Abstract
Blooms of the freshwater cyanobacterium Anabaena circinalis are recognized as an important health risk worldwide due to the production of a range of toxins such as saxitoxin (STX) and its derivatives. In this study we used HIP1 octameric-palindrome repeated-sequence PCR to compare the genomic structure of phylogenetically similar Australian isolates of A. circinalis. STX-producing and nontoxic cyanobacterial strains showed different HIP1 (highly iterated octameric palindrome 1) DNA patterns, and characteristic interrepeat amplicons for each group were identified. Suppression subtractive hybridization (SSH) was performed using HIP1 PCR-generated libraries to further identify toxic-strain-specific genes. An STX-producing strain and a nontoxic strain of A. circinalis were chosen as testers in two distinct experiments. The two categories of SSH putative tester-specific sequences were characterized by different families of encoded proteins that may be representative of the differences in metabolism between STX-producing and nontoxic A. circinalis strains. DNA-microarray hybridization and genomic screening revealed a toxic-strain-specific HIP1 fragment coding for a putative Na+-dependent transporter. Analysis of this gene demonstrated analogy to the mrpF gene of Bacillus subtilis, whose encoded protein is involved in Na+-specific pH homeostasis. The application of this gene as a molecular probe in laboratory and environmental screening for STX-producing A. circinalis strains was demonstrated. The possible role of this putative Na+-dependent transporter in the toxic cyanobacterial phenotype is also discussed, in light of recent physiological studies of STX-producing cyanobacteria.
During the Australian drought of 1991, blooms of neurotoxic Anabaena circinalis strains infested nearly 1,000 km of the Darling River in New South Wales (7). The neurotoxins of A. circinalis remained uncharacterized until the recently identification of paralytic shellfish poisoning (PSP) toxins in several Australian isolates by Humpage et al. (15). PSP is a life-threatening affliction that results from the consumption of contaminated seafood (20). PSP toxins, of which the most potent representative is saxitoxin (STX), are a class of neurotoxic alkaloids that selectively block voltage-gated Na+ channels in excitable cells (9). The ensuing effect on impulse generation in animals can lead, in extreme cases, to death (9, 44). Apart from A. circinalis, saxitoxin and its analogue compounds have been reported to occur naturally in other filamentous cyanobacteria (2, 8, 21, 31) as well as in certain heterotrophic bacteria (10), although these toxins are more commonly associated with red tides and the presence of marine dinoflagellates (12, 26, 36, 44).
Although A. circinalis is a species found worldwide, there is a geographical segregation of toxin-producing strains. While this cyanobacterium is known in other continents for the production of the tropane-related neurotoxic alkaloids anatoxin-a and anatoxin-a(s) (38), only Australian isolates produce PSP toxins. The reason for this geographical segregation of neurotoxin production is not known. However, adaptation to specific environmental pressures or genetic heterogeneity within the species A. circinalis is a possible explanation. Recently, the phylogenetic structure of this species was determined by analyzing the 16S rRNA gene sequences (5). A. circinalis strains were found to form a monophyletic group of worldwide distribution. However, the PSP toxin- and non-PSP toxin-producing isolates formed two clusters according to the 16S rRNA gene tree, with most of the toxic and nontoxic strains clustering separately, with few exceptions (5). These data suggested that a certain degree of genomic divergence is present among toxic and nontoxic strains of this species. A phylogenetic analysis targeting STX-encoding genes in A. circinalis would be more precise and ideal for both toxigenicity identification and population analysis. Unfortunately, DNA sequence information regarding these biosynthesis genes is presently unavailable.
In this study we used HIP1 (highly iterated octameric palindrome 1) repeated-sequence PCR to compare the genomes of phylogenetically closely related isolates of toxic and nontoxic A. circinalis strains. This technique is performed on the basis of the genetic polymorphisms within defined cyanobacterial repetitive elements (11, 32) and utilizes HIP1-directed PCR primers. HIP1 is an octameric sequence (5′-GCGATCGC-3′) abundant in the coding regions of cyanobacterial genomes (32). HIP1 PCR has been previously used to demonstrate genetic diversity among strains of the genera Anabaena and Nostoc (39, 43) and to distinguish between Cylindrospermopsis raciborskii isolates (14, 24, 34). The aim here was to identify genomic differences that correlated with STX production in Australian strains of A. circinalis. Additionally, HIP1-generated libraries from STX-producing and nontoxic cyanobacterial isolates were investigated by suppression subtractive hybridization (SSH) to further recover possible toxin- and non-toxin-specific sequences (1). DNA fragments from both the HIP1 and SSH analyses were screened by DNA microarray hybridization with labeled toxic and nontoxic A. circinalis genomic DNA. A single, toxic-strain-specific gene was identified by this multistaged process. The application of this gene as a molecular probe for routine assessment of the potential risk associated with the presence of PSP toxin-producing A. circinalis in water reservoirs was also demonstrated in this study.
MATERIALS AND METHODS
Cyanobacteria.
The cyanobacterial strains investigated in this study are listed in Table 1. PSP toxin-producing and nontoxic isolates of A. circinalis were obtained from the Australian Water Quality Centre (Adelaide) and maintained in Jaworski's medium (15). All cyanobacterial cultures were grown without agitation or aeration in 250-ml glass flasks at a constant temperature of 26°C under continuous irradiance of cool white light at an intensity of 15 μmol of photons m−2 s−1. Environmental samples of PSP toxin-producing and nontoxic cyanobacterial blooms in the Sydney Catchment Area were kindly provided by the Department of Land and Water Conservation.
TABLE 1.
Cyanobacterial strains used in this study
DNA extraction.
Cyanobacterial cultures were filtered through a 3.0-μm-pore-size filter (Millipore, Billerica, Mass.), and cells were washed twice with sterile water. This method has been previously demonstrated to be effective in the removal of possible contaminating heterotrophic bacteria (4, 5, 19). Genomic DNA was extracted from filtered and washed cyanobacterial cells and environmental bloom samples by the use of the XS procedure as described previously (42) and resuspended in TE buffer (10 mM Tris-HCl, pH 7.4; 1 mM EDTA, pH 8).
PCR amplifications and DNA sequencing.
HIP1 PCR amplifications were performed using both primer Hip-CA and primer Hip-TG (34, 39). The reaction volume (20 μl) contained 200 μM deoxynucleoside triphosphates (dNTP), 2.5 mM MgCl2, Taq polymerase buffer, 5 pmol of each primer, 100 ng of DNA template, and 1 U of Taq polymerase. Reactions were cycled using a temperature profile of 95°C for 5 min followed by 30 cycles of 95°C for 10 s, 40°C for 20 s, and 72°C for 90 s and concluded with 1 cycle of 72°C for 5 min. HIP1 PCR products were separated by 4% polyacrylamide gel electrophoresis in Tris-borate-EDTA buffer according to standard protocols (35). DNA was extracted from 4% polyacrylamide gel electrophoresis bands by the standard crush and soak procedure (35) and reamplified by HIP1 PCR. PCR products were ethanol precipitated and cloned into pGEM-TE vector. Clones were amplified using pGEM-TE vector-specific primers (mpf and mpR) and sequenced by PRISM automated BigDye terminator sequencing with an ABI 373 sequencer (PE Applied Biosystems, Foster City, Calif.), with reactions performed using 3 μl (∼150 ng) of each PCR product and 10 pmol of each appropriate primer in a half-scale reaction as specified by the manufacturer.
Amplification of the Na+-dependent transporter (NaDT) gene sequences was performed using 20 pmol each of the degenerate primers NaTf (5′-AT[ATC]AT[ATC]ATG[TC]TNGGNATGGG-3′) and NaTR (5′-ATNGCNGCAGGAATNGCCAT-3′) in a 20-μl reaction volume containing 200 μM dNTP, 2.5 mM MgCl2, Taq polymerase buffer, 100 ng of DNA template, and 0.25 U of Taq polymerase. PCRs were cycled using a temperature profile of 94°C for 3 min followed by 30 cycles of 94°C for 10 s, 50 ± 5°C for 20 s, and 72°C for 50 s and concluded with 1 cycle of 72°C for 5 min. YZf (5′-AGCTGTGGCCATTGGCTTAA-3′) and YZR (5′-GCAATACAGATTTGCTGACG-3′), specific primers for the NaDT, were also designed, and 10 pmol of each was utilized in 20-μl reaction volumes as described above. The following protocol was used: 94°C for 3 min followed 30 cycles of 94°C for 10 s, 55°C for 20 s, and 72°C for 50 s, concluding with 1 cycle of 72°C for 5 min. To simultaneously detect the presence of cyanobacteria and the gene encoding the NaDT, a multiplex PCR was performed. This involved combining the primers YZf and YZR with the cyanobacterium-specific 16S rRNA gene primers 27f (5′-AGAGTTTGATTTACGCGACA-3′) and 809R (5′-GCTTCGGCACGGCTCGGGTCGATA-3′) in the same PCR. Primer concentrations and PCR cycling conditions were as described above. All results were visualized by 1.5 or 2% agarose gel electrophoresis in Tris-acetate-EDTA buffer according to standard protocols (35). PCR products were either ethanol precipitated or extracted from agarose with a QIAquick gel extraction kit (QIAGEN, Germantown, Md.) and sequenced as mentioned above. Phylogenetic tree construction and sequence alignments were performed using version 1.8 ClustalX software (41).
SSH.
DNA libraries were generated from the HIP1 PCR products as reported above and with the addition of fresh Pfu polymerase (Promega, Madison, Wis.) at 0.5 U per 20-μl reaction volume at the end of the temperature cycles. Pfu polymerase reactions were carried out at 72°C for 20 min, and the DNA was precipitated with ethanol and resuspended in water. Subtraction of HIP1 PCR libraries was achieved by means of a modified PCR-based subtractive hybridization protocol (1). Subtraction experiments were carried out using A. circinalis strains 332H (nontoxic) as the driver and 134C (toxic) as the tester in one experiment and A. circinalis strains 131C (toxic) as the driver and 306A (nontoxic) as the tester in another experiment. Briefly, for each experiment, 1 μg of HIP1-generated DNA library from each strain (driver and tester) was digested with RsaI and two different PCR adaptors were ligated to two different aliquots of the tester DNA. Two hybridizations were then performed. In the first hybridization, an excess of driver DNA was added to each of the adaptor-ligated tester DNAs. Each sample mixture was then denatured at 98°C for 2 min and allowed to anneal at 63°C for 90 min. After this hybridization, single-stranded DNA was enriched for tester-specific DNA, as DNA fragments that are not tester specific will form hybrid molecules with the driver DNA. In the second hybridization, the two primary hybridization reaction mixtures were combined without denaturing and allowed to anneal at 63°C overnight. Only the subtracted single-stranded tester-specific DNA should reassociate to make hybrids with the two different terminal adaptors. Molecules with different adaptors at each end were amplified exponentially using PCR primers for the two adaptor sequences. PCR products obtained after SSH were cloned into the pGEM-TE vector, and clones were amplified using the vector-specific primers (mpf and mpR) and sequenced as mentioned above.
Microarray design and production.
Positive-testing SSH clones, together with the cloned NaDT fragment from A. circinalis 134C, were amplified and purified using 96-well multiscreening membrane plates (Millipore) and resuspended in 70 μl of water. Clone and purified PCR products corresponding to the NaDT and the 50 probes from SSH libraries were spotted in duplicate with more than 300 other DNA fragments, including 16S rRNA of the investigated strains included as housekeeping genes (BGGM1 microarray), as reported elsewhere (27; http://149.171.168.73/microarray/arraylist.html).
Genomic DNA labeling and hybridization.
Fluorescently labeled DNA was prepared indirectly by incorporating amino-allyl (aa) dUTP followed by coupling with the fluorescent dyes. Genomic A. circinalis DNA (1 μg) from strains 332H and 306A (nontoxic) and strains 134C and 131C (toxic) was digested with RsaI, extracted with phenol, and precipitated with ethanol. DNA was resuspended into 40 μl of H2O, heat denatured at 99°C for 10 min, and chilled on ice. A total of 5 μl of 10× NEB labeling buffer (New England Biolabs, Inc., Beverly, Mass.), 3 μl of aa dNTP mix (3 mM dGTP, dATP, dCTP, 1.8 mM aa-dUTP, 1.2 mM dTTP) and 2 μl (10 U) of the large Klenow fragment of DNA polymerase I (Promega) was added to a final reaction volume of 50 μl. The reaction mixture was incubated at 37°C for 2 h, and unincorporated amines were removed by purification using a QIAquick PCR purification kit (QIAGEN). DNA samples were dried in a Speed-Vac, resuspended in 9 μl of 0.1 M NaHCO3 (pH 9.0), and added to 2 μl of prepared Cy3 (for A. circinalis 332H and 131C DNA) or Cy5 (for A. circinalis 134C and 306A DNA) dye aliquots (Amersham Pharmacia Biotech, Piscataway, N.J.). Reactions were incubated for 60 min at room temperature in the dark and purified (QIAquick PCR kit). Labeled DNA samples were dried to ∼20 μl, combined (134C with 332H and 306A with 131C), and evaporated to dryness.
Fluorescently labeled DNA was resuspended in 20 μl of hybridization solution containing DIG Easy buffer (Roche Applied Science, Penzberg, Germany) and 500 μg of yeast tRNA (Sigma-Aldrich Co., Dorset, United Kingdom), denatured for 2 min at 99°C, cooled to the ambient temperature, and applied to the microarray slide. Glass 22- by 22-mm coverslips were placed over the solution, and hybridization was performed overnight at 37°C in a water-tight humidified hybridization chamber. Array slides were washed in two stages: three washes at 50°C for 15 min with preheated 1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) buffer-0.1% sodium dodecyl sulfate followed by three rinses at room temperature with 1× SSC buffer. Slides were spun dry at 500 × g for 5 min and kept in the dark prior to scanning. Each microarray hybridization was performed in duplicate.
Microarray scanning, data acquisition, and statistical analyzes.
Clean slides were scanned with an ArrayWorx “e” microarray scanner from Applied Precision, Inc. (Issaquah, Wash.). Scanned slide images were generated automatically with ArrayWorx software provided with the ArrayWorx Scanner. Images were quantified using GenePix Pro software from Axon Instruments (Foster City, Calif.). Erroneous spots were manually flagged and removed from the final data set. All microarray data were filtered to remove any spots in which less than 60% of the signal pixels exceeded the local background value for both lasers (595 and 685 nm). The median Cy5/Cy3 ratio automatically generated by the GenePix Pro software for the filtered data for each spot was used for subsequent analysis. The ratio of medians was normalized to give a ratio measurement of 1 for the control sequences corresponding to the 16S rRNA gene of the tested strains.
RESULTS
HIP1 genomic polymorphism.
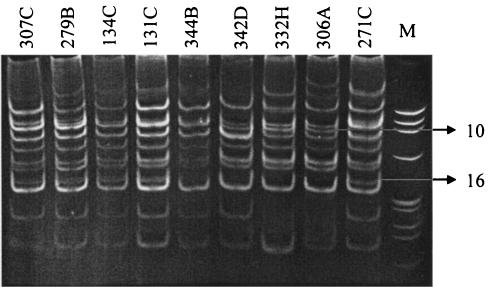
The HIP1 PCR technique showed clear differences in banding patterns between toxic and nontoxic isolates of A. circinalis (Fig. 1). By visual analysis of DNA bands, each of the two groups of A. circinalis strains, nontoxic and toxic, was delineated by the presence of a single unique DNA band; these bands were denoted 10 and 16, respectively (Fig. 1). Acrylamide bands were excised, and the DNA was extracted, cloned, and sequenced. Band 10, characteristic of the majority of nontoxic strains, was extracted from the A. circinalis 306A electrophoresis profile. The DNA fragment (clone 47; GenBank accession number AY326671) encoded an unknown conserved hypothetical protein, with the best BLASTX (3) sequence analysis scores of 77% identity and 89% similarity to locus ZP_00112177 from Nostoc punctiforme (strain ATCC 29133-PCC 73102). Band 16, characteristic of the majority of STX-producing strains, was extracted from the A. circinalis 134C banding pattern. The recovered DNA fragment (clone 219) encoded a putative NaDT protein, with the best BLASTX score of 71% identity and 85% similarity to open reading frame (ORF) alr5254 (accession number BAB76953) from Nostoc sp. strain PCC7120 (Table 2). Clone 219 was spotted onto the DNA microarray for further investigation.
FIG. 1.
Electrophoretic comparison of the PCR products formed in reactions primed with Hip-CA and Hip-TG primers for the nine A. circinalis isolates. M, φX174 HaeIII DNA marker. Band 10, characteristic of the majority of nontoxic strains, and band 16, characteristic of the majority of STX-producing strains, are indicated.
TABLE 2.
A. circinalis 134C putative specific sequences with significant protein matches in The National Center for Biotechnology Information protein database
| Clone | GenBanka accession no. | Best BLASTX hit | Organism | % Identity | % Similarity | Microarray hybridization ratio (avg ± SE)b |
|---|---|---|---|---|---|---|
| 219 | AY326655 | Putative NaDT | Nostoc sp. strain PCC7120 | 71 | 85 | 2.42 ± 0.12 |
| 194 | AY326662 | Hypothetical protein | Cytophaga hutchinsonii | 41 | 61 | 1.45 ± 1.03 |
| 147 | AY326665 | Hypothetical protein | Magnetospirillum magnetotacticum | 47 | 50 | 1.17 ± 0.11 |
| 146 | AY326664 | Succinate dehydrogenase flavoprotein | Nostoc sp. strain PCC7120 | 93 | 96 | 1.15 ± 0.01 |
| 149 | AY326666 | Hypothetical protein | Burkholderia fungorum | 42 | 62 | 1.11 ± 0.03 |
| 153 | AY326668 | Carbamoyl phosphate synthase, pyrimidine-specific, large chain | Synechocystis sp. strain PCC6803 | 96 | 98 | 1.03 ± 0.02 |
| 154 | AY326669 | Hypothetical ORF | Saccharomyces cerevisiae | 37 | 55 | 0.90 ± 0.15 |
| 129 | AY326659 | Mucin 1 precursor | Mus musculus | 36 | 56 | 0.85 ± 0.12 |
| 130 | AY326656 | Dolichol-phosphate mannosyltransferase | Bacteriodes thetaiotaomicron VPI-5482 | 80 | 90 | 0.76 ± 0.10 |
| 155 | AY326670 | Putative phosphoglycerate dehydrogenase | B. thetaiotaomicron VPI 5482 | 50 | 72 | 0.61 ± 0.04 |
| 145 | AY326663 | Inorganic pyrophosphatase | Acetabacteria mediterranea | 54 | 74 | 0.60 ± 0.02 |
| 150 | AY326667 | Hypothetical protein | Vibrio parahaemolyticus RIMD | 39 | 69 | 0.58 ± 0.03 |
| 122 | AY326657 | Oligopeptide binding protein | Nostoc sp. strain PCC7120 | 31 | 63 | 0.50 ± 0.16 |
| 190 | AY326660 | Peptidoglycan anchored protein | Listeria monocytogenes EGD-e | 29 | 50 | 0.50 ± 0.14 |
| 123 | AY326658 | Hypothetical protein | Trichodesmium erythraeum IMS101 | 44 | 62 | NDc |
| 192 | AY326661 | S-adenosyl-methyltransferase mraW | B. thetaiotaomicron VPI-5482 | 45 | 59 | ND |
Nucleotide sequence database (http://www.ncbi.nlm.nih.gov/Genbank).
Values are the result of two independent hybridizations and are expressed as averages ± standard errors of Cy5/Cy3 normalized ratios.
ND, not detected.
SSH of HIP1 genomic libraries.
DNA sequences unique to the toxic 134C strain and the nontoxic 306A strain of A. circinalis were tentatively identified by the SSH of HIP1 PCR-generated libraries. Strains 332H and 131C were used as the driver genomes. DNA containing putative tester-specific products was cloned, and the insert sizes of 50 randomly selected clones per library were estimated by PCR amplification. Insert sizes ranged from 0.15 to 0.5 kb; for each experiment, a total of 25 cloned DNA fragments with different sizes were purified and sequenced. Of the 50 clones analyzed, 9 sequences were encountered more than once (18%) whereas 10 showed no significant similarity with any entry in the databases. General features of the putative tester-specific sequences detected in this study for A. circinalis 134C (toxic) and 306A (nontoxic) are reported in Table 2 and Table 3, respectively. The library of possible toxic-strain sequences contained both hypothetical proteins of unknown function and membrane proteins as well as enzymes that could be involved in both primary and secondary metabolic pathways, such as carbamoyl-phosphate synthase and methyltransferase (Table 2). The putative library of non-toxic-strain-specific fragments comprised mostly hypothetical proteins of unknown function and three defined enzymes, two of which were protein kinases (Table 3).
TABLE 3.
A. circinalis 306A putative specific sequences with significant protein matches in The National Center for Biotechnology Information protein database
| Clone | GenBanka accession no. | Best BLASTX hit | Organism | % Identity | % Similarity | Microarray hybridization ratio (avg ± SE)b |
|---|---|---|---|---|---|---|
| 208 | AY326680 | Serine/threonine kinase | O. javanicus | 31 | 56 | 2.28 ± 0.22 |
| 158 | AY326683 | Hypothetical protein | Nostoc punctiforme | 59 | 76 | 1.86 ± 0.12 |
| 205 | AY326677 | Splicing factor Prp8 | Guillardia theta | 31 | 53 | 1.59 ± 0.01 |
| 163 | AY326685 | Hypothetical protein | Rhodobacter sphaeroides | 43 | 56 | 1.41 ± 0.26 |
| 210 | AY326681 | Hypothetical protein | Nostoc sp. strain PCC7120 | 37 | 68 | 1.11 ± 0.05 |
| 166 | AY326687 | Locus CG32796-PB | Drosophila melanogaster | 40 | 65 | 1.04 ± 0.06 |
| 162 | AY326684 | Hypothetical protein | N. punctiforme | 47 | 54 | 1.00 ± 0.02 |
| 164 | AY326686 | Two-component sensor histidine kinase | Nostoc sp. strain PCC71120 | 75 | 91 | 0.92 ± 0.09 |
| 143 | AY326673 | DNA polymerase III alpha subunit | Bacteriodes thetaiotaomicron VPI-5482 | 80 | 85 | 0.66 ± 0.02 |
| 137 | AY326674 | Hypothetical protein | Cytophaga hutchinsonii | 33 | 61 | 0.65 ± 0.02 |
| 140 | AY326672 | Conserved hypothetical protein | B. thetaiotaomicron VPI-5482 | 41 | 66 | 0.55 ± 0.01 |
| 207 | AY326679 | Conserved hypothetical protein | Yersinia pestis KIM | 47 | 65 | 0.51 ± 0.03 |
| 202 | AY326675 | Hypothetical protein | N. punctiforme | 55 | 77 | 0.48 ± 0.18 |
| 206 | AY326678 | Hypothetical protein | C. hutchinsonii | 34 | 57 | 0.37 ± 0.05 |
| 204 | AY326676 | Hypothetical protein | C. hutchinsonii | 44 | 67 | 0.16 ± 0.03 |
| 213 | AY326682 | cylM protein | Enterococcus faecalis V583 | 42 | 57 | NDc |
Nucleotide sequence database (http://www.ncbi.nlm.nih.gov/Genbank).
Values are the result of two independent hybridizations and are expressed as averages ± standard errors of Cy5/Cy3 normalized ratios.
ND, not detected.
Microarray hybridization.
The array was hybridized with labeled genomic DNA of the SSH testers and drivers. By microarray hybridization, only one tester-specific sequence was identified for each strain (Table 2 and Table 3). The only fragment with a Cy5/Cy3 means ratio higher than 2 arising from toxic strain 134C was the HIP1 band corresponding to the NaDT. On the other hand, the nontoxic 306A strain revealed a candidate gene similar to a serine/threonine kinase (locus BAB17220) from Oryzias javanicus (ratio of 2.28; Table 3).
NaDT.
Degenerate primers (Natf/R) were designed on the basis of the conserved regions among the translated sequence of clone 219 from A. circinalis 134C and putative NaDT protein homologues from Nostoc sp. strain PCC7120 and Synechocystis sp. strain PCC6803. By Natf/R PCR, amplicons of the expected size (602 bp) were produced from toxic A. circinalis strains. PCR products amplified from strains 134C and 279B were purified and sequenced. The data confirmed that the DNA fragment corresponded to the same putative NaDT. BLAST analysis also indicated the relationship of this sequence to members of the sodium bile acid symporter family (SBF) and arsenical resistance protein (ACR).
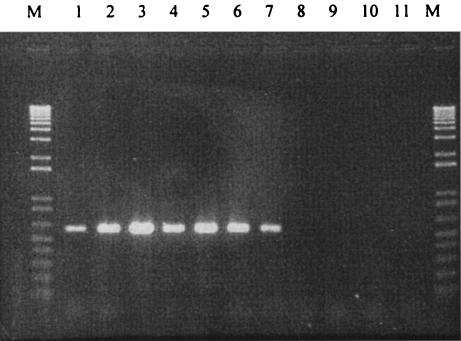
On the basis of sequences from strains 134C and 279B, the specific primers YZf/R were designed to amplify a 704-bp DNA fragment of the A. circinalis putative NaDT. As seen in Fig. 2, the primer set amplified NaDT gene sequences only from toxic isolates of A. circinalis, with the exception of the nontoxic strain 271C. Different stringencies for the PCR were tested on the basis of the use of YZf/R, showing no product of the expected size being amplified from the other nontoxic DNA templates even for the lowest stringency employed, corresponding to a primer annealing temperature of 45°C (data not shown).
FIG. 2.
Detection of NaDT genes in toxic and nontoxic strains of A. circinalis by PCR amplification with YZf- and YZR-specific primers. Lane 1, strain 344B; lane 2, 131C; lane 3, 279B; lane 4, 150A; lane 5, 307C; lane 6, 134C; lane 7, 271C; lane 8, 306A; lane 9, 342D; lane 10, 332H; lane 11, no-DNA control. PCR products were loaded at a total of 4 μl per sample and run with a 1-kb Plus DNA ladder (lanes M) as a standard.
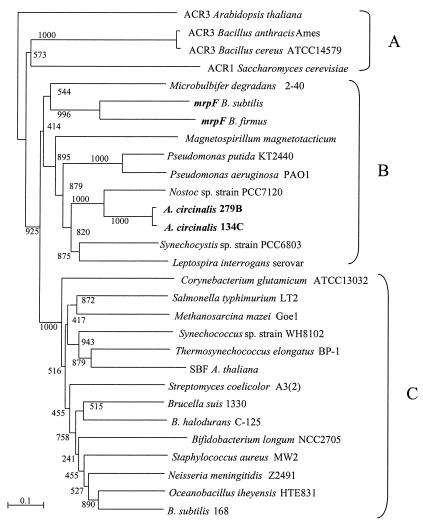
Phylogenetic tree reconstruction was performed using several NaDT proteins from the databases to define possible functional homologies of the A. circinalis gene (Fig. 3). Three distinct phylogenetic groups were recognized. All the ACR-like proteins clustered (Fig. 3A), with no cyanobacterial counterparts. The second group (Fig. 3B) clustered with proteins encoded by mrpF-like genes, including the Bacillus subtilis and B. firmus mrpF genes, and the NaDT genes from A. circinalis 134C and 279B, Synechocystis sp. strain PCC6803 (ORF sll1428), and Nostoc sp. strain PCC7120 (ORF arl5254). A third group (Fig. 3C), comprising various bacterial and cyanobacterial putative sequences, clustered with sequences of genes encoding an SBF protein from Arabidopsis thaliana. Partial alignment comparison (Fig. 4) of the NaDT gene sequences of A. circinalis 279B, Synechocystis sp. strain PCC6803, and Nostoc sp. strain PCC7120, together with the sequences of mrpF genes from B. subtilis and B. firmus, showed that loci of major similarity for the two Bacillus sequences were also conserved in the sequences of genes encoding the three cyanobacterial proteins.
FIG. 3.
Phylogenetic affiliations of prokaryotic homologues of A. circinalis 134C and 279B NaDTs. A. thaliana ACR3 and SBF proteins, together with the protein encoded by the Saccharomyces cerevisiae ACR1 gene, are included as references. The phenogram was reconstructed from a pairwise distance matrix (18) by the neighbor-joining method (33). The scale represents one substitution per 100 amino acid positions. Local bootstrap values (1,000 resampling cycles) are shown. (A) ACR transporters; (B) mrpF-like proteins; (C) other NaDTs.
FIG. 4.
Alignment of partial amino acid sequences corresponding to the cyanobacterial NaDTs of A. circinalis 279B, Synechocystis sp. strain PCC6803, and Nostoc sp. strain PCC7120, together with mrpF gene products of B. subtilis and B. firmus. Regions of identity and similarity are shown as follows: stars, fully conserved positions; stacked double dots, strongly conserved residues; single dots, weakly conserved residues.
Screening for STX-producing A. circinalis strains by multiplex PCR.
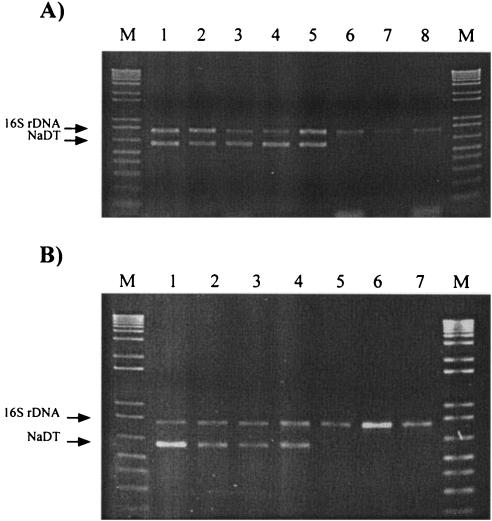
A multiplex PCR assay was developed with the aim of detecting the presence of potential PSP toxin-producing A. circinalis strains in the environment. Figure 5 shows the validation of this method for toxic and nontoxic cyanobacterial isolates (Fig. 5A) and its application to natural samples of PSP toxin-producing and nontoxic blooms (Fig. 5B). The 16S rDNA band (782 bp) identified the occurrence of cyanobacteria in the sample, while the YZf/R band (602 bp) indicated the presence of the toxic-strain-specific NaDT and the potentiality of STX-producing A. circinalis.
FIG. 5.
Electrophoretic comparison of the PCR products formed in reactions primed with the 16S rDNA primers 27f/809R and the toxic-strain-specific NaDT primers YZf/R. (A) Multiplex PCR screening of PSP toxin-producing and nontoxic A. circinalis isolates. Lane 1, isolate 131C; lane 2, 134C; lane 3, 279B; lane 4, 118C; lane 5, 150A; lane 6, 332H; lane 7, 306A; lane 8, 342D. (B) Multiplex PCR screening of PSP toxin-producing and nontoxic cyanobacterial blooms. Lane 1, strain 118C (positive control); lanes 2 to 4, PSP toxin-producing cyanobacterial blooms; lanes 5 to 7, nontoxic cyanobacterial blooms. PCR products were loaded at a total of 6 μl per sample and run with a 1-kb Plus DNA ladder (lanes M) as a standard.
DISCUSSION
For the present study we report the identification of genomic differences between toxic and nontoxic strains of A. circinalis. This was achieved by the application of HIP1 repeated-sequence PCR. Since the HIP1 element is present in more than 50% of cyanobacterial ORFs and is absent from rRNA and tRNA genes (6), it can be used to identify gene content and genomic structure in this phylum of bacteria.
The differences in HIP1 genomic profiles among isolates of A. circinalis were first indicated by the presence of a single unique DNA band for both toxic and nontoxic strains. This suggested consistent differences in composition and genomic organization between the two groups or the evolutionary divergence of toxic strains from the nontoxic congeners. These bands were characterized as encoding an NaDT and a conserved hypothetical protein in the STX-producing and the nontoxic isolates 134C and 306A, respectively.
Genomic heterogeneity between toxic and nontoxic strains of A. circinalis was further explored using the SSH technique for subtraction of HIP1-generated DNA pools. The aim here was to highlight the diversity in HIP1 libraries that could not be identified or resolved by gel electrophoresis. SSH has been used for the rapid identification of genetic differences between pathogenic bacteria (1, 13) as well as to derive information on ecologically relevant genetic adaptations in closely related prokaryotes (23, 25). The two groups of clones obtained here, though not exhaustive of the entire pool, were sufficiently representative of the whole SSH libraries compared to those used in previous studies (1), since an average of 18% of the sequences were encountered more than once. However, SSH did not detect the two HIP1 PCR products found to be characteristic of the toxic and nontoxic DNA banding patterns, possibly as a consequence of the number of clones analyzed.
Among the two categories of SSH putative tester-specific DNA fragments, the genes coding for carbamoyl-phosphate synthase and S-adenosyl-methyltransferase (clones 153 and 192, isolated from the toxic strain 134C) were of particular interest. These two enzymes could be potentially involved in regulation or production of STX in cyanobacteria (37, 40). Clones 153 and 192, however, were found both by microarray (Table 2) and by further PCR analyses (data not shown) to have no toxic-strain specificity. The specificity of tester-recovered fragments was verified by microarray hybridization, indicating that only one HIP1-SSH sequence was tester specific: a sequence encoding a putative serine/threonine kinase, clone 208 of the nontoxic strain 306A (Table 3).
One toxic-strain-specific gene was, however, indicated by microarray hybridization: the gene encoding NaDT that was originally cloned from the HIP1 PCR banding pattern of strain 134C. With the exception of the nontoxic strain 271C, this gene was found by the use of either or both degenerate and specific PCR only in STX-producing A. circinalis isolates (Fig. 2). Strain 271C gave the only false-positive toxin result observed by the use of YZf/R PCR amplification as well as HIP1 typing in this study. It is possible that this nontoxic isolate, previously shown by 16S rRNA analysis to cluster with toxic strains (5), could be a spontaneous natural mutant with regard to STX biosynthesis.
The putative function of the recovered NaDT was investigated by comparing the translated amino acid sequences from A. circinalis 134C and 279B with sequences encoding several other similar proteins; the results showed A. circinalis genes clustering in a group characterized by other bacterial and cyanobacterial NaDT genes (Fig. 3). Together with conservation of particular peptide motifs (Fig. 4), these data suggested functional homology between cyanobacterial NaDT genes and mrpF genes of Bacillus strains.
The mrp (multiple resistance and pH adaptation) operon and its homologues are distributed among diverse prokaryotic genera and function in multiple processes involving ion-coupled transport reactions, including Na+-specific pH homeostasis. Thus far, only the mrp operon of B. subtilis has been studied in detail (16, 17). The mrpF gene has been found to encode a protein functioning in cholate and Na+ efflux, with MrpF activity independent of the expression of any other additional mrp gene product (17). The Na+ efflux catalyzed by the independent transporter MrpF, coupled to solute efflux (e.g., endogenous cholate-like substrate and/or exogenous cholate-like compounds), has been suggested to be particularly important for coordinating a full Na+ cycle and achieving both substrate uptake and cytoplasmic pH regulation under alkaline pH conditions (17). Apart from that of N. punctiforme (http://www.jgi.doe.gov/JGI_microbial/html/), homologues of mrpF are present as single-copy genes in the known cyanobacterial genomes (Cyanobase: http://www.kazusa.or.jp/cyano/) and none are organized in a cluster similar to the Bacillus mrp operon. The putative transposases upstream and/or downstream of the coding region for the cyanobacterial NaDT homologues may indicate the possibility of a mobile genomic region comprising this transporter protein. Taken together, these data may suggest an essential difference between STX-producing and nontoxic isolates of A. circinalis in terms of Na+-dependent pH homeostasis and the Na+ cycle in these two groups of strains.
Previous studies have demonstrated an intrinsic association between the variation of cellular Na+ levels and the regulation of STX metabolism in the freshwater cyanobacterium C. raciborskii T3 (28, 30). STX production was strongly induced by alkaline pH and salt stress, while STX inhibited sodium uptake in C. raciborskii and A. circinalis (29). These results suggested a potential advantage for PSP toxin-producing microorganisms over other nontoxic species under conditions of high pH or salt stress. Differences in this Na+-dependent pH homeostasis may also be the reason for the geographic segregation of STX production in cyanobacteria, as an adaptation to specific environmental pressures such as natural cycles of flood and drought periods. Similar environmental conditions correlated with the dominance of STX-producing A. circinalis blooms in Australian freshwaters (7).
Additionally, it has been previously shown by 16S rDNA sequencing and phylogeny that most toxic and nontoxic A. circinalis strains cluster separately (5). Our results suggest that the NaDT gene did not specifically coevolve with 16S rDNA. The toxic strain 134C, which clustered within the nontoxic branch by 16S phylogeny, also contained the NaDT gene. This evidence further suggested an association of the NaDT sequence with the STX-producing genotype.
Therefore, we utilized the toxic-strain-specific gene NaDT to develop a PCR-based screening assay to detect the presence of potential STX-producing A. circinalis in the environment. The multiplex PCR was optimized to have an internal positive control (16S rDNA) to assess the occurrence of cyanobacterial DNA as well as the YZf/R probe to indicate the presence of PSP toxin-producing A. circinalis (Fig. 5). The YZf/R primers were designed to specifically differentiate A. circinalis NaDT sequences from other cyanobacterial (including Nostoc and Synechocystis) and bacterial (such as Bacillus) sequences from the databases. There is no evidence of false-positive PCRs from this study other than with A. circinalis 271C. Compared to previous approaches (5), this NaDT-based multiplex technique has the advantage of returning no false-negative toxin results, which represent a serious problem in water management strategies. Due to the sensitivity of the PCR (detection limit of ∼100 cells), this test may prove invaluable in the early detection of potentially toxic blooms. As bloom treatment can be environmentally and economically costly, the rapid and accurate nature of this test would allow for more logical assessment of bloom toxicity before treatment. In addition, by performing these predictive tests on sediment during the winter months areas of concern can be highlighted and thus treated prior to bloom proliferation in the warmer months.
In summary, HIP1 PCR was shown to be a valid tool for the investigation of the toxigenicity of related A. circinalis strains and allowed the identification of a toxic-strain-specific gene corresponding to NaDT. This discovery further supports a link between the production of STX and the maintenance of sodium and pH homeostasis in cyanobacteria. Specific primers were designed and applied to laboratory and environmental screening of STX-producing A. circinalis strains. As these tests are based on PCR, the combination of HIP1 typing and SSH requires only a small amount of original template DNA. Together with microarray technology, this method may also represent in the future an advantageous procedure for investigating heterogeneity in gene structures and metabolism between closely related slow-growing or environmental isolates.
Acknowledgments
We are grateful to R. Kellmann for advice, T. Salmon for assistance in Web publishing, and R. Cavalieri for technical assistance.
Research scholarships to F.P. from the University of New South Wales and the School of Biotechnology and Biomolecular Sciences are also acknowledged.
REFERENCES
- 1.Akopyants, N. S., A. Fradkov, L. Diatchenko, J. E. Hill, P. D. Siebert, S. A. Lukyanov, E. D. Sverdlov, and D. E. Berg. 1998. PCR-based subtractive hybridization and differences in gene content among strains of Helicobacter pylori. Proc. Natl. Acad. Sci. USA 95:13108-13113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alam, M., M. Ikawa, J. J. Sasner, Jr., and P. J. Sawyer. 1973. Purification of Aphanizomenon flos-aquae toxin and its chemical and physiological properties. Toxicon 11:65-72. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beltran, E. C. 2001. Taxonomy and molecular genetics of the saxitoxin producing cyanobacterium Anabaena circinalis. Ph.D. thesis. School of Microbiology and Immunology, The University of New South Wales, Sydney, Australia.
- 5.Beltran, E. C., and B. A. Neilan. 2000. Geographical segregation of the neurotoxin-producing cyanobacterium Anabaena circinalis. Appl. Environ. Microbiol. 66:4468-4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhaya, D., D. Vaulot, P. Amin, A. W. Takahashi, and A. R. Grossman. 2000. Isolation of regulated genes of the cyanobacterium Synechocystis sp. strain PCC6803 by differential display. J. Bacteriol. 182:5692-5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bowling, L. C., and P. D. Baker. 1996. Major cyanobacterial bloom in the Barwon-Darling River, Australia, in 1991, and underlying limnological conditions. Mar. Freshwater Res. 47:643-657. [Google Scholar]
- 8.Carmichael, W. W., W. R. Evans, Q. Q. Yin, P. Bell, and E. Moczydlowsky. 1997. Evidence of paralytic shellfish poisons in the freshwater cyanobacterium Lyngbya wollei (Farlow ex Gomont) comb. nov. Appl. Environ. Microbiol. 63:3104-3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Catterall, W. A. 1980. Neurotoxins that act on voltage-sensitive sodium channels in excitable membranes. Annu. Rev. Pharmacol. Toxicol. 20:15-43. [DOI] [PubMed] [Google Scholar]
- 10.Gallacher, S., and E. A. Smith. 1999. Bacteria and paralytic shellfish toxins. Protist 150:245-255. [DOI] [PubMed] [Google Scholar]
- 11.Gupta, A., A. P. Morby, J. S. Turner, B. A. Whitton, and N. J. Robinson. 1993. Deletion within the metallothionein locus of cadmium tolerant Synechococcus PCC6301 involving a highly iterated palindrome (Hip1). Mol. Microbiol. 5:825-834. [DOI] [PubMed] [Google Scholar]
- 12.Harada, T., Y. Oshima, and T. Yasumoto. 1982. Structure of two paralytic shellfish toxins, Gonyautoxins V and VI, isolated from a tropical dinoflagellate, Pyrodinium bahamense var. compressa. Agric. Biol. Chem. 46:1861-1864. [Google Scholar]
- 13.Harakava, R., and D. W. Gabriel. 2003. Genetic differences between two strains of Xylella fastidiosa revealed by suppression subtractive hybridization. Appl. Environ. Microbiol. 69:1315-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hawkins, P. R., N. R. Chandrasena, G. J. Jones, A. R. Humpage, and I. R. Falconer. 1997. Isolation and toxicity of Cylindrospermopsis raciborskii from an ornamental lake. Toxicon 35:341-346. [DOI] [PubMed] [Google Scholar]
- 15.Humpage, A. R., J. Rositano, A. Bretag, R. Brown, P. Baker, B. C. Nicholson, and D. A. Steffensen. 1994. Paralytic shellfish poisons from Australian cyanobacterial blooms. Aust. J. Mar. Freshwater Res. 45:761-771. [Google Scholar]
- 16.Ito, M., A. A. Guffanti, B. Oudega, and T. A. Krulwich. 1999. mrp, a multigene, multifunctional locus in Bacillus subtilis with roles in resistance to cholate and to Na+ and in pH homeostasis. J. Bacteriol. 181:2394-2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ito, M., A. A. Guffanti, W. Wang, and T. A. Krulwich. 2000. Effects of nonpolar mutations in each of the seven Bacillus subtilis mrp genes suggest complex interactions among the gene products in support of Na+ and alkali but not cholate resistance. J. Bacteriol. 182:5663-5670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jukes, T. H., and C. R. Cantor. 1969. Evolution of protein molecules. p. 21-132. In H. N. Munro (ed.), Mammalian protein metabolism, vol. 3. Academic Press Inc., New York, N.Y. [Google Scholar]
- 19.Kaebernick, M. 2001. Regulation and function of microcystin production in Microcystis aeruginosa. Ph.D. thesis. School of Microbiology and Immunology, The University of New South Wales, Sydney, Australia.
- 20.Kao, C. Y. 1993. Paralytic shellfish poisoning, p. 75. In E. R. Falconer (ed.), Algal toxins in seafood and drinking water. Academic Press, London, United Kingdom.
- 21.Lagos, N., H. Onodera, P. A. Zagatto, D. Andrinolo, S. M. F. Q. Azevedo, and Y. Oshima. 1999. The first evidence of paralytic shellfish toxins in the freshwater cyanobacterium Cylindrospermopsis raciborskii, isolated from Brazil. Toxicon 37:1359-1373. [DOI] [PubMed] [Google Scholar]
- 22.Llewellyn, L. E., A. P. Negri, J. Doyle, P. D. Baker, E. C. Beltran, and B. A. Neilan. 2001. Radioreceptor assays for sensitive detection and quantitation of saxitoxin and its analogues from strains of the freshwater cyanobacterium, Anabaena circinalis. Environ. Sci. Technol. 35:1445-1451. [DOI] [PubMed] [Google Scholar]
- 23.Mavrodi, D. V., O. V. Mavrodi, B. B. McSpadden-Gardener, B. B. Landa, D. M. Weller, and L. S. Thomashow. 2002. Identification of differences in genome content among phlD-positive Pseudomonas fluorescens strains by using PCR-based subtractive hybridization. Appl. Environ. Microbiol. 68:5170-5176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neilan, B. A., M. L. Saker, J. Fastner, A. Torokne, and B. P. Burns. 2003. Phylogeography of the invasive cyanobacterium Cylindrospermopsis raciborskii. Mol. Ecol. 12:133-140. [DOI] [PubMed] [Google Scholar]
- 25.Nesbø, C. L., K. E. Nelson, and W. F. Doolittle. 2002. Suppressive subtractive hybridization detects extensive genomic diversity in Thermotoga maritima. J. Bacteriol. 184:4475-4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oshima, Y., M. Hasegawa, T. Yasumoto, G. Hallegaeff, and S. Blackburn. 1987. Dinoflagellate Gimnodium catenatum as the source of paralytic shellfish toxins in Tasmanian shellfish. Toxicon 25:1105-1111. [DOI] [PubMed] [Google Scholar]
- 27.Pomati, F., and B. A. Neilan. 2004. PCR-based positive hybridization to detect genomic diversity associated with bacterial secondary metabolism. Nucleic Acid Res. 32:e7. [Online.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pomati, F., B. A. Neilan, G. Manarolla, T. Suzuki, and C. Rossetti. 2003. Enhancement of intracellular saxitoxin accumulation by lidocaine hydrochloride in the cyanobacterium Cylindrospermopsis raciborskii T3 (Nostocales). J. Phycol. 39:535-542. [Google Scholar]
- 29.Pomati, F., C. Rossetti, D. Calamari, and B. A. Neilan. 2003. Effects of saxitoxin and veratridine on bacterial Na+/K+ fluxes: a prokaryote-based STX bioassay. Appl. Environ. Microbiol. 69:7371-7376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pomati, F., C. Rossetti, G. Manarolla, and B. A. Neilan. Interactions between intracellular Na+ levels and saxitoxin production in Cylindrospermopsis raciborskii T3. Microbiology 150:455-461. [DOI] [PubMed]
- 31.Pomati, F., S. Sacchi, C. Rossetti, S. Giovannardi, H. Onodera, Y. Oshima, and B. A. Neilan. 2000. The freshwater cyanobacterium Planktothrix sp. FP1: molecular identification and detection of paralytic shellfish poisoning toxins. J. Phycol. 36:553-562. [DOI] [PubMed] [Google Scholar]
- 32.Robinson, N. J., P. J. Robinson, A. Gupta, A. J. Bleasby, B. A. Whitton, and A. P. Morby. 1995. Singular overrepresentation of an octameric palindrome, HIP1, in DNA from many cyanobacteria. Nucleic Acids Res. 23:729-735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saitou, N., and M. Nei. 1987. The neighbour-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 34.Saker, M. L., and B. A. Neilan. 2001. Varied diazotrophies, morphologies, and toxicities of genetically similar isolates of Cylindrospermopsis raciborskii (Nostocales, Cyanophyceae) from northern Australia. Appl. Environ. Microbiol. 67:1839-1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 36.Shimizu, Y. 1977. Chemistry and distribution of deleterious dinoflagellate toxins, p. 261-269. In D. J. Faulkner and W. H. Fenical (ed.), Marine Natural Products Chemistry. Plenum, New York, N.Y.
- 37.Shimizu, Y. 1993. Microalgal metabolites. Chem. Rev. 93:1685-1698. [Google Scholar]
- 38.Sivonen, K., and G. Jones. 1999. Cyanobacterial toxins, p. 41-111. In I. Chorus and J. Bartram (ed.), Toxic cyanobacteria in water. E & FN Spon Publishers, London, United Kingdom.
- 39.Smith, J. K., J. D. Parry, J. G. Day, and R. J. Smith. 1998. A PCR technique based on the Hip1 interspersed repetitive sequence distinguishes cyanobacterial species and strains. Microbiology 144:2791-2801. [DOI] [PubMed] [Google Scholar]
- 40.Taroncher-Oldenburg, G., and D. M. Anderson. 2000. Identification and characterization of three differentially expressed genes, encoding S-adenosylhomocysteine hydrolase, methionine aminopeptidase, and a histone-like protein, in the toxic dinoflagellate Alexandrium fundyense. Appl. Environ. Microbiol. 66:2105-2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tillett, D., and B. A. Neilan. 2000. Xanthogenate nucleic acid isolation from cultured and environmental cyanobacteria. J. Phycol. 35:1-8. [Google Scholar]
- 43.Zheng, W., T. Song, X. Bao, B. Bergman, and U. Rasmussen. 2002. High cyanobacterial diversity in coralloid roots of cycads revealed by PCR fingerprinting. FEMS Microbiol. Ecol. 40:215-222. [DOI] [PubMed] [Google Scholar]
- 44.Zingone, A., and H. O. Enevoldsen. 2000. The diversity of harmful algal blooms: a challenge for science and management. Ocean & Coastal Management 43:725-748. [Google Scholar]