Abstract
A set of proteins that changed their levels of synthesis during growth of Acidithiobacillus ferrooxidans ATCC 19859 on metal sulfides, thiosulfate, elemental sulfur, and ferrous iron was characterized by using two-dimensional polyacrylamide gel electrophoresis. N-terminal amino acid sequencing and mass spectrometry analysis of these proteins allowed their identification and the localization of the corresponding genes in the available genomic sequence of A. ferrooxidans ATCC 23270. The genomic context around several of these genes suggests their involvement in the energetic metabolism of A. ferrooxidans. Two groups of proteins could be distinguished. The first consisted of proteins highly upregulated by growth on sulfur compounds (and downregulated by growth on ferrous iron): a 44-kDa outer membrane protein, an exported 21-kDa putative thiosulfate sulfur transferase protein, a 33-kDa putative thiosulfate/sulfate binding protein, a 45-kDa putative capsule polysaccharide export protein, and a putative 16-kDa protein of unknown function. The second group of proteins comprised those downregulated by growth on sulfur (and upregulated by growth on ferrous iron): rusticyanin, a cytochrome c552, a putative phosphate binding protein (PstS), the small and large subunits of ribulose biphosphate carboxylase, and a 30-kDa putative CbbQ protein, among others. The results suggest in general a separation of the iron and sulfur utilization pathways. Rusticyanin, in addition to being highly expressed on ferrous iron, was also newly synthesized, as determined by metabolic labeling, although at lower levels, during growth on sulfur compounds and iron-free metal sulfides. During growth on metal sulfides containing iron, such as pyrite and chalcopyrite, both proteins upregulated on ferrous iron and those upregulated on sulfur compounds were synthesized, indicating that the two energy-generating pathways are induced simultaneously depending on the kind and concentration of oxidizable substrates available.
Acidithiobacillus ferrooxidans (formerly Thiobacillus ferrooxidans) is a chemolithoautotrophic bacterium that obtains its energy from the oxidation of ferrous iron, elemental sulfur, or partially oxidized sulfur compounds (6, 16, 26, 30, 41). The ability of this and other microorganisms present in its habitat to solubilize metal sulfides is successfully applied in biomining operations (30, 41).
The reactions involved in ferrous iron oxidation have been studied in detail (5, 8, 21, 26, 30, 41). Nevertheless, the electron pathway from ferrous iron to oxygen has not been entirely established (44). The terminal electron acceptor is assumed to be a cytochrome oxidase anchored to the cytoplasmic membrane. The transfer of electrons would occur through several periplasmic carriers, including at least the blue copper protein rusticyanin and cytochrome c552. Recently, a high-molecular-weight c-type cytochrome, Cyc2, has been suggested to be the prime candidate for the initial electron acceptor in the respiratory pathway between ferrous iron and oxygen. This pathway would be Cyc2→rusticyanin→Cyc1(c552)→cytochrome aa3 oxidase (44).
The aerobic oxidation of elemental sulfur by A. ferrooxidans and other microorganisms is carried out by a sulfur dioxygenase (31, 36-38). Thiosulfate has been postulated as a key compound in the oxidation of the sulfur moiety of pyrite (34). All reactions comprising this oxidation have been shown to occur chemically (32, 33). However, sulfur compound-oxidizing enzymes such as the tetrathionate hydrolase of A. ferrooxidans, A. thiooxidans, or Acidiphilium acidiphilum may also be involved in the process (9, 11, 12, 22, 39). In addition, enzymes for thiosulfate or sulfite oxidation of A. ferrooxidans or A. thiooxidans may successfully compete with the chemical reactions with iron(III) ions as an oxidizing agent (34). A rhodanese activity has been described previously for A. ferrooxidans (40). This enzyme is a thiosulfate sulfur transferase (TST) which breaks the S-S bond present in thiosulfate, generating sulfur and sulfite. Other enzymes may also participate in the thiosulfate mechanism, such as the thiosulfate-oxidizing enzyme of A. ferrooxidans (36). Ramírez et al. have recently described an exported TST-like protein in A. ferrooxidans that is highly expressed when the bacterium is grown on pyrite and sulfur, but not on ferrous iron (29), and which is possibly involved in sulfur metabolism.
To further study some of the components involved in the oxidation of sulfur and metal sulfides, we analyzed by expression proteomics a subset of proteins synthesized by the bacterium when grown on Fe(II) ions, S0, thiosulfate, pyrite, chalcopyrite, or iron-free metal sulfides. Our results indicate that synthesis of several proteins related to sulfur metabolism, including oxidation and acquisition, was induced (or derepressed) when the microorganisms grew on reduced inorganic sulfur forms. By growth of the microorganism on ferrous iron, most of these proteins were downregulated, while proteins of the ferrous iron oxidation pathway were greatly induced, supporting the existence of separate regulation for the iron and sulfur oxidation pathways which depends on the substrate being oxidized.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
A. ferrooxidans strain ATCC 19859 was grown in ferrous iron-containing modified 9K medium as described previously (14, 29). Growth on elemental sulfur was carried out by using sulfur prills (3). Pyrite (40% [wt/wt] Fe, found to pass a 75-μm-mesh sieve) and a chalcopyrite concentrate were kindly given to us by Paul Norris (University of Warwick, Coventry, United Kingdom). ZnS and CuS were purchased from Aldrich and contained 28 and 125 ppm of trace iron, respectively. Growth of cells in these substrates was carried out in modified 9K medium (pH 2.5) in which ferrous iron was replaced with 1% (wt/vol) of the corresponding metal sulfide. Growth of A. ferrooxidans in thiosulfate was carried out in DSMZ medium 71 containing 20 mM thiosulfate and the following components (in grams per liter): KH2PO4, 3.0; MgSO4 · 7H2O, 0.5; (NH4)2SO4, 3.0; CaCl2 · 2H2O, 0.25. The pH was kept at about 4.6 by addition of 1 M NaOH. For radioactive labeling of total-cell proteins, washed cells (1010) were resuspended in 10 ml of the corresponding medium, 0.1 mCi (specific activity, 1,087 Ci/mmol) of [35S]methionine was added, and cells were incubated with or without additions (where indicated) under the same growth conditions for 30 to 40 h.
2-D NEPHGE, SDS-PAGE, and autoradiography.
Total-cell proteins were separated by two-dimensional nonequilibrium pH polyacrylamide gel electrophoresis (2-D NEPHGE) (28), performed as described previously for A. ferrooxidans (29), by using ampholytes (pH 3 to 10) from Bio-Rad. Cell samples (4 mg [wet weight] of unlabeled cells, or 500,000 cpm contained in cells labeled by growth on [35S]methionine) or cell-free fractions to be analyzed were resuspended in 80 μl of sonication buffer (10 mM Tris-HCl [pH 7.4], 5 mM MgCl2, and 50 μg of pancreatic RNase per ml), sonicated, and treated with DNase (final concentration, 50 μg per ml). The mixture was then lyophilized and dissolved in lysis buffer as described previously (29). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (25) gels consisted of 12.5% polyacrylamide or 5-to-20% polyacrylamide gradients. Protein arrays were visualized by staining with Coomasie blue or by autoradiography as described previously (29). Proteins containing heme groups were stained by their peroxidase activity in the presence of H2O2 with 3,3′,5′,5′-tetramethyl benzidine as the electron donor. By this method, the proteins with noncovalently bound heme groups are much less intensely stained in the presence of β-mercaptoethanol (10). Since this thiol is present under our conditions for 2-D PAGE, we are most likely detecting only the covalently bound heme groups. The relative levels of synthesis of radioactive proteins were determined from the autoradiograms by using the Scion Image analysis program (www.scioncorp.com).
Microsequencing and MS analysis of proteins extracted from 2-D gels.
Proteins of interest were recovered from Coomassie blue-stained and heat-dried 2-D gels by excising the protein spots. After rehydration and concentration of the spots by SDS-PAGE, the proteins were electroblotted onto a polyvinylidene difluoride membrane (27) and were subjected to microsequencing by the Laboratoire de Microséquençage des Protéines of the Institut Pasteur, Paris, France. Alternatively, silver-stained proteins were isolated from 2-D gels, and in-gel trypsin digestion and protein extraction were performed. The peptide solutions were analyzed by electron spray tandem ionization-mass spectrometry (ESI-MS-MS). The results obtained were analyzed with MASCOT software (http://www.matrixscience.com/index.html) by using the MS/MS Ion search. The entire MS analysis was performed at the Cambridge Center for Proteomics, University of Cambridge, Cambridge, United Kingdom.
Western immunoblotting.
The proteins synthesized by A. ferrooxidans grown on different oxidizable substrates were separated by SDS-PAGE and electrotransferred to a polyvinylidene difluoride membrane as described previously (14). For the antigen-antibody reaction, membranes containing the transferred proteins were treated with an antiserum against rusticyanin as the primary antibody (dilution, 1:5,000) and monoclonal anti-mouse antibodies conjugated with peroxidase (Amersham) as the secondary antibodies (dilution, 1:2,500). A colorimetric method was used to develop Western blots as recommended by Promega. The relative intensities of the Rus protein bands in Western blots were determined by using a scanner and the Scion Image analysis program.
Preparation of cell-free fractions from A. ferrooxidans.
A. ferrooxidans was separated into outer membrane, soluble, and inner membrane fractions as previously described (14). The soluble fraction was the supernatant obtained after the first centrifugation at 100,000 × g for 2 h to pellet the total membrane fraction. The inner membrane fraction was the last supernatant obtained after treatment of the total membrane fraction in the presence of 2% sodium laurylsarcosinate to separate the final outer membrane preparation.
Oligonucleotides.
The following synthetic oligonucleotides were used: for reverse transcription (RT) experiments, 5′-CCTGCGGGTTGTCATACT-3′; for PCRs, a (5′-CGTCTGGTTGGATAGCTG-3′), b (5′-GTCGTCGGCGGAGGCCACCGA-3′), c (5′-CATCGCTCAAAGGCAAAG-3′), d (5′-ATGGCAACGACCATGTCA-3′), e (5′-CAACGTCCCGATCCCCGCGAA-3′), and f (5′-GTAAAACGCTGCAAGTCG-3′).
RT-PCR.
A. ferrooxidans ATCC 23270 total RNA was prepared from a sulfur-grown culture by a modified hot phenol method as described previously (15). After the phenol extraction, the fraction containing RNA was extracted twice with chloroform. The RNA was precipitated overnight with ethanol at −20°C, washed with 70% ethanol, and resuspended in 60 μl of nuclease-free water. Four micrograms of total RNA was treated with 30 U of RNase-free DNase I (Promega) and 5 mM MgCl2 in a final volume of 10 μl for 1 h at 37°C. Finally, the DNase I was inactivated by incubating at 65°C for 10 min.
RT-PCR was carried out with 2 μg of DNase I-treated total RNA of A. ferrooxidans. The RNA was denatured in the presence of 400 ng of the RT primer at 70°C for 5 min and then cooled on ice. The RT reaction was carried out at 42°C for 1 h in a final volume of 25 μl according to the manufacturer′s instructions (Promega). After the RT reaction, the enzyme was inactivated by heating at 65°C for 10 min. PCR was performed using 3 μl of the RT reaction product in a final volume of 50 μl containing Taq DNA polymerase reaction buffer (Promega), 2.5 mM MgCl2, 100 μM each deoxynucleoside triphosphate, 5% (vol/vol) dimethyl sulfoxide, 250 ng of each primer, and 1 U of Taq DNA polymerase (Promega). For the PCR, the following program was used: an initial denaturing step at 95°C for 3 min; 40 cycles of 30 s at 95°C, 30 s at 50°C, and 45 s at 72°C; and a final extension step at 72°C for 3 min.
For each RT-PCR experiment, a control RT reaction without reverse transcriptase was carried out to check for the absence of genomic DNA contaminations in the RNA preparations used. RT-PCR products were checked by electrophoresis in a 1% agarose gel in 0.5× TAE buffer.
Sequence analysis.
Identity and similarity searching in databases was done by using the BlastP program (1) of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov). We used the available A. ferrooxidans ATCC 23270 genomic sequence, which is finished but not yet annotated (http://www.tigr.org). Molecular masses and isoelectric points of open reading frame (ORF) products were obtained by using the ProtParam tool (http://www.expasy.ch/cgi-bin/protparam). The possible presence of transmembrane domains in the ORFs analyzed was predicted by TMpred (http://www.ch.embnet.org/software/TMPRED_form.html.
RESULTS AND DISCUSSION
Patterns of global protein synthesis of A. ferrooxidans grown with different oxidizable substrates.
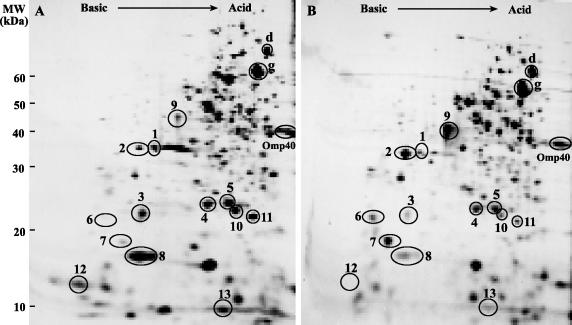
To characterize some of the proteins differentially synthesized by A. ferrooxidans when grown on elemental sulfur or ferrous iron, we used 2-D NEPHGE analysis of the total-cell proteins labeled with radioactive methionine followed by autoradiography. The general pattern of protein synthesis for ferrous iron-grown A. ferrooxidans showed several differences from that observed for the sulfur-grown bacterium (Fig. 1). The differences involved both upregulation and downregulation of the synthesis of numerous polypeptides. We decided to study in detail the group of proteins indicated in the gels of Fig. 1, since all these polypeptides were well stained by Coomassie blue or silver and therefore could be subjected to an efficient N-terminal sequencing or MS analysis. Some of the less abundant proteins (such as spots 6 and 3) were concentrated after isolation of their spots from several equivalent 2-D gels. Other proteins were not isolated directly from the 2-D gels of total-cell proteins for N-terminal sequencing but from subproteomes consisting of outer membrane or soluble fractions from the cells analyzed (see below).
FIG. 1.
2-D protein arrays of A. ferrooxidans grown on ferrous iron or elemental sulfur. Total proteins from cells grown in the presence of [35S]methionine and ferrous iron (A) or elemental sulfur (B) were separated by 2-D NEPHGE with a pH gradient between 3.0 (right side of the gel) and 10.0 (left side of the gel). Spots were detected by autoradiography. Numbered spots are those characterized in this study. As internal controls for proteins that do not greatly change their levels of synthesis during growth under these conditions, we used GroEL (g), DnaK (d,) and Omp40 proteins previously identified in our laboratory (14, 42). Positions of molecular mass standards (in kilodaltons) are given on the left.
Amino acid sequences of proteins and their identification by use of databases.
The chosen proteins, which in general were upregulated or downregulated more than twofold, were isolated from 2-D gels and subjected to microsequencing or MS analysis. Using these sequences, we performed the corresponding tBlastN analysis and identified those proteins described in Table 1. All these sequences were also present in the genome sequence of A. ferrooxidans strain ATCC 23270.
TABLE 1.
A. ferrooxidans protein spots upregulated or downregulated by growth on sulfur and identified by N-terminal microsequencing or MS analysis
| Characteristic of protein changing its level of synthesis
|
Characteristic of matching protein from NCBI databasea
|
||||||
|---|---|---|---|---|---|---|---|
| Spotb | SPc | Mr/pId | S0e | Amino acid sequence | Homologous protein | Organism | Accession no. |
| 1 | + | 32,975/8.58 | ↓2.4 | APTISLLETGSTLLYPLFNLAV | Phosphate binding protein PstS | Mycobacterium tuberculosis | AAK45208 |
| 2 | + | 33,984/8.77 | ↑1.9 | ADMGWNGKAEAPRYQEQVFPPW | Sulfate/thiosulfate binding protein | Nostoc sp. strain PCC7120 | NP_484366 |
| 3 | + | 19,950/8.86 | ↓3.4 | AVGSADAPAPYRVVIXAM | Cytochrome c552 | Acidithiobacillus ferrooxidans | P74917 |
| 4 | − | 22,377/5.69 | ↓1.6 | AVLVGKAADDFVAPAVMPDN | Thioredoxin peroxidase | Rickettsia prowazekii | CAA14787 |
| 5 | − | 22,948/6.05 | ↓2.1 | SEYSVREELKPSGLDGISDA | Superoxide dismutase | Thermoplasma volcanium | BAB59201 |
| 6 | + | 20,915/9.18 | ↑∞ | DDSGNQAAQQVLNARMEKFFADQKPYG | Thiosulfate sulfur transferase | Acidithiobacillus ferrooxidans | CAC43401 |
| 7 | + | 16,322/8.84 | ↑7.7 | AEAPSGATGSTAGAPMAPPAF | Unknown | Aquifex aeolicus | NP_213005 |
| 8 | + | 16,550/8.00 | ↓5.9 | GTLDTTWKEATLPQVKAMLEKD | Rusticyanin | Acidithiobacillus ferrooxidans | P24930 |
| 9 | + | 45,137/7.85 | ↑4.6 | VTMQISRAGK | Capsule polysaccharide export WcbC | Burkholderia mallei | AAK26456 |
| 10 | − | 22,673/5.54 | ↓5.7 | MAGLASDGLDLLDR | Putative protein | Acidithiobacillus ferrooxidans | |
| 11 | − | 20,371/5.25 | ↓3.5 | GGQDSDQLGPPPAADPR | Conserved hypothetical protein | Aquifex aeolicus | NP_213857 |
| 12 | − | 11,332/9.00 | ↓∞ | EQVDYPEAVAGGK | Bacterial nucleoid DNA-binding protein | Ralstonia metallidurans | ZP_23168 |
| 13 | − | 12,877/5.90 | ↓6.5 | LIGYDNYSQSQGSAFVVFR | Small RuBisCo subunit CbbS | Acidithiobacillus ferrooxidans | AAD30509 |
| OMP44 | + | 44,556/6.55 | ↑∞ | TDGYQLIGIGQYAVGMAGAV | FadL outer membrane protein | Pseudomonas putida | CAC86824 |
| P30 | − | 29,662/5.30 | ↓∞ | MSPEIDQYLVRNNPYYRTVA | CbbQ | Acidithiobacillus ferrooxidans | CAB45260 |
| P51 | − | 52,793/5.71 | ↓3.5 | AVKKEAGVKEYRQTYWAPE | Large RuBisCo subunit CbbL | Acidithiobacillus ferrooxidans | AA30508 |
Spot designations are from Fig. 1 except for OMP44, P30, and P51 (analyzed by 2-D PAGE of outer membrane [OMP44] or soluble fractions). Spots 9 to 13 were analyzed by ESI-MS-MS.
The presence (+) or absence (−) of putative signal peptides (SP) was determined by using the SignalP 3.0 server (available at http://www.cbs.dtu.dk/services/SignalP).
Obtained by using the ProtParam tool (available at http://www.expasy.ch/cgi-bin/protparam).
Fold increase (↑) or decrease (↓) in cells grown on S0 relative to levels in cells grown on Fe2+.
Proteins upregulated by growth on sulfur (and downregulated by growth on ferrous iron).
Protein spot 7 was highly upregulated by growth on sulfur (more than sevenfold). Searches in sequence databases showed that its N-terminal amino acid sequence was 40% identical to that of a conserved putative protein of unknown function present in the genome of Aquifex aeolicus (accession no. NP_213005). By comparison of the amino acid sequence of protein spot 7 against the genome of A. ferrooxidans, we found a gene for a putative basic protein which contained the original N-terminal sequence after a putative signal peptide. The possibly exported protein of 16 kDa contains about 10% methionine residues (including its signal peptide).
Spot 2 in Fig. 1 was upregulated almost twofold by growth on sulfur. Its N-terminal amino acid sequence showed extended similarity to that of a sulfate/thiosulfate binding protein (SBP). By searching for its amino acid sequence in the A. ferrooxidans genome, we found a gene coding for a deduced SBP also containing a predicted signal peptide. The N terminus of the theoretically processed protein was identical to the sequence of spot 2. This putative SBP corresponds to SBP1, one of the two putative SBPs deduced to be present in the genomic context of the exported rhodanese-like protein P21 (29). The sulfate binding proteins are induced in heterotrophic bacteria when sulfate is limiting (less than 500 μM) (20). A. ferrooxidans usually encounters high sulfate anion concentrations in its environment compared to heterotrophic bacteria, and therefore it is difficult to assess the effect of sulfate concentration changes on the expression of SBPs. Nevertheless, if the synthesis of an SBP is repressed by higher sulfate concentrations, its synthesis would be expected to increase both on sulfur and on thiosulfate, as we have found, since the total sulfate concentrations of these cultures are much lower than those in ferrous iron-grown cells (see Fig. 1 and 3 below).
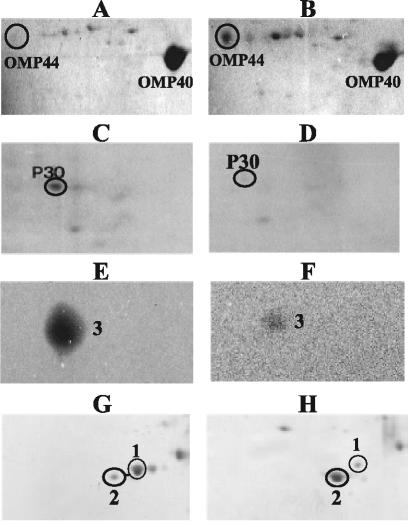
FIG. 3.
Characterization of some proteins from total cells or different fractions from A. ferrooxidans. Omp44 was identified in outer membrane-enriched fractions from cells grown on ferrous iron (A) or sulfur (B). P30 was identified in soluble fractions obtained from cells grown on ferrous iron (C) or sulfur (D). Cytochrome c552 was identified in total-cell proteins from cells grown on ferrous iron (E) or sulfur (F). Sulfate binding protein (spot 2) and PstS (spot 1) were identified in total proteins from cells grown on ferrous iron (G) or thiosulfate (H). All proteins were separated by 2-D NEPHGE, and spots were stained with Coomassie blue (A, B, C, D, G, and H) or with heme-staining reagent (E and F).
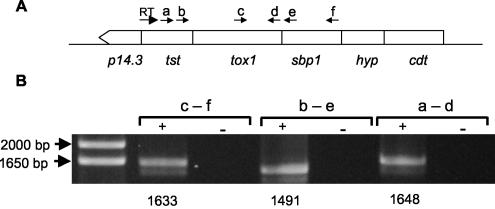
Figure 1 and Table 1 show that as described previously (29), spot 6, a putative TST, was greatly induced (much more than fivefold) when the cells grew on sulfur (Fig. 1B) and was entirely absent when A. ferrooxidans was grown on ferrous iron (Fig. 1A; Table 1). By performing RT-PCR experiments with the appropriate primers, we found that the genes coding for SBP1, Tox1, and TST may form part of a polycistronic cluster with three other putative genes having the same orientation (Fig. 2).
FIG. 2.
Analysis of cotranscription of the tst and sbp1 genes of A. ferrooxidans by RT-PCR. (A) Schematic map of the genetic context around the tst and sbp1 genes of A. ferrooxidans ATCC 23270. Arrows indicate locations of primers used for the RT reaction and for PCR (a through f). The genes present in the region analyzed were p14.3 and tst, encoding TST-like proteins; tox1, encoding a terminal oxidase subunit; sbp1, encoding an SBP; hyp, encoding a hypothetical protein; and cdt, encoding a C4-dicarboxylate transporter. (B) Agarose gel electrophoresis of RT-PCR products obtained for the tst, tox1, and sbp1 intergenic regions. The primers used in each reaction are indicated (a through f). The RT reaction was carried out on 2 μg of total RNA obtained from sulfur-grown cells of A. ferrooxidans. RT reactions with (+ lanes) and without (− lanes) the Moloney murine leukemia virus reverse transcriptase enzyme were carried out in order to exclude amplification due to genomic DNA contamination. Sizes of DNA markers are shown on the left. Expected sizes (in base pairs) for the corresponding RT-PCR products are given below the gel.
Spot 9 (Fig. 1 and Table 1) was highly induced in sulfur-grown cells. This protein showed homology to WcbC, a bacterial capsule polysaccharide export protein. A. ferrooxidans is known to adhere to solid mineral surfaces such as the elemental sulfur used here by forming an extracellular polysaccharide substance (13). Therefore, it will be of great interest to further study the possible role of this protein in the generation of these polymers.
Figure 3A and B show that Omp44 was dramatically upregulated by growth on sulfur (see Table 1). This outer membrane protein is present in much smaller amounts than Omp40, which is the major outer membrane protein in A. ferrooxidans (14). The sequence and putative structure of Omp44 differ from those of the 42-kDa outer membrane protein previously described by Buonfiglio et al. (7), which is also induced by growth of A. ferrooxidans on sulfur. Comparison with databases shows the similarity of Omp44 to a family of proteins which includes FadL, a protein involved in the translocation of long-chain fatty acids across the outer membrane (4). The relationship of Omp44 to sulfur metabolism is not clear at present. Growth of A. ferrooxidans on elemental sulfur requires attachment of the cells to the solid substrate, and this interaction could induce the changes we observed and those reported by Buonfiglio et al. (7) in the outer membrane proteins of this microorganism.
Proteins downregulated by growth on sulfur (and upregulated by growth on ferrous iron).
Table 1 shows that protein spot 13, which was identified as the small subunit of ribulose bisphosphate carboxylase (RuBisCo), was downregulated on sulfur compared with its levels of synthesis on ferrous iron. The same happened in the case of the large subunit of RuBisCo identified under this growth condition (P51 [Table 1]). A. ferrooxidans possesses two forms of RuBisCo with their corresponding sets of genes (24). These are cbbLS1 and cbbLS2. The RuBisCo subunits that we identified belong to cbbLS1.
Table 1 and Fig. 3C and D show that protein P30 was also greatly downregulated in sulfur. P30 presented amino acid squence similarity to the related proteins CbbQ, NirQ, and NorQ (17). In A. ferrooxidans a cbbQ gene has been initially described upstream of a cbbO gene and downstream of the second set of RuBisCo genes, cbbLS2 (19). Both the cbbQ and cbbO genes are involved in maintaining the conformational states and activity of RuBisCo (18). Our DNA sequencing of the genomic context of the p30 gene in A. ferrooxidans ATCC 19859 and the equivalent context in the genome of A. ferrooxidans ATCC 23270 showed the following gene order: cbbR-cbbL1-cbbS1(spot 13)-the carboxysome genes (csoS2, csoS3, orfA, orfB, csoS1C, csoS1A, csoS1B)-orf1-orf2-orf3-orf4-p30-cbbO. Therefore, P30 most likely corresponds to a second CbbQ protein, since its contiguous cbbO-like gene is cotranscribed with p30, as we determined by RT-PCR experiments (data not shown).
Autotrophy in A. ferrooxidans is driven by the Calvin cycle. The expression of Calvin cycle enzymes varies considerably in different chemoautotrophic microorganisms depending on the substrate being oxidized (35). Expression of the Calvin cycle genes is regulated by the transcriptional activator CbbR [activated by the binding of NAD(P)H] and by a possible repressor protein that may respond to the intracellular concentration of an intermediary metabolite such as phosphoenolpyruvate in the general model for the regulation of expression of the Calvin cycle proposed by Shively et al. (35). CbbR has been identified and shown to control the expression of cbb genes in A. ferrooxidans (23).
Figure 1 and Table 1 show that protein spots 4 and 5 were downregulated only 1.6- to 2-fold in cells grown on sulfur. Protein spot 4 showed similarity to thioredoxin peroxidase, and protein spot 5 showed similarity to superoxide dismutase. This suggests a possibly higher-stress condition when A. ferrooxidans is grown on iron. Protein spot 1 in Fig. 1 was found to be the putative phosphate-binding protein that is induced in A. ferrooxidans by the lack of phosphate and that is apparently part of a Pho regulon in this microorganism (43). PstS levels decreased more than twofold by growth on sulfur or thiosulfate (Fig. 3G and H), suggesting a possible limitation of phosphate during growth in ferrous iron medium.
Protein spots 10 and 11 were downregulated by growth on sulfur (Fig. 1; Table 1). These two proteins corresponded, respectively, to a putative protein from A. ferrooxidans and to a conserved hypothetical protein from A. aeolicus, both of unknown function. Spot 12 was dramatically downregulated by growth of A. ferrooxidans on sulfur. This protein was homologous to a bacterial nucleoid DNA binding protein. The possible role of a protein of this kind in A. ferrooxidans is actually unknown.
Protein spot 3 was highly downregulated by growth of cells on sulfur (Fig. 1). The staining of this protein with 3,3′,5′,5′-tetramethyl benzidine (Fig. 3E and F) suggested the presence of heme groups and a possible cytochrome. The protein was identified as cytochrome c552 by its N-terminal sequence, as seen in Table 1.
Spot 8 in Fig. 1 was identified as rusticyanin (Rus), a protein known to be induced mainly by growth of A. ferrooxidans on ferrous iron (8, 21, 45). We found that Rus was downregulated dramatically (more than fivefold) by growth on sulfur compared to its levels in ferrous iron-grown cells. Nevertheless, the protein was still synthesized de novo in sulfur-grown cells (see below).
Effects of ferrous iron on the expression of sulfur-upregulated proteins.
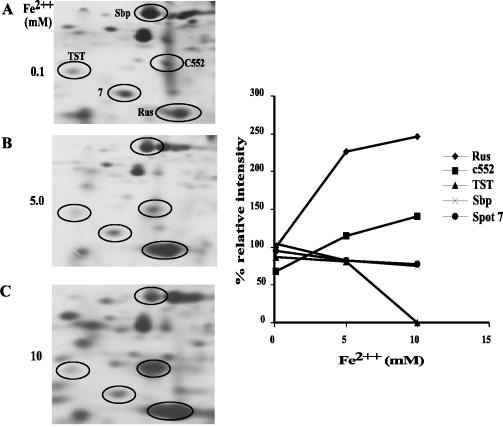
When small, increasing ferrous iron concentrations were added to A. ferrooxidans grown on elemental sulfur, the synthesis of proteins such as spots 6 (TST), 2 (SBP), and 7 was downregulated by the presence of iron in the growth medium. Synthesis of TST was strongly inhibited by increasing ferrous iron concentrations, while the synthesis of SBP and spot 7 showed small reductions (Fig. 4). This suggests a possible functional role for TST, SBP, and spot 7 in sulfur metabolism, and the negative regulation of the expression of these proteins by the ferrous ion concentration present in the growth medium.
FIG. 4.
Effect of ferrous iron concentration on the synthesis of some proteins in A. ferrooxidans grown on elemental sulfur. Cells were grown on elemental sulfur medium, to which the indicated ferrous iron concentrations and [35S]methionine were added. After 30 h of incubation under these conditions, total-cell proteins were separated by 2-D PAGE (left) and subjected to autoradiography, and the intensities of the indicated spots were determined by using an image analysis program (graph on the right). The positions of the indicated spots were identified previously by their coordinates in the 2-D gels and by N-terminal amino acid sequence analysis of the isolated spots.
On the other hand, we observed a marked concomitant increase in the synthesis of both rusticyanin and cytochrome c552 in sulfur-grown cells when the concentration of ferrous iron in the medium was increased, indicating a coordinated regulation at the protein synthesis level. Cytochrome c552 is known to participate in ferrous iron oxidation (2). The genes coding for rusticyanin, Cyc1 (c552) Cyc2 together with ORF1, coxB, coxA, coxC, and coxD, form an operon; they are all transcribed in A. ferrooxidans cells grown on ferrous iron as well as in those grown on sulfur, suggesting that the proteins encoded by this operon play a role in both growing conditions (2).
Synthesis of rusticyanin in A. ferrooxidans grown on different metal sulfides.
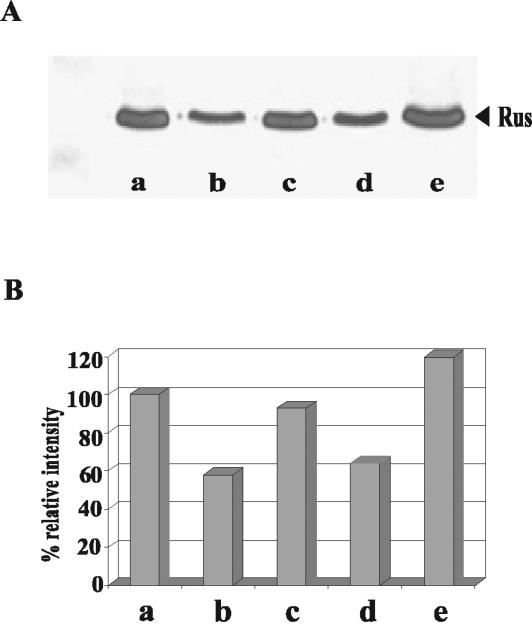
To determine whether Rus synthesis was restricted only to growth on ferrous iron and elemental sulfur, we determined the levels of the protein in cells grown on different metal sulfides. Total-cell proteins were separated by SDS-PAGE followed by Western blotting with an antiserum against Rus, as shown in Fig. 5. Clearly, there was a great increase in Rus levels in cells grown on ferrous iron or pyrite (Fig. 5A, lanes a and e). Rus was also present in cells grown on elemental sulfur, copper sulfide, or zinc sulfide (Fig. 5A, lanes b, c, and d, respectively), where no ferrous iron was present, in agreement with our results obtained with elemental sulfur by using radioactively labeled cells (Fig. 1).
FIG. 5.
Western blot analysis of rusticyanin in A. ferrooxidans grown under different conditions. (A) Equivalent amounts of total-cell proteins from A. ferrooxidans grown on ferrous iron (lane a), elemental sulfur (lane b), copper sulfide (lane c), zinc sulfide (lane d), or pyrite (lane e) were used for immunodetection with anti-Rus antibodies. (B) Quantification of the bands shown in panel A by using the Scion Image processing program. Values shown are relative to those obtained for ferrous iron-grown cells.
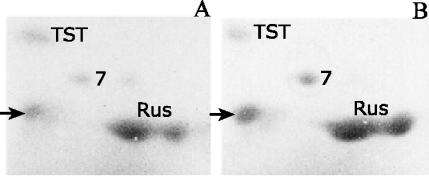
The iron- and sulfur-oxidizing activities of biomining microorganisms such as A. ferrooxidans are important for the bioleaching of ores containing copper, such as chalcopyrite. There is no evidence in the literature as to which bacterial proteins might be involved during the attack of these minerals. To find out if some of the proteins upregulated in sulfur and those upregulated in iron are synthesized during growth on minerals, we grew the bacteria on pyrite or chalcopyrite and determined the levels of synthesis of the TST, Rus, and spot 7 proteins, as shown in Fig. 6. All these proteins were synthesized when A. ferrooxidans was grown on pyrite or chalcopyrite. The presence of TST during growth on pyrite or chalcopyrite and the lack of it during growth on ferrous iron could be explained by assuming that ferrous iron is the agent inhibiting the expression of TST. During pyrite or chalcopyrite attack, only ferric iron would be produced, suggesting that this ion is inactive in the repression of TST synthesis. The results in Fig. 6 indicate that when both types of electron donors (the sulfide moiety and ferrous iron) are present simultaneously, both sulfur-upregulated and ferrous iron-upregulated genes are expressed, their expression being dependent on the relative concentration of each oxidizable component, as we have found out. Although it has recently been proposed that the oxidation of reduced inorganic sulfur compounds by nonacidophilic bacteria may use a common mechanism involving the sox gene cluster (12), no genes related to this cluster were evident when we searched for them in the available A. ferrooxidans genomic sequence.
FIG. 6.
Synthesis of rusticyanin, putative protein 7, and TST during growth of A. ferrooxidans on pyrite or chalcopyrite. Cells were grown on pyrite (A) or chalcopyrite (B), total-cell proteins were separated by 2-D PAGE, and the proteins were stained with Coomassie blue. Only the section of the gel containing the proteins analyzed is shown. Arrow indicates the spot corresponding to pancreatic RNase, used to treat the samples for 2-D PAGE.
In conclusion, the partial proteomic analysis presented in this work constitutes a first insight into the global understanding of the physiology of A. ferrooxidans and the adaptive responses that it uses to attack mineral ores. The lack of a reproducible genetic system in which to perform functional genomics in A. ferrooxidans does not allow us yet to assign definitive roles to the proteins described here in the energetic metabolism of this microorganism. Nevertheless, once the A. ferrooxidans strain 23270 genomic sequence is annotated and made available to the scientific community, extended proteomic and transcriptional analysis will greatly improve understanding of the energetic metabolism of this biomining bacterium.
Acknowledgments
This work was supported in part by a grant from FONDECYT (1030767), a CONICYT doctoral fellowship (to L.V.), and ICM grant P99-031-F. P.R. was the recipient of a DAAD Ph.D. scholarship.
We acknowledge Violaine Bonnefoy for supplying the anti-Rus serum.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. L. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Appia-Ayme, C., N. Guiliani, J. Ratouchniak, and V. Bonnefoy. 1999. Characterization of an operon encoding two c-type cytochromes, an aa3-type cytochrome oxidase, and rusticyanin in Thiobacillus ferrooxidans ATCC 33020. Appl. Environ. Microbiol. 65:4781-4787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arredondo, R., A. García, and C. A. Jerez. 1994. The partial removal of lipopolysaccharide from Thiobacillus ferrooxidans affects its attachment to solids. Appl. Environ. Microbiol. 60:2846-2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Black, P. N., S. F. Kianian, C. C. DiRusso, and D. N. William. 1985. Long-chain fatty acid transport in Escherichia coli. Cloning, mapping, and expression of the fadL gene. J. Biol. Chem. 260:1780-1789. [PubMed] [Google Scholar]
- 5.Blake, R. C., II, and E. A. Shute. 1994. Respiratory enzymes of Thiobacillus ferrooxidans: kinetic properties of an acid-stable iron:rusticyanin oxidoreductase. Biochemistry 33:9220-9228. [DOI] [PubMed] [Google Scholar]
- 6.Brierley, C. L. 1978. Bacterial leaching. Crit. Rev. Microbiol. 6:207-262. [DOI] [PubMed] [Google Scholar]
- 7.Buonfiglio, V., M. Polidoro, F. Soyer, P. Valenti, and J. Shively. 1999. A novel gene encoding a sulfur-regulated outer membrane protein in Thiobacillus ferrooxidans. J. Biotechnol. 72:85-93. [DOI] [PubMed] [Google Scholar]
- 8.Cox, J. C., and D. H. Boxer. 1986. The role of rusticyanin, a blue copper protein, in the electron transport chain of Thiobacillus ferrooxidans grown on iron or thiosulfate. Biotechnol. Appl. Biochem. 8:269-275. [Google Scholar]
- 9.De Jong, G. A. H., W. Hazeu, P. Bos, and G. Kuenen. 1997. Polythionate degradation by tetrathionate hydrolase of Thiobacillus ferrooxidans. Microbiology 143:499-504. [DOI] [PubMed] [Google Scholar]
- 10.Francis, R. T., and R. R. Becker. 1984. Specific indication of hemoproteins in polyacrylamide gels using a double-staining process. Anal. Biochem. 136:509-514. [DOI] [PubMed] [Google Scholar]
- 11.Friedrich, C. G. 1998. Physiology and genetics of sulfur-oxidizing bacteria. Adv. Microb. Physiol. 39:235-289. [DOI] [PubMed] [Google Scholar]
- 12.Friedrich, C. G., D. Rother, F. Bardischewsky, A. Quentmeier, and J. Fischer. 2001. Oxidation of reduced inorganic sulfur compounds by bacteria: emergence of a common mechanism? Appl. Environ. Microbiol. 67:2873-2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gehrke, T., J. Telegdi, D. Thierry, and W. Sand. 1998. Importance of extracellular polymeric substances from Thiobacillus ferrooxidans for bioleaching. Appl. Environ. Microbiol. 64:2743-2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guiliani, N., and C. A. Jerez. 2000. Molecular cloning, sequencing, and expression of omp-40, the gene coding for the major outer membrane protein from the acidophilic bacterium Thiobacillus ferrooxidans. Appl. Environ. Microbiol. 66:2318-2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guiliani, N., A. Bengrine, F. Borne, M. Chippaux, and V. Bonnefoy. 1997. Alanyl-tRNA synthetase gene of the extreme acidophilic chemolithoautotrophic Thiobacillus ferrooxidans is highly homologous to alaS genes from all living kingdoms but cannot be transcribed from its promoter in Escherichia coli. Microbiology 143:2179-2187. [DOI] [PubMed] [Google Scholar]
- 16.Harrison, A. P., Jr. 1984. The acidophilic thiobacilli and others acidophilic bacteria that share their habitat. Annu. Rev. Microbiol. 38:265-292. [DOI] [PubMed] [Google Scholar]
- 17.Hayashi, N. R., and Y. Igarashi. 2002. ATP binding and hydrolysis and autophosphorylation of CbbQ encoded by the gene located downstream of RubisCO genes. Biochem. Biophys. Res. Commun. 290:1434-1440. [DOI] [PubMed] [Google Scholar]
- 18.Hayashi, N. R., H. Arai, T. Kodama, and Y. Igarashi. 1997. The novel genes, cbbQ and cbbO, located downstream from the RubisCO genes of Pseudomonas hydrogenothermophila, affect the conformational states and activity of RubisCO. Biochem. Biophys. Res. Commun. 241:565-569. [DOI] [PubMed] [Google Scholar]
- 19.Heinhorst, S., S. H. Baker, D. R. Johnson, P. S. Davies, G. C. Cannon, and J. M. Shively. 2002. Two copies of form I RuBisCO genes in Acidithiobacillus ferrooxidans ATCC 23270. Curr. Microbiol. 45:115-117. [DOI] [PubMed] [Google Scholar]
- 20.Hummerjohann, J., E. Küttel, M. Quadroni, J. Ragaller, T. Leisinger, and M. A. Kertesz. 1998. Regulation of the sulfate starvation response in Pseudomonas aeruginosa: role of cysteine biosynthetic intermediates. Microbiology 144:1375-1386. [DOI] [PubMed] [Google Scholar]
- 21.Ingledew, W. J. 1982. Thiobacillus ferrooxidans. The bioenergetics of an acidophilic chemolitotroph. Biochim. Biophys. Acta 683:89-117. [DOI] [PubMed] [Google Scholar]
- 22.Kelly, D. P., J. K. Shergill, W.-P. Lu, and A. P. Wood. 1997. Oxidative metabolism of inorganic sulfur compounds by bacteria. Antonie Leewenhoek 71:95-107. [DOI] [PubMed] [Google Scholar]
- 23.Kusano, T., and K. Sugawara. 1993. Specific binding of Thiobacillus ferrooxidans RbcR to the intergenic sequence between the rbc operon and the rbcR gene. J. Bacteriol. 175:1019-1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kusano, T., T. Takeshima, C. Inoue, and K. Sugawara. 1991. Evidence for two sets of structural genes coding for ribulose bisphosphate carboxylase in Thiobacillus ferrooxidans. J. Bacteriol. 173:7313-7323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 26.Lundgren, D. G. 1980. Ore leaching by bacteria. Annu. Rev. Microbiol. 34:263-283. [DOI] [PubMed] [Google Scholar]
- 27.Matsudaira, P. T. 1989. A practical guide to protein and peptide purification for microsequencing. Academic Press, Inc., San Diego, Calif.
- 28.O'Farrel, P. Z., H. M. Goodman, and P. H. O′Farrel. 1977. High resolution two-dimensional electrophoresis of basic as well as acidic proteins. Cell 12:1133-1142. [DOI] [PubMed] [Google Scholar]
- 29.Ramírez, P., H. Toledo, N. Guiliani, and C. A. Jerez. 2002. An exported rhodanese-like protein is induced during growth of Acidithiobacillus ferrooxidans in metal sulfides and different sulfur compounds. Appl. Environ. Microbiol. 68:1837-1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rawlings, D. E. 2002. Heavy metal mining using microbes. Annu. Rev. Microbiol. 56:65-91. [DOI] [PubMed] [Google Scholar]
- 31.Rohwerder, T., and W. Sand. 2003. The sulfane sulfur of persulfides is the actual substrate of the sulfur-oxidizing enzymes from Acidithiobacillus and Acidiphilium spp. Microbiology 149:1699-1709. [DOI] [PubMed] [Google Scholar]
- 32.Sand, W., T. Gehrke, P. G. Jozsa, and A. Schippers. 2001. (Bio)chemistry of bacterial leaching—direct vs. indirect bioleaching. Hydrometallurgy 59:159-175. [Google Scholar]
- 33.Sand, W., T. Gehrke, R. Hallmann, and A. Schippers. 1995. Sulfur chemistry, biofilm, and the (in)direct attack mechanism—a critical evaluation of bacterial leaching. Appl. Microbiol. Biotechnol. 43:961-966. [Google Scholar]
- 34.Schippers, A., and W. Sand. 1999. Bacterial leaching of metal sulfides proceeds by two indirect mechanisms via thiosulfate or via polysulfides and sulfur. Appl. Environ. Microbiol. 65:319-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shively, J. M., G. van Keulen, and W. G. Meijer. 1998. Something from almost nothing: carbon dioxide fixation in chemoautotrophs. Annu. Rev. Microbiol. 52:191-230. [DOI] [PubMed] [Google Scholar]
- 36.Silver, M., and D. G. Lundgren. 1968. The thiosulfate-oxidizing enzyme of Ferrobacillus ferrooxidans (Thiobacillus ferrooxidans). Can. J. Biochem. 46:1215-1220. [DOI] [PubMed] [Google Scholar]
- 37.Silver, M., and D. G. Lundgren. 1968. Sulfur-oxidizing enzyme of Ferrobacillus ferrooxidans (Thiobacillus ferrooxidans). Can. J. Biochem. 46:457-461. [DOI] [PubMed] [Google Scholar]
- 38.Sugio, T., W. Mizunashi, K. Inagaki, and T. Tano. 1987. Purification and some properties of sulfur:ferric ion oxidoreductase from Thiobacillus ferrooxidans. J. Bacteriol. 169:4916-4922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suzuki, I. 1999. Oxidation of inorganic sulfur compounds: chemical and enzymatic reactions. Can. J. Microbiol. 45:97-105. [Google Scholar]
- 40.Tabita, R., M. Silver, and D. G. Lundgren. 1969. The rhodanese enzyme of Ferrobacillus ferrooxidans (Thiobacillus ferrooxidans). Can. J. Biochem. 47:1141-1145. [DOI] [PubMed] [Google Scholar]
- 41.Tuovinen, O. 1990. Biological fundamentals of mineral leaching processes, p. 55-77. In H. L. Ehrlich and C. L. Brierley (ed.), Microbial mineral recovery. McGraw-Hill Book Co., New York, N.Y.
- 42.Varela, P., and C. A. Jerez. 1992. Identification and characterization of GroEL and DnaK homologues in Thiobacillus ferrooxidans. FEMS Microbiol. Lett. 98:149-154. [DOI] [PubMed] [Google Scholar]
- 43.Vera, M., N. Guiliani, and C. A. Jerez. 2003. Proteomic and genomic analysis of the phosphate starvation response of Acidithiobacillus ferrooxidans. Hydrometallurgy 71:125-132. [Google Scholar]
- 44.Yarzábal, A., G. Brasseur, J. Ratouchniak, K. Lund, D. Lemesle-Meunier, J. A. DeMoss, and V. Bonnefoy. 2002. The high-molecular-weight cytochrome c Cyc2 of Acidithiobacillus ferrooxidans is an outer membrane protein. J. Bacteriol. 184:313-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yarzábal, A., K. Duquesne, and V. Bonnefoy. 2003. Rusticyanin gene expression of Acidithiobacillus ferrooxidans ATCC 33020 in sulfur- and in ferrous iron media. Hydrometallurgy 71:107-114. [Google Scholar]