysis of the admixture was performed using Bayesian method with the software STRUCTURE 2.3.4 (Pritchard, Stephens & Donnelly, 2000). The estimation of the number of populations was calculated based on the second order rate of change of the likelihood (Evanno, Regnaut & Goudet, 2005).
Results
Incidence study in maize
The two year maize sampling resulted in a collection of 1,761 Fusarium isolates which were identified to species from the sampling of 2,970 maize stalk pieces (967 Fusarium isolates) and 9,900 maize kernels (794 Fusarium isolates). There were only three ears and one stalk sample that had observable Fusarium mycelium out of all of the samples evaluated. This study identified fifteen Fusarium species in maize with the predominant species being: F. graminearum (32.9%), F. culmorum (34.1%), F. solani (19.8%), F. cerealis (3.5%), F. avenaceum (3.0%) and F. poae (2.8%). Other Fusarium species identified in the collection were: F. oxysporum (1.2%), F. verticillioides (1.1%), F. proliferatum (0.4%), F. tricinctum (0.3%), F. subglutinans (0.3%), F. langsethiae (0.2%), F. napiforme (0.2%), F. equiseti (0.2%) and one isolate of F. scripi (Table 2). The species-specific assay for F. avenaceum (Turner et al., 1998) may not differentiate between F. avenaceum and the closely related F. tricinctum (Kulik, 2008; Tan & Niessen, 2003). Differentiation of F. avenaceum and F. tricinctum was not always conclusive with macroconidia, therefore we confirmed the species based on differences in the formation of microconidia.
Table 2. Percentage incidence of Fusarium species recovered from UK maize in 2011 and 2012.
| UK maize region | Sample site | Year | F. avenaceum | F. cerealis | F. culmorum | F. equiseti | F. graminearum | F. langsethiae | F. napiforme | F. oxysporum | F. poae | F. proliferatum | F. solani | F. subglutinans | F. tricinctum | F. verticillioides |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| North | 1 | 2011K | 1.5 | 1.5 | 6.1 | – | 15.2 | – | 1.5 | 1.5 | 6.1 | – | 15.2 | – | – | – |
| 2011S | 9.1 | 1.5 | 22.7 | – | 66.7 | – | – | – | – | – | 22.7 | – | – | – | ||
| 2012K | – | – | – | – | 4.5 | – | – | – | – | – | 15.2 | – | – | – | ||
| 2012S | 3.0 | 7.6 | 34.8 | – | 27.3 | – | – | – | – | – | 24.2 | – | – | – | ||
| North Central | 2 | 2011K | 6.1 | 1.5 | 39.4 | – | 48.5 | – | – | – | 6.1 | – | 19.7 | – | – | – |
| 2011S | 6.1 | – | 39.4 | – | 16.7 | – | – | 1.5 | – | – | 15.2 | – | – | – | ||
| 3 | 2011K | 7.6 | 1.5 | 16.7 | – | 34.8 | – | 1.5 | – | 1.5 | – | 6.1 | – | – | – | |
| 2011S | 6.1 | 3.0 | 25.8 | – | 24.2 | – | – | 1.5 | – | – | 24.2 | – | 1.5 | 1.5 | ||
| 4 | 2011K | – | 16.7 | 51.5 | – | 6.1 | – | – | – | – | – | 3.0 | 1.5 | – | — | |
| 2011S | – | 6.1 | 54.5 | – | 4.5 | – | – | 1.5 | – | – | 9.1 | – | – | – | ||
| 2012K | – | – | 7.6 | – | 6.1 | – | – | – | – | – | 45.5 | – | – | – | ||
| 2012S | – | 1.5 | 34.8 | – | 19.7 | – | – | – | – | – | 54.5 | – | – | – | ||
| South East | 5 | 2011K | – | 1.5 | 18.2 | – | 28.8 | – | – | – | 1.5 | – | 9.1 | – | – | – |
| 2011S | 1.5 | 3.0 | 28.8 | 1.5 | 48.5 | – | – | – | 3.0 | – | 7.6 | – | – | – | ||
| 6 | 2012K | – | – | – | – | 6.1 | – | – | – | – | – | 6.1 | – | – | – | |
| 2012S | – | – | 10.6 | – | 37.9 | – | – | – | – | – | 36.4 | – | – | – | ||
| South West | 7 | 2011K | 3.0 | 3.0 | 13.6 | – | 10.6 | 1.5 | – | – | 9.1 | 1.5 | 6.1 | – | – | – |
| 2011S | – | 1.5 | 27.3 | – | 22.7 | – | – | – | – | – | 12.1 | – | – | 1.5 | ||
| 8 | 2011K | 1.5 | 1.5 | 37.9 | – | 33.3 | – | – | – | 3.0 | – | 9.1 | – | – | – | |
| 2011S | 1.5 | 3.0 | 31.8 | – | 39.4 | – | – | – | – | – | 7.6 | – | – | – | ||
| 2012K | – | – | – | – | – | – | – | – | – | – | – | – | – | – | ||
| 2012S | – | 3.0 | 9.1 | – | 25.8 | – | – | – | – | 1.5 | 43.9 | – | – | – | ||
| West Central | 9 | 2011K | – | 1.5 | 10.6 | – | 54.5 | – | – | 1.5 | 13.6 | 3.0 | 18.2 | – | – | – |
| 2011S | 3.0 | 4.5 | 28.8 | – | 47.0 | 1.5 | – | – | – | – | 10.6 | – | – | – | ||
| 10 | 2011K | – | 6.1 | 10.6 | 1.5 | 18.2 | – | – | – | 3.0 | 1.5 | 4.5 | – | – | – | |
| 2011S | 1.5 | 4.5 | 16.7 | 0.0 | 33.3 | – | – | – | – | 1.5 | 1.5 | – | – | – | ||
| 11 | 2011K | 4.5 | 4.5 | 31.8 | – | 6.1 | 1.5 | 1.5 | 1.5 | 4.5 | – | 9.1 | – | – | 1.5 | |
| 2011S | 10.6 | 1.5 | 45.5 | – | 3.0 | – | – | 1.5 | – | – | 19.7 | – | 1.5 | – | ||
| 2012K | 1.5 | 1.5 | 3.0 | 1.5 | – | – | – | – | – | – | 7.6 | – | – | – | ||
| 2012S | 4.5 | – | 31.8 | – | 30.3 | – | – | – | – | – | 27.3 | – | – | – | ||
| Total 2011K | 2.4 | 3.9 | 23.6 | 0.2 | 25.6 | 0.3 | 0.5 | 0.5 | 4.8 | 0.6 | 10.0 | 0.2 | 0.2 | 0.2 | ||
| Total 2011S | 3.9 | 2.9 | 32.1 | 0.2 | 30.6 | 0.2 | – | 0.6 | 0.3 | 0.2 | 13.0 | – | 0.9 | 0.3 | ||
| Total 2012K | 0.3 | 0.3 | 2.1 | 0.3 | 3.3 | – | – | – | – | – | 14.8 | – | – | – | ||
| Total 2012S | 1.5 | 2.4 | 24.2 | – | 28.2 | – | – | – | – | 0.3 | 37.3 | – | – | – | ||
Notes.
- K
- Kernel tissue
- S
- Stalk tissue
- –
- no Fusarium spp
1 Yorkshire; 2 Nottinghamshire; 3 Yorkshire; 4 Shropshire; 5 Sussex; 6 Kent; 7 Somerset; 8 Devon; 9 Oxfordshire; 10 Herefordshire; 11 Gloucestershire.
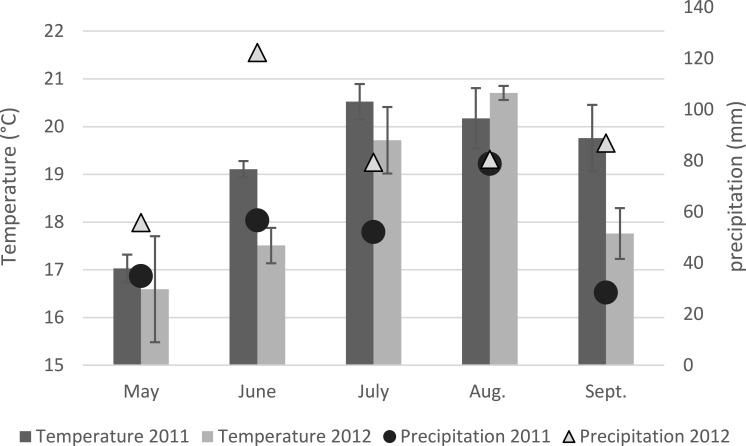
Across all fields sampled in the UK, the incidence of Fusarium colonized maize plants with at least one Fusarium isolate recovered in either stalk of kernels was 85% ± 2.7 SD for 2011 and was 86% ± 0.08 SD for 2012. F. solani regularly recovered as a saprophyte was found to have co-occurred with another Fusarium species for 85% ± 0.18 SD of the samples for both years. Of the 990 maize plants sampled, F. graminearum (44.0% ± 0.14 SD) or F. culmorum (31.0% ± 0.16 SD) were present in 63.9% ± 0.12 SD of the plants. The incidence of F. graminearum and F. culmorum between fields was insignificant within the same year. The incidence per field of F. graminearum was significantly (P = 0.002) greater in 2011 than in 2012 as was the incidence of both F. graminearum and/or F. culmorum (P < 0.001). The incidence per field by F. culmorum was not significantly (P = 0.139) different between 2011 and 2012. The mean maximum temperatures and mean precipitation for the sites in Yorkshire, Shropshire, Devon and Gloucestershire are shown in Fig. 1 for the growing seasons in 2011 and 2012.
Figure 1. Mean maximum temperatures and precipitation on the four fields sampled in the UK in 2011 and 2012.
Trichothecenes for F. graminearum and F. culmorum
Trichothecene types for F. graminearum and F. culmorum isolates in maize had consistent results for all isolates in both multiplex PCR assays. The F. culmorum and the F. graminearum isolates from maize chemotypes of DON (3AcDON-type and 15AcDON-type) and NIV-type are shown in Table 3. The F. graminearum isolates were predominantly the 15AcDON-type (84.1%) followed by NIV-type (15.0%) and three isolates of 3AcDON-type. The F. culmorum isolates were predominantly the NIV-type (75.1%) followed by 3AcDON-type (24.0%) and three isolates of 15AcDON-type. The F. graminearum isolates from wheat used in the population genetic study were all determined to be 15AcDON-type.
Table 3. Geographical origin, year, mean number of isolates per site, species and trichothecene genotype of Fusarium species from maize.
| Maize growing area | Year | Mean isolates/field | F. culmorum | Mean isolates/field | F. graminearum | ||||
|---|---|---|---|---|---|---|---|---|---|
| 3AcDON | 15AcDON | NIV | 3AcDON | 15AcDON | NIV | ||||
| North | 2011a | 19 | 8 | 0 | 11 | 48 | 0 | 47 | 1 |
| 2012a | 23 | 0 | 0 | 23 | 21 | 0 | 20 | 1 | |
| North Central | 2011c | 50.7 | 11.7 | 0.3 | 38.7 | 28.7 | 0.3 | 24.7 | 3.3 |
| 2012a | 28 | 6 | 0 | 22 | 16 | 1 | 16 | 0 | |
| South East | 2011a | 32 | 9 | 0 | 23 | 42 | 1 | 37 | 4 |
| 2012a | 7 | 1 | 0 | 6 | 29 | 0 | 26 | 3 | |
| South West | 2011b | 36 | 15.5 | 0 | 20.5 | 33.5 | 0 | 14 | 19.5 |
| 2012a | 6 | 0 | 0 | 6 | 17 | 0 | 17 | 0 | |
| West Central | 2011c | 31.7 | 6 | 0.7 | 25 | 35.3 | 0 | 32.3 | 3 |
| 2012a | 25 | 3 | 0 | 22 | 21 | 0 | 19 | 2 | |
| All regions | 2011d | 37 | 10.1 | 0.3 | 26.6 | 34.8 | 0.2 | 28.3 | 6.3 |
| 2012d | 17.8 | 2 | 0 | 15.8 | 21 | 0.2 | 19.6 | 1.2 | |
Notes.
One field sampled.
Two fields sampled.
Three fields sampled.
Mean all fields for year.
- 3AcDON
- 3,acetyldeoxynivalenol
- 15AcDON
- 15,acetyldeoxynivalenol
- NIV
- Nivalenol
Population structure and diversity of F. graminearum populations
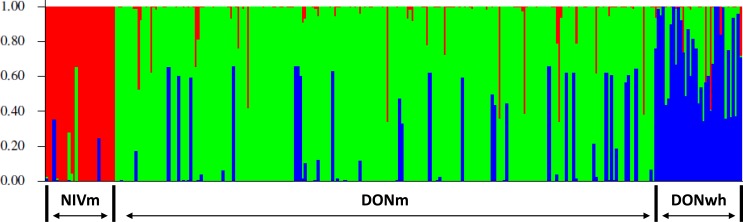
Three genetic clusters (K = 3) was determined to capture the major genetic structure in our data set based on the highest (<10). The three clusters were determined from the evaluation of the genetic variation based on the VNTR loci utilizing a Bayesian clustering method for admixture in the 245 (217 DON-type and 28 NIV-type) F. graminearum isolates from maize and the 32 DON-type isolates from wheat. The clusters corresponded and were organized based on the host species and the TCT-type of the isolates. The three groups were NIV-type from maize (NIVm), 15AcDON-type from maize (DONm) and 15AcDON-type from wheat (DONwh). The admixture estimates for each isolate is shown in Fig. 2. The pairwise FST distance estimates indicated significant differences between the groups based on trichothecene chemotype; NIVm and DONm (FST =0.2207, P < 0.001) and NIVm and DONwh (FST =0.2038, P < 0.001). There were also differences between host species with the same trichothecene chemotype; DONm and DONwh (FST =0.033, P < 0.05).
Figure 2. Representation of admixture estimates based on VNTR data for F. graminearum isolates from UK wheat and maize.
Red, NIV maize; green, 15AcDON maize; blue, 15AcDON wheat.
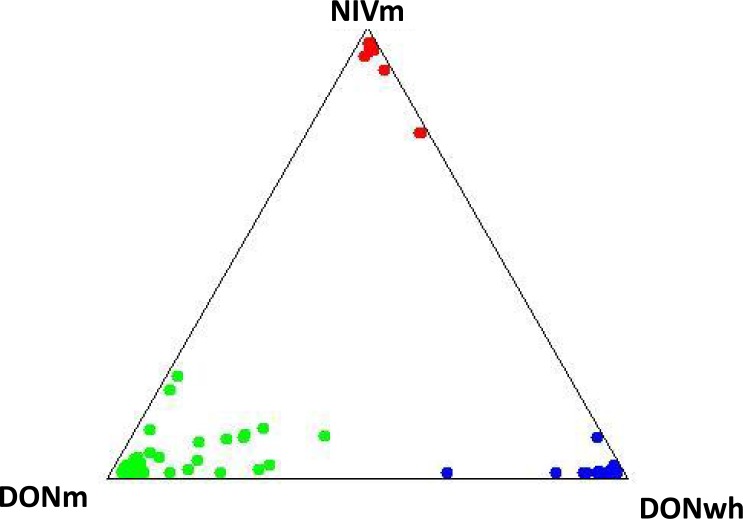
Admixture probabilities greater than 95% were obtained for 39 of the 277 isolates analyzed. Bayesian posterior mean estimates for the proportion of the genome for the DONm isolates that is derived from NIVm ranged from 13 to 98%. DONwh isolates had a significant (P < 0.05) admixture with genetic material shared with DONm isolates. The DONm isolates with high admixture shared a significant (P < 0.05) amount of genetic material with NIVm isolates. A visual representation of isolate distribution based on VNTR data for the F. graminearum populations minus the isolates with admixture probabilities greater than 95% is shown in Fig. 3.
Figure 3. Isolate distribution for F. graminearum isolates.
Red, NIV maize; green, 15AcDON maize; blue, 15AcDON wheat. Does not include isolates with admixture probabilities greater than 95%.
Discussion
Fusarium is a major pathogen of maize and wheat responsible for severe crop loss and mycotoxin contaminations (Charmley et al., 1995; Logrieco et al., 2003). In the present study, we identified the biodiversity of Fusarium species from maize in the UK. An extensive collection of 1,761 isolates identified 15 species and characterized the mycotoxin profiles of the predominant species of F. graminearum and F. culmorum. The prevalent TCT-types of the predominant species were 15AcDON-type and NIV-type for F. graminearum and 3AcDON-type and NIV-type for F. culmorum. We also identified unique isolates of F. culmorum with the 15AcDON-type, F. graminearum with the 3AcDON-type and F. verticillioides. F. verticillioides is a species that produces the mycotoxin T2 and is typically found in warmer maize growing regions in Europe. Additionally, we determined three subpopulations of F. graminearum isolates from maize and wheat based on trichothecene profile and host from population genetic analysis approaches. We believe that this study provides comprehensive information on important Fusarium species and their associated mycotoxins for maize and wheat grown in the UK.
Environmental factors and host species have a strong impact on the occurrence of a specific chemotype and the incidence of Fusarium species. The distribution of Fusarium species in maize is influenced by optimal climatic conditions, pathogenicity and competition between other fungi (Doohan, Brennan & Cooke, 2003). Year-to-year variability is one type of environmental factor identified in the incidence of Fusarium species as demonstrated in recent EU maize surveys (Dorn et al., 2009; Goertz et al., 2008; Scauflaire et al., 2011). In those studies the prevalence of species varied year-to-year and was believed to be associated with the differences in climatic conditions between years. In this study the warm and drier conditions in June, July and September in 2011 compared to the same fields in 2012 may have contributed to year-to-year incidence differences for F. graminearum and other Fusarium species in those fields. The growing conditions in 2011 were characterized by dry conditions in the early stages of maize development with moderate precipitation and warm weather in the later stages of plant development until anthesis. In 2012, moderate temperatures and precipitation was present in early stages of maize development with heavy precipitation and cooler temperatures in later stages of plant development. As was reported by Doohan, Brennan & Cooke (2003), F. graminearum is more prevalent when at anthesis there is warm weather with moderate precipitation as was present in the 2011 sampling. Reis (1990) determined that the dispersal of ascospores is halted in heavy rain as was present in 2012 and could have been an additional factor for the reduced presence of F. graminearum in that year compared to the previous year. The variation in climatic conditions present in the two years of the study probably contributed to the year-to-year differences in the incidence of F. graminearum and could also be extended to the other Fusarium species and strain types identified in this study.
In maize, wheat and barley in European countries the dominant FHB species has been identified as F. graminearum (Eckard et al., 2011; Goertz et al., 2010; Prodi et al., 2009; Talas, Parzies & Miedaner, 2011). An evaluation of Italian Fusarium species indicates that climatic conditions influence the prevalence of 15AcDON-type and 3AcDON-type strains (Covarelli et al., 2015). The 15AcDON-type is believed to be prevalent in regions with cool winter climates (Prodi et al., 2009) such as the UK, which is supported in a previous study of UK chemotypes of F. graminearum (Jennings et al., 2004). The prevalence of 15AcDON-types of F. graminearum from maize and wheat in this study are further support of this chemotype for this climate. In Norway, the 3AcDON-type of F. graminearum was identified as the predominant chemotype (Aamot et al., 2015). In North America the increased presence of 3AcDON-type since 1998 is suggested to be from shifts in agricultural practices or environmental conditions (Ward et al., 2008). The presence of the 3AcDON-type in the UK may be attributed to similar changes in the landscape with isolates originating from Norway where this is the predominant chemotype (Aamot et al., 2015). In Europe the DON chemotype of the F. culmorum population (Pasquali et al., 2016) has been exclusively identified as the 3AcDON-type but the species is known to produce 15AcDON (Mirocha et al., 1994). The presence of the 15AcDON-type isolates of F. culmorum in this study may be a subpopulation in Europe not yet identified but would require further evidence in additional studies to confirm this subpopulation. The sampling method used for this study was intended to capture the presence of species present at greater than 5% in each field which would also capture novel characteristics of chemotypes of subpopulations. New emergence, climate change, sampling method and host species could be contributing factors for the identification of the 3AcDON-type of F. graminearum isolates and 15AcDON-type of F. culmorum isolates found in this study.
The oxygenation or acetylation of TCT, including 15AcDON and NIV, alters the toxicity and bioactivity (Kimura et al., 1998) and may affect the frequency of a specific TCT-type. The 15AcDON-type and 3AcDON-type chemotypes are acetylated derivatives of DON (Miller et al., 1991) while NIV is an oxygenated C-4 (Kimura et al., 2007). Modification of the bioactivity occurs from the structural differences of oxygenation and acetylation which may affect strain fitness (Ward et al., 2002). Studies in terms of fecundity, growth rates and toxicity have identified the 3AcDON-type isolates as the dominant population type over the NIV-type (Zhang et al., 2012) and 15AcDON-type (Ward et al., 2008). However, the NIV-type is believed to be a virulence factor in maize (Carter et al., 2002; Maier et al., 2006) and is not affected by climatic conditions (Covarelli et al., 2015). The presence of the NIV-type F. graminearum and F. culmorum isolates found in this maize sampling may be an indicator of the virulence factor of NIV producing strains in maize. However, additional study of this potential of NIV being a virulence factor would require greater depth of investigation than addressed in this study.
The association of chemotypes with distinct F. graminearum populations has been found in wheat (Karugia et al., 2009; Ward et al., 2008), barley (Burlakoti et al., 2011) and maize (Qiu & Shi, 2014). While this study evaluated the population genetics of F. graminearum isolates from a single year, multiple year population studies identified shifts in isolates based on chemotypes (Guo, Fernando & Seow-Brock, 2008; Karugia et al., 2009; Ward et al., 2008). Population genetic studies of F. graminearum in wheat have indicated that the direction of gene flow was going from 3AcDON-type towards NIV-type (Zhang et al., 2012). In this study of population genetics of F. graminearum there were NIV-type and 15AcDON-type isolates of F. graminearum from maize and 15AcDON-type isolates from wheat. Isolates were represented by a VNTR haplotype and assigned to a cluster based on chemotype and host species of each isolate. The three clusters were identified as: maize 15AcDON-type, maize NIV-type and wheat 15AcDON-type. Fourteen percent of the isolates demonstrated strong admixture probabilities with other genetic clusters than their assigned cluster. The wheat DON-types with high admixture shared significant genetic material with the maize DON-types while the maize DON-types with high admixture shared significant genetic material with the maize NIV-types. The prevalence of sexual reproduction of field populations of F. graminearum populations and spore dispersal may explain the gene flow between chemotypes identified in other studies (Guo, Fernando & Seow-Brock, 2008; Karugia et al., 2009; Ward et al., 2008) and explain the high admixtures in this study.
F. graminearum is an important etiological agent on wheat and maize. The sexual ascospores and asexual macroconidia of F. graminearum overwinters in maize crop residues with dispersal favored by moist warm conditions (Schmale, Arntsen & Bergstrom, 2005; Paul et al., 2004). While subpopulations of the species are restricted by natural barriers, which includes geographical distances, sea and mountains, Karugia et al., (2009) studies on the long-distance dispersal of ascospores (Maldonado-Ramirez et al., 2005) have found that ascospores are present at high altitudes which would overcome those natural barriers. The high precipitation in the UK, the increase in maize production and the high admixture rates between F. graminearum populations in maize and wheat presents an opportunity to introduce a pathogen population that may be more toxigenic and more vigorous which would have significant implications for food safety in the UK.
In conclusion, this study provides the important first description of Fusarium species in UK maize. The identification of the predominant Fusarium species, F. graminearum and F. culmorum, along with potentially emerging species (F. verticillioides) and subpopulations of species (3AcDON F. graminearum; 15AcDON F. culmorum) in UK maize will enable maize breeding programs to focus their resistance strategies on these pathogens. The identification of subpopulations within F. graminearum associated with their host species and chemotype profile provides an important informative baseline for future epidemiological surveillances. Given that wheat is a significantly greater economical crop compared to maize in the UK, continued monitoring and evaluation of the TCT-type populations of F. graminearum population in both crops will be of great benefit for breeders and growers. We believe that future epidemiological surveys should continue to monitor the NIV-type populations of F. graminearum in both maize and wheat.
Supplemental Information
Acknowledgments
The laboratory research for this study was undertaken at the National Institute of Agricultural Botany in Cambridge, UK. The gamma irradiated carnation leaves used in CLA were provided by the Fusarium Research Center at Penn State in Pennsylvania, USA.
Funding Statement
Funding for this research was provided by the Felix Thornley Cobbold Trust and the John Oldacre Foundation. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Additional Information and Declarations
Competing Interests
The author declares there are no competing interests.
Author Contributions
Ryan Basler conceived and designed the experiments, performed the experiments, analyzed the data, contributed reagents/materials/analysis tools, wrote the paper, prepared figures and/or tables, reviewed drafts of the paper.
Data Availability
The following information was supplied regarding data availability:
The raw data has been supplied as Data S1.
References
- Aamot et al. (2015).Aamot HU, Ward TJ, Brodal G, Vrålstad T, Larsen GB, Klemsdal SS, Elameen A, Uhlig S, Hofgaard IS. Genetic and phenotypic diversity within the Fusarium graminearum species complex in Norway. European Journal of Plant Pathology. 2015;142:501–519. doi: 10.1007/s10658-015-0629-4. [DOI] [Google Scholar]
- Arino et al. (2007).Arino A, Juan T, Estopanan G, Gonzalez-Cabo JF. Natural occurrence of Fusarium species, fumonisin production by toxigenic strains, and concentrations of fumonisins B-1 and B-2 in conventional and organic maize grown in Spain. Journal of Food Protection. 2007;70:151–156. doi: 10.4315/0362-028x-70.1.151. [DOI] [PubMed] [Google Scholar]
- Assemat et al. (1996).Assemat P, Dawson M, Quenin H, Tregou M. Fusarioses des épis de blé en France. Phytoma. 1996;479:22–24. [Google Scholar]
- Burlakoti et al. (2011).Burlakoti RR, Neate SM, Adhikari TB, Gyawali S, Salas B, Steffenson BJ, Schwarz PB. Trichothecene profiling and population genetic analysis of Gibberella zeae from barley in North Dakota and Minnesota. Phytopathology. 2011;101:687–695. doi: 10.1094/PHYTO-04-10-0101. [DOI] [PubMed] [Google Scholar]
- Carter et al. (2002).Carter JP, Rezanoor HN, Holden D, Desjardins AE, Plattner RD, Nicholson P. Variation in pathogenicity associated with the genetic diversity of Fusarium graminearum. European Journal of Plant Pathology. 2002;108:573–583. doi: 10.1023/A:1019921203161. [DOI] [Google Scholar]
- Charmley et al. (1995).Charmley LL, Trenholm HL, Prelusky DB, Rosenberg A. Economic losses and decontamination. Natural Toxins. 1995;3:199–203. doi: 10.1002/nt.2620030406. [DOI] [PubMed] [Google Scholar]
- Covarelli et al. (2015).Covarelli L, Beccari G, Prodi A, Generotti S, Etruschi F, Juan C, Ferrer E, Mañes J. Fusarium species, chemotype characterisation and trichothecene contamination of durum and soft wheat in an area of central Italy. Journal of the Science of Food and Agriculture. 2015;95:540–551. doi: 10.1002/jsfa.6772. [DOI] [PubMed] [Google Scholar]
- DEFRA (2014).DEFRA Agriculture in the United Kingdom 2014. 2014. https://www.gov.uk/government/statistics/agriculture-in-the-united-kingdom-2014 https://www.gov.uk/government/statistics/agriculture-in-the-united-kingdom-2014
- Desjardins (2006).Desjardins A. Fusarium mycotoxins, chemistry, genetics, and biology. APS Press; St Paul: 2006. [Google Scholar]
- Doohan, Brennan & Cooke (2003).Doohan FM, Brennan J, Cooke BM. Influence of climatic factors on Fusarium species pathogenic to cereals. European Journal of Plant Pathology. 2003;109:755–768. doi: 10.1023/A:1026090626994. [DOI] [Google Scholar]
- Dorn et al. (2009).Dorn B, Forrer H-R, Schuerch S, Vogelgsang S. Fusarium species complex on maize in Switzerland: occurrence, prevalence, impact and mycotoxins in commercial hybrids under natural infection. European Journal of Plant Pathology. 2009;125:51–61. doi: 10.1007/s10658-009-9457-8. [DOI] [Google Scholar]
- Eckard et al. (2011).Eckard S, Wettstein FE, Forrer H-R, Vogelgsang S. Incidence of Fusarium species and mycotoxins in silage maize. Toxins. 2011;3:949–967. doi: 10.3390/toxins3080949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evanno, Regnaut & Goudet (2005).Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Excoffier, Laval & Schneider (2005).Excoffier L, Laval G, Schneider S. Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evolutionary Bioinformatics Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- FAO (2013).FAO FAOSTAT database collections. Food and Agriculture Organization of the United Nations; Rome: 2013. [10 December 2015]. [Google Scholar]
- GenStat (2011).GenStat GenStat for windows. 14th edition. VSN International; Hemel Hempstead: 2011. [Google Scholar]
- Goertz et al. (2008).Goertz A, Oerke E-C, Steiner U, Waalwijk C, De Vries I, Dehne H-W. Biodiversity of Fusarium species causing ear rot of maize in Germany. Cereal Research Communications. 2008;36:617–622. doi: 10.1556/CRC.36.2008.Suppl.B.51. [DOI] [Google Scholar]
- Goertz et al. (2010).Goertz A, Zuehlke S, Spiteller M, Steiner U, Dehne HW, Waalwijk C, de Vries I, Oerke EC. Fusarium species and mycotoxin profiles on commercial maize hybrids in Germany. European Journal of Plant Pathology. 2010;128:101–111. doi: 10.1007/s10658-010-9634-9. [DOI] [Google Scholar]
- Guo, Fernando & Seow-Brock (2008).Guo XW, Fernando WGD, Seow-Brock HY. Population structure, chemotype diversity, and potential chemotype shifting of Fusarium graminearum in wheat fields of Manitoba. Plant Disease. 2008;92:756–762. doi: 10.1094/PDIS-92-5-0756. [DOI] [PubMed] [Google Scholar]
- Ioos, Belhadj & Menez (2004).Ioos R, Belhadj A, Menez M. Occurrence and distribution of Microdochium nivale and Fusarium species isolated from barley, durum and soft wheat grains in France from 2000 to 2002. Mycopathologia. 2004;158:351–362. doi: 10.1007/s11046-004-2228-3. [DOI] [PubMed] [Google Scholar]
- Jennings et al. (2004).Jennings P, Coates ME, Walsh K, Turner JA, Nicholson P. Determination of deoxynivalenol- and nivalenol-producing chemotypes of Fusarium graminearum isolated from wheat crops in England and Wales. Plant Pathology. 2004;53:643–652. doi: 10.1111/j.0032-0862.2004.01061.x. [DOI] [Google Scholar]
- Karugia et al. (2009).Karugia GW, Suga H, Gale LR, Nakajima T, Ueda A, Hyakumachi M. Population structure of Fusarium asiaticum from two Japanese regions and eastern China. Journal of General Plant Pathology. 2009;75:110–118. doi: 10.1007/s10327-009-0153-5. [DOI] [Google Scholar]
- Kimura et al. (1998).Kimura M, Kaneko I, Komiyama M, Takatsuki A, Koshino H, Yoneyama K, Yamaguchi I. Trichothecene 3-O-acetyltransferase protects both the producing organism and transformed yeast from related mycotoxins. Cloning and characterization of Tri101. Journal of Biological Chemistry. 1998;3:1654–1661. doi: 10.1074/jbc.273.3.1654. [DOI] [PubMed] [Google Scholar]
- Kimura et al. (2007).Kimura M, Tokai T, Takahashi-Ando N, Ohsato S, Fujimura M. Molecular and genetic studies of Fusarium trichothecenebiosynthesis: pathways, genes, and evolution. Bioscience Biotechnology and Biochemistry. 2007;71:2105–2123. doi: 10.1271/bbb.70183. [DOI] [PubMed] [Google Scholar]
- Kulik (2008).Kulik T. Detection of Fusarium tricinctum from cereal grain using PCR assay. Journal of Applied Genetics. 2008;49:305–311. doi: 10.1007/BF03195628. [DOI] [PubMed] [Google Scholar]
- Leslie & Summerell (2006).Leslie JF, Summerell BA. The Fusarium laboratory manual. Blackwell Publishing; Ames: 2006. [Google Scholar]
- Logrieco et al. (2003).Logrieco A, Bottalico A, Mule G, Moretti A, Perrone G. Epidemiology of toxigenic fungi and their associated mycotoxins for some Mediterranean crops. European Journal of Plant Pathology. 2003;109:645–667. doi: 10.1023/A:1026033021542. [DOI] [Google Scholar]
- Maier et al. (2006).Maier FJ, Miedaner T, Hadeler B, Felk A, Salomoni S, Lemmens M, Kassner H, Schafer W. Involvement of trichothecenes in fusarioses of wheat, barley and maize evaluated by gene disruption of the trichodiene synthase (Tri5) gene in three field isolates of different chemotype and virulence. Molecular Plant Pathology. 2006;7:449–461. doi: 10.1111/j.1364-3703.2006.00351.x. [DOI] [PubMed] [Google Scholar]
- Maldonado-Ramirez et al. (2005).Maldonado-Ramirez SL, Schmale DG, Jr, Shields EJ, Bergstrom GC. The relative abundance of viable spores of Gibberella zeae in the planetary boundary layer suggests the role of long-distance transport in regional epidemics of Fusarium head blight. Agricultural and Forest Meteorology. 2005;132:20–27. doi: 10.1016/j.agrformet.2005.06.007. [DOI] [Google Scholar]
- Meissle et al. (2010).Meissle M, Mouron P, Musa T, Bigler F, Pons X, Vasileiadis VP, Otto S, Antichi D, Kiss J, Palinkas Z, Dorner Z, Van der Weide R, Groten J, Czembor E, Adamczyk J, Thibord JB, Melander B, Nielsen GC, Poulsen RT, Zimmermann O, Verschwele A, Oldenburg E. Pests, pesticide use and alternative options in European maize production: current status and future prospects. Journal of Applied Entomology. 2010;134:357–375. doi: 10.1111/j.1439-0418.2009.01491.x. [DOI] [Google Scholar]
- Miller et al. (1991).Miller JD, Greenhalgh R, Wang YZ, Lu M. Trichothecene chemotypes of 3 Fusarium species. Mycologia. 1991;83:121–130. doi: 10.2307/3759927. [DOI] [Google Scholar]
- Mirocha et al. (1994).Mirocha CJ, Xie W, Xu Y, Wilcoxson RD, Woodward RP, Etebarian RH, Behele G. Production of trichothecene mycotoxins by Fusarium graminearumand Fusarium culmorum on barley and wheat. Mycopathologia. 1994;128:19–23. doi: 10.1007/BF01104274. [DOI] [PubMed] [Google Scholar]
- National Centre for Atmospheric Science (2012).National Centre for Atmospheric Science . MIDAS land surface stations data (1853-current) Leeds: NCAS; 2012. [5 January 2014]. [Google Scholar]
- OERKE (2006).OERKE E-C. Crop losses to pests. The Journal of Agricultural Science. 2006;144:31–43. doi: 10.1017/S0021859605005708. [DOI] [Google Scholar]
- Osborne & Stein (2007).Osborne LE, Stein JM. Epidemiology of Fusarium head blight on small-grain cereals. International Journal of Food Microbiology. 2007;119:103–108. doi: 10.1016/j.ijfoodmicro.2007.07.032. [DOI] [PubMed] [Google Scholar]
- Pasquali et al. (2011).Pasquali M, Beyer M, Bohn T, Hoffmann L. Comparative analysis of genetic chemotyping methods for Fusarium: Tri13 polymorphism does not discriminate between 3- and 15-acetylated deoxynivalenol chemotypes in Fusarium graminearum. Journal of Phytopathology. 2011;159:700–704. doi: 10.1111/j.1439-0434.2011.01824.x. [DOI] [Google Scholar]
- Pasquali et al. (2010).Pasquali M, Giraud F, Brochot C, Cocco E, Hoffmann L, Bohn T. Genetic Fusarium chemotyping as a useful tool for predicting nivalenol contamination in winter wheat. International Journal of Food Microbiology. 2010;137:246–253. doi: 10.1016/j.ijfoodmicro.2009.11.009. [DOI] [PubMed] [Google Scholar]
- Pasquali et al. (2016).Pasquali M, Beyer M, Logrieco A, Audenaert K, Balmas V, Basler R, Boutigny AL, Chrpová J, Czembor E, Gagkaeva T, González-Jaén MT, Hofgaard IS, Köycü ND, Hoffmann L, Lević J, Marín P, Miedaner T, Migheli Q, Moretti A, Müller ME, Munaut F, Parikka P, Pallez-Barthel M, Piec J, Scauflaire J, Scherm B, Stanković S, Thrane U, Uhlig S, Vanheule A, Yli-Mattila T, Vogelgsang S. A European database of Fusarium graminearum and F. culmorum trichothecene genotypes. Frontiers in MicroBiology. 2016;7:406. doi: 10.3389/fmicb.2016.00406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul et al. (2004).Paul PA, El-Allaf SM, Lipps PE, Madden LV. Rain splash dispersal of Gibberella zeae within wheat canopies in Ohio. Phytopathology. 2004;94:1342–1349. doi: 10.1094/PHYTO.2004.94.12.1342. [DOI] [PubMed] [Google Scholar]
- Pritchard, Stephens & Donnelly (2000).Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prodi et al. (2009).Prodi A, Tonti S, Nipoti P, Pancaldi D, Pisi A. Identification of deoxynivalenol and nivalenol producing chemotypes of Fusarium graminearum isolates from durum wheat in a restricted area of northern Italy. Journal of Plant Pathology. 2009;91:727–731. [Google Scholar]
- Qiu & Shi (2014).Qiu J, Shi J. Genetic relationships, Carbendazim sensitivity and mycotoxin production of the Fusarium graminearumpopulations from maize, wheat and rice in eastern China. Toxins. 2014;6:2291–2309. doi: 10.3390/toxins6082291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quarta et al. (2006).Quarta A, Mita G, Haidukowski M, Logrieco A, Mule G, Visconti A. Multiplex PCR assay for the identification of nivalenol, 3- and 15-acetyl-deoxynivalenol chemotypes in Fusarium. FEMS Microbiology Letters. 2006;259:7–13. doi: 10.1111/j.1574-6968.2006.00235.x. [DOI] [PubMed] [Google Scholar]
- Reis (1990).Reis EM. Effects of rain and relative humdity on the release of ascospores and on the infection of wheat heads by Gibberella zeae. Fitopatologia Brasileira. 1990;15:339–343. [Google Scholar]
- Sampietro et al. (2011).Sampietro DA, Díaz CG, Gonzalez V, Vattuone MA, Ploper LD, Catalan CAN, Ward TJ. Species diversity and toxigenic potential of Fusarium graminearum complex isolates from maize fields in northwest Argentina. International Journal of Food Microbiology. 2011;145:359–364. doi: 10.1016/j.ijfoodmicro.2010.12.021. [DOI] [PubMed] [Google Scholar]
- Scauflaire et al. (2011).Scauflaire J, Mahieu O, Louvieaux J, Foucart G, Renard F, Munaut F. Biodiversity of Fusarium species in ears and stalks of maize plants in Belgium. European Journal of Plant Pathology. 2011;131:59–66. doi: 10.1007/s10658-011-9787-1. [DOI] [Google Scholar]
- Schmale, Arntsen & Bergstrom (2005).Schmale DG, Arntsen QA, Bergstrom GC. The forcible discharge distance of ascospores of Gibberella zeae. Canadian Journal Plant Pathology. 2005;27:376–382. doi: 10.1080/07060660509507235. [DOI] [Google Scholar]
- Slatkin (1995).Slatkin M. A measure of population subdivision based on microsatellite allele frequencies. Genetics. 1995;139:457–462. doi: 10.1093/genetics/139.1.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suga, Gale & Kistler (2004).Suga H, Gale LR, Kistler HC. Development of VNTR markers for two Fusarium graminearum clade species. Molecular Ecology Notes. 2004;4:468–470. doi: 10.1111/j.1471-8286.2004.00703.x. [DOI] [Google Scholar]
- Talas, Parzies & Miedaner (2011).Talas F, Parzies HK, Miedaner T. Diversity in genetic structure and chemotype composition of Fusarium graminearumsensu stricto populations causing wheat head blight in individual fields in Germany. European Journal of Plant Pathology. 2011;131:39–48. doi: 10.1007/s10658-011-9785-3. [DOI] [Google Scholar]
- Tan & Niessen (2003).Tan MK, Niessen LM. Analysis of rDNA its sequences to determine genetic relationships among, and provide a basis for simplified diagnosis of Fusarium species causing crown rot and head blight of cereals. Mycological Research. 2003;107:811–821. doi: 10.1017/S0953756203008013. [DOI] [PubMed] [Google Scholar]
- Turner et al. (1998).Turner AS, Lees AK, Rezanoor HN, Nicholson P. Refinement of PCR-detection of Fusarium avenaceum and evidence from DNA marker studies for phenetic relatedness to Fusarium tricinctum. Plant Pathology. 1998;47:278–288. [Google Scholar]
- Waalwijk et al. (2003).Waalwijk C, Kastelein P, De Vries I, Kerenyi Z, Van der Lee T, Hesselink T, Kohl J, Kema G. Major changes in Fusarium spp. in wheat in the Netherlands. European Journal of Plant Pathology. 2003;109:743–754. doi: 10.1023/A:1026086510156. [DOI] [Google Scholar]
- Ward et al. (2002).Ward TJ, Bielawski JP, Kistler HC, Sullivan E, O’Donnell K. Ancestral polymorphism and adaptive evolution in the trichothecene mycotoxin gene cluster of phytopathogenic Fusarium. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:9278–9283. doi: 10.1073/pnas.142307199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward et al. (2008).Ward TJ, Clear RM, Rooney AP, O’Donnell K, Gaba D, Patrick S, Starkey DE, Gilbert J, Geiser DM, Nowicki TW. An adaptive evolutionary shift in Fusarium head blight pathogen populations is driving the rapid spread of more toxigenic Fusarium graminearum in North America. Fungal Genetics and Biology. 2008;45:473–484. doi: 10.1016/j.fgb.2007.10.003. [DOI] [PubMed] [Google Scholar]
- Zhang et al. (2012).Zhang H, Van der Lee TV, Waalwijk C, Chen W, Xu J, Xu J, Zhang Y, Feng J. Population analysis of the Fusarium graminearum species complex from wheat in China show a shift to more aggressive isolates. PLoS ONE. 2012;7(2):e2143. doi: 10.1371/journal.pone.0031722. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The following information was supplied regarding data availability:
The raw data has been supplied as Data S1.