Abstract
The fraction of planktonic heterotrophic bacteria capable of incorporating dissolved dimethylsulfoniopropionate (DMSP) and leucine was determined at two coastal sites by microautoradioagraphy (AU). In Gulf of Mexico seawater microcosm experiments, the proportion of prokaryotes that incorporated sulfur from [35S]DMSP ranged between 27 and 51% of 4′,6-diamidino-2-phenylindole (DAPI)-positive cells, similar to or slightly lower than the proportion incorporating [3H]leucine. In the northwest Mediterranean coast, the proportion of cells incorporating sulfur from [35S]DMSP increased from 5 to 42% from January to March, coinciding with the development of a phytoplankton bloom. At the same time, the proportion of cells incorporating [3H]leucine increased from 21 to 40%. The combination of AU and fluorescence in situ hybridization (FISH) revealed that the Roseobacter clade (α-proteobacteria) accounted for 13 to 43% of the microorganisms incorporating [35S]DMSP at both sampling sites. Significant uptake of sulfur from DMSP was also found among members of the γ-proteobacteria and Cytophaga-Flavobacterium groups. Roseobacter and γ-proteobacteria exhibited the highest percentage of DAPI-positive cells incorporating 35S from DMSP (around 50%). Altogether, the application of AU with [35S]DMSP combined with FISH indicated that utilization of S from DMSP is a widespread feature among active marine bacteria, comparable to leucine utilization. These results point toward DMSP as an important substrate for a broad and diverse fraction of marine bacterioplankton.
The great majority of dimethylsulfoniopropionate (DMSP) produced by phytoplankton is rapidly cycled within the upper ocean and may be a major carrier for transferring sulfur and carbon among microorganisms in the marine food web (2, 18, 26, 35). Recent studies have shown that DMSP can account for ≥50% of the sulfur flux and approximately 10% of the carbon flux through various trophic levels in microbial food webs dominated by DMSP-producing phytoplankton (4, 35). Only a minor fraction of the gaseous by-products of DMSP degradation, mainly dimethylsulfide (DMS), escapes a tight cycling in the water column and vents to the atmosphere (2, 16, 24, 37). Despite being a small leakage from a much larger flux, sea-to-air DMS emission is enough to constitute the largest natural source of tropospheric sulfur at a global scale (1).
DMSP is an intracellular reduced sulfur compound produced by a wide variety of unicellular algae to fulfill a number of physiological functions, mainly as a compatible solute and oxidant scavenger (40, 41). It is released into the dissolved organic matter pool through algal autolysis, viral lysis, and grazing by zooplankton (34, 43). Bacterioplankton are the main agents in the turnover of dissolved DMSP through two main pathways (18, 43): direct cleavage to DMS mediated by the enzyme DMSP lyase, and demethylation-demethiolation to methanethiol (MeSH), a key intermediate for the assimilation of S into protein (19, 20). As a substrate for heterotrophic bacterioplankton, DMSP has been shown to supply 1 to 15% of the carbon demand and 50 to 100% of the sulfur demand (16, 34, 45).
Kiene et al. (18) hypothesized that bacteria exert a control on DMSP dynamics (including DMS production) by preferentially using the demethylation/demethiolation pathway over the lyase pathway when the dissolved DMSP concentration is low relative to the S demand. Crucial to this “bacterial switch” hypothesis (34) is whether DMSP utilization is widespread among members of the bacterioplankton, allowing many bacterial taxa to participate in DMSP dynamics, or whether it is characteristic of only a few phylotypes, potentially with more limited distribution and ecological impact.
Few studies have tried to link the activity and phylogeny of DMSP-degrading bacteria in marine environments (9, 10, 23, 44, 45). The bacterial assemblage associated with a bloom of the coccolithophore Emiliania huxleyi (an alga producing high amounts of DMSP) in the North Atlantic was dominated by Roseobacter, and a correlation was found between the abundance of this phylogroup and the concentration of DMSP (10). Similarly, in the northern North Sea, a single Roseobacter species dominated the bacterioplankton assemblage associated with a bloom of DMSP-producing phytoplankton, and a close correlation between the abundance of this species and the loss rate of dissolved DMSP was found (45). These field studies suggested a prominent role of the Roseobacter group among DMSP-degrading bacteria in the ocean, a capability that had been already demonstrated by experimental work with cultures of marine isolates (9, 19, 23, 28).
Despite the important body of evidence generated by this work, neither field studies nor culture work provide direct observations of the link between DMSP degradation and the Roseobacter group in the sea. First, spatial correlation between substrate concentrations or activity rates with the abundance of a particular phylotype does not definitely prove that the process is carried out by this phylotype. Second, culturable marine prokaryotes, even if members of major marine lineages, do not always represent the species- and subspecies-level taxa that dominate in situ (7, 8).
The extent of DMSP utilization among marine bacterioplankton, the phylogenetic composition of bacterial DMSP consumers, and the quantitative importance of DMSP consumers in the bacterial assemblage have not been determined. Single-cell analysis methods which rely on interrogation of individual cells for information on identity and activity are the best experimental approach to answer these questions. Microautoradiography (3, 31) provides information on the percentage of cells that take up a given compound by visualizing radioactivity incorporated into individual cells. The powerful combination of microautoradiography (AU) with fluorescence in situ hybridization (FISH) (MicroFISH or STARFISH methods) (6, 25, 29) allows individual active cells to be assigned to phylogenetic groups.
We have developed protocols for AU and MicroFISH analyses of the extent of [35S]DMSP utilization activity in marine prokaryotes. Here we describe experiments carried out for optimization of methods and report on the application of these protocols to marine bacterioplankton communities from two sampling sites at different periods of the year. Throughout the study, [35S]DMSP incorporation was compared to [3H]leucine incorporation, since leucine is considered one of the most universal substrates for heterotrophic bacterioplankton (22).
MATERIALS AND METHODS
Sampling.
On 8 June 2001, surface seawater was collected in clear waters off Mobile Bay in the vicinity of Dauphin Island, in the northern Gulf of Mexico (30° 15′ N, 88° 05′ W) by submerging rinsed polycarbonate carboys to a depth of 0.5 m. Carboys were kept at ambient temperature in closed coolers until use in a microcosm experiment (see details below).
On 13 and 28 January and on 4 March 2003, surface seawater was collected in the same manner approximately 1 mile offshore in the Bay of Blanes (41° 40′ N, 2° 48′ E), 70 km north of Barcelona (NW Mediterranean). In this case, we sampled typical winter conditions and the onset of a natural phytoplankton bloom. Characteristics of the water samples are detailed in Table 1.
TABLE 1.
Characteristics of the water samples used
| Sample | Temp (°C) | Salinity (psu) | Chl a (μg liter−1) | Prokaryotic cells (105) ml−1 | Bacterial production (pmol ·liter−1 · h−1) | [DMSPd] (nM) | [DMSPp] (nM) | DMSPp/chla (nmol μg−1) | AU-positive cells
|
|
|---|---|---|---|---|---|---|---|---|---|---|
| [35S]DMSP (%) | [3H]leucine (%) | |||||||||
| DIME, Gulf of Mexicoa (8-16 June) | ||||||||||
| Nutrient amended, day 2 | 27 | 33.3 | 1.00 | 13.6 | 64 | 5.2 | 24.4 | 24.4 | 50 | 70 |
| Nutrient amended, day 5 | 27 | 33.3 | 1.67 | 13.6 | 378 | 5.0 | 46.1 | 27.6 | 51 | 45 |
| Nutrient amended, day 8 | 27 | 33.3 | 1.39 | 20.0 | 468 | 4.4 | 73.8 | 53.1 | 51 | 63 |
| Control, day 2 | 27 | 33.3 | 0.39 | 15.1 | 58 | 4.0 | 18.5 | 47.4 | NDb | 71 |
| Control, day 5 | 27 | 33.3 | 0.27 | 10.0 | 41 | 2.5 | 13.0 | 48.1 | 38 | 47 |
| Control, day 8 | 27 | 33.3 | 0.40 | 9.2 | 39 | 3.4 | 15.6 | 39.0 | 27 | 55 |
| Blanes Bay, NW Mediter- ranean Sea | ||||||||||
| 13 January | 13.5 | 37.4 | 1.35 | 6.1 | 3.8 | 2.7 | 8.8 | 6.5 | 5 | 21 |
| 28 January | 14 | 37.5 | 1.51 | 8.2 | 2.6 | 2.2 | 7.8 | 5.2 | 10 | 25 |
| 4 March | 11 | 36.1 | 2.21 | 8.9 | 42 | 11.6 | 42.4 | 19.2 | 42 | 40 |
Each data point is the average of two replicate carboys. Amended, spiked with P and N at Redfield ratio.
ND, not determined.
The Dauphin Island Microcosm Experiment (DIME).
Gulf of Mexico water (salinity, 33.3‰) was prefiltered through a 130-μm-pore-size mesh to exclude large zooplankton and was partitioned between four 25-liter polycarbonate carboys. Carboys were placed in a climate-controlled chamber set at 27°C (in situ temperature at the time of sampling) with an artificial light source (12 h light and 12 h dark, 200 μE cm−2). Two carboys were amended with 10 μM NO3− and 0.6 μM PO43− (labeled NUT1 and NUT2), and two carboys were maintained as controls (no amendments, labeled C1 and C2). The carboys were sampled daily at 10 a.m., immediately before the light was turned on. Two liters of water were collected into polycarbonate bottles by siphon with an acid-rinsed pipette connected to silicon tubing. Gentle mixing of the carboys was provided once daily by turning them upside down twice before sampling. The microcosm experiment lasted for 8 days.
Chemical analyses.
Chlorophyll a (Chl a) was measured by fluorometry in 90% acetone extracts (30). Dissolved DMSP (DMSPd, Whatman glass fiber filter GF/F filtrate) and particulate DMSP (DMSPp, retained by GF/F) were measured as DMS after alkaline hydrolysis. The evolved DMS was determined by gas chromatography, either by the headspace sweeping method (14) for DIME or by water purge and trap for the NW Mediterranean samples (36).
Bacterial numbers and heterotrophic production.
Samples for enumeration of bacteria were preserved with 0.2-μm-pore-size-filtered formaldehyde (final concentration, 2% [wt/vol]). Within 48 h, total cell counts of bacteria were determined by epifluorescence microscopy after staining of cells with 4′,6-diamidino-2-phenylindole (DAPI) (2 μg ml−1 for 10 min) and filtering them onto black 0.2-μm-pore-size polycarbonate filters at a vacuum pressure of 100 to 200 mm Hg (33).
Bacterial production was determined by incorporation of [3H]leucine using the method of Kirchman et al. (22) with the modifications of Smith and Azam (39). For each seawater sample, live and killed (5% trichloroacetic acid) controls were incubated with [3H]leucine (final concentration, 20 nM for DIME; 40 nM for NW Mediterranean) for 1 to 2 h, at in situ temperature, in the dark.
Microautoradiography.
Incubations for AU were carried out with samples from the DIME and from the NW Mediterranean (Table 1). Two radioactive substrates were used: [3H]leucine (specific activity: 98.84 Ci mmol−1 in DIME, 161 Ci/mmol−1 in the NW Mediterranean) and [35S]DMSP (specific activity: 3.3 Ci mmol−1 in DIME, 203.1 Ci/mmol−1 in the NW Mediterranean). Aliquots of 5 ml from each time point in DIME and 30-ml samples from the NW Mediterranean were incubated in the dark at in situ temperature with [3H]leucine for ∼5 h or [35S]DMSP for ∼13 h. A formaldehyde-killed control was prepared for each incubation. Incubations were stopped with formaldehyde (final concentration, 4%). Microautoradiograms were prepared by filtering 1-ml (DIME) or 5-ml (NW Mediterranean) aliquots through 0.2-μm-pore-size polycarbonate filters (Nuclepore), thus providing five or six replicates per sample. The filters were rinsed twice with 0.2-μm-pore-size-filtered seawater to remove unincorporated radiolabel and were removed from the filtration apparatus without disconnecting the vacuum. Filters were air dried and stored at −20°C until processed.
Microautoradiography was carried out essentially as described by Pedrós-Alió and Newell (31). In the darkroom, slides were dipped in melted NTB-2 nuclear track emulsion (diluted 1:1 with filter-sterilized deionized water; Kodak). The filter was carefully placed face down on the emulsion. Slides were kept for 10 min on a metal tray in contact with ice. The slides were dried and exposed in the dark at 4°C for 20 days ([35S]DMSP) or 10 days ([3H]leucine).
The microautoradiograms were developed for 4 min in Kodak D19 developer (diluted 1:1 with distilled water), followed by a 30-s stop rinse in deionized water and a 4-min soak in Kodak fixer. Slides were washed in tap water for 10 min, dipped in glycerol (1%) for 2 min, and stored inside a desiccator overnight, protected from light. The filter was gently peeled off, and cells in the emulsion were stained with a drop of mounting solution (Vecta:Citifluor solution, 4:1 [vol/vol], plus DAPI at 1 μg ml−1) and covered with a coverslip. Total DAPI-positive and radiolabeled cell counts were determined with an Olympus BH microscope under simultaneous UV radiation epifluorescence and visible light-transmitted illumination. Following this protocol, between 75 and 98% of the cells were transferred from the filter to the emulsion.
Optimization of the incubation and exposure times is described in Results.
MicroFISH.
The MicroFISH method consists of the combination of AU and FISH (6, 25, 29). FISH was carried out on sections of the filter after incubation with radiolabel but before the AU step. Cells on filter sections were hybridized with the group-specific oligonucleotide probes Eub338 (5′-GCTGCCTCCCGTAGGAGT-3′) for eubacteria, Ros536 (5′-CAACGCTAACCCCCTCCG-3′) for Roseobacter, Alf968 (5′-GGTAAGGTTCCGCGCGTT-3′) for α-proteobacteria, Gam42a (5′-GCCTTCCCACATCGTTT-3′) for γ-proteobacteria, CF319a (5′-TGGTCCGTGTCTCAGTAC-3′) for Cytophaga-Flavobacterium, and Non338 (5′-ACTCCTACGGGAGGCAGC-3′) as a negative control. Probes labeled with the cyanine dye CY3 at the 5′ end were purchased from Thermo Hybaid GmbH. Hybridization was carried out by using the method of Pernthaler et al. (32).
35S labeling of DMSP.
[35S]DMSP for DIME (Gulf of Mexico) was synthesized biologically using the alga Platymonas subcordiformis and following the culture and extraction procedures of Kiene et al. (15). For the NW Mediterranean, [35S]DMSP was synthesized chemically from l-[35S]methionine as described elsewhere (27). In both cases the [35S]DMSP product was partially purified by solid-phase, ion-exchange extraction onto a Dowex-50 (H+ resin). Further purification was achieved by evaporating the Dowex-50 eluate to dryness, reconstituting in 100 μl of pure water, and injecting the sample into a high-performance liquid chromatographer (HPLC) equipped with a Whatman Partisil SCX cation-exchange column. The DMSP peak fraction was collected and subjected to a final Dowex-50 solid-phase extraction to remove phosphate from the HPLC eluent. Radiochemical purity was >98% as judged by HPLC analysis and trapping of volatile 35S after conversion of the [35S]DMSP to [35S]DMS by alkaline hydrolysis.
RESULTS
Setting conditions for AU with [35S]DMSP.
To optimize [35S]DMSP incorporation by the water samples for subsequent AU, we carried out seawater incubations for different lengths of time and with different amounts of [35S]DMSP. Results are shown in Fig. 1. The water sample used for these tests was from nutrient-enriched carboy NUT2 on day 2 of the DIME (Gulf of Mexico). Uptake and incorporation of 35S into filterable material was monitored at several time points over 13 h by counting the filters in liquid scintillation cocktail (Ecolume; ICN Biomedicals). At selected time points, AU was performed on parallel filters. Since the full AU procedure took several weeks, optimization was based on the conditions which gave maximal radioactivity incorporated into filterable material. In Fig. 1A, total radioactivity on the filters is plotted as a function of time for various concentrations of substrate. Maximal total uptake was reached after 10 to 14 h. In Fig. 1B the same data set is plotted as a function of the added substrate concentration. Based on radioactivity incorporated into filterable material, we chose an incubation time of 13 h and a DMSP addition of 5.6 · 104 dpm ml−1, which was close to saturation of DMSP uptake. To optimize the exposure time of the emulsion, replicate slides from nutrient-amended carboy NUT2 on day 5 of DIME (with a [35S]DMSP level of 5.6 · 104 dpm/filter) were exposed at 4°C in the dark for various times. The optimal exposure time for the AU was determined to be about 20 days (Fig. 2). In the case of [3H]leucine, the exposure time was optimal at 10 days.
FIG. 1.
Incorporation of [35S]DMSP plotted as a function of incubation time (A) and substrate addition (B). The boxed numbers indicate the percent of cells incorporating DMSP at that time point and substrate concentration, if determined. The samples were from a nutrient-enriched carboy on day 2 of the DIME (Gulf of Mexico, June 2000).
FIG. 2.
Percentage of labeled cells as a function of exposure time for AU with [35S]DMSP (filled symbols) and [3H]Leu (empty symbols). Error bars show the standard deviation of three replicates. The samples were from a nutrient-enriched carboy on day 5 of the DIME (Gulf of Mexico, June 2000).
Under these experimental conditions, AU of marine bacterioplankton cells produced a well-defined crown of exposed silver grains with [3H]leucine labeling, making identification of positive cells easy (Fig. 3A). In the case of 35S AU, the exposed grains were slightly more irregularly distributed around the cells, likely due to the higher energy of the beta particles emitted by 35S (Fig. 3B). Nonetheless, identification of 35S-positive cells was also straightforward.
FIG. 3.
Examples of microautoradiograms of [35S]DMSP-labeled (A) and [3H]Leu-labeled (B) bacteria observed under transmitted light (for exposed silver grains) and epifluorescence microscopy (for DAPI stain). White arrows point to DAPI-stained nonradiolabeled cells, while black arrows point to DAPI-stained radiolabeled cells. Note morphological diversity of labeled cells and low background of exposed silver grains compared to labeled cells. The [35S]DMSP and [3H]Leu microautoradiograms are from a nutrient-enriched sample on days 2 and 8 of the DIME (Gulf of Mexico, June 2000).
The variability associated with AU counts was assessed by comparing replicate sets of different samples. Four samples of [35S]DMSP AU with two to four replicates of each sample gave coefficients of variation in AU-positive counts of 7 to 20% (average, 12%). For [3H]leucine AU, 11 samples with 2 to 4 replicates each gave coefficients of variation of ≤12% (average, 6%).
DIME.
In response to the nutrient amendment, a phytoplankton bloom was induced that reached a peak Chl a concentration of ∼3 μg liter−1 after 4 days of incubation (Fig. 4A). In the control carboys, Chl a remained constant around 0.5 μg liter−1. These dynamics were accompanied by increased DMSPp concentrations, up to 52 to 96 nM, in the nutrient-amended microcosms on day 8, while DMSPp concentrations remained constant at about 15 nM in the controls (Table 1). The DMSPp/Chl ratio remained around 40 to 50 nmol μg−1 in the control microcosms throughout the experiment. In the amended microcosms, this ratio decreased to about 10 in the peak of the phytoplankton bloom (data not shown) and increased, as Chl a decreased, to eventually reach values slightly higher than to those in the controls on day 8 (Table 1).
FIG. 4.
Microbial dynamics in the DIME study (Gulf of Mexico, June 2000). (A) Changes in Chl a with time in control microcosms (filled symbols, carboys C1 and C2) and nutrient-amended microcosms (empty symbols, carboys NUT1 and NUT2). Arrows indicate time points samples were incubated for AU. (B) Changes in leucine incorporation with time. Each error bar shows standard error of three replicates.
As a consequence of the phytoplankton bloom and its decay, bacterial leucine incorporation rates increased to around 600 pmol liter−1 h−1 on day 6 and thereafter (Fig. 4B). In the control carboys, leucine incorporation remained below 100 pmol liter−1 h−1. Bacterial numbers increased very slowly throughout the experiment, averaging 1.40 × 106 cells ml−1 in the control carboys and 2.0 × 106 cells ml−1 in the bloom carboys. A more detailed description of the microcosm bacterial communities is reported elsewhere (J. Pinhassi, R. Simó, R. Kiene, C. Pedrós-Alió, M. Vila, L. Alonso, J. M. González, and M. A. Moran, unpublished data).
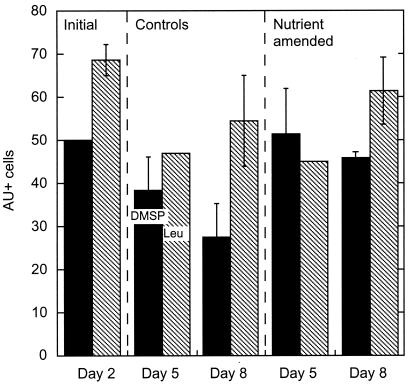
[35S]DMSP AU was carried out after the first 48 h of the microcosm experiment (day 2), 1 day after the peak of Chl in the enriched carboys (day 5), and when the DMSPp/Chl ratio was highest (day 8) (Fig. 4). At the beginning of the experiment, the percentage of cells assimilating [35S]DMSP was 50%. A small decrease in positive cells occurred in the controls during the course of the experiment (down to 27% on day 8), but not in the nutrient-amended samples, where it remained approximately 50% (Fig. 5).
FIG. 5.
Percentage of cells taking up DMSP (black bars) and leucine (striped bars) in the DIME (Gulf of Mexico, June 2000). Error bars show standard deviation of two replicates.
At the beginning of the experiment (day 2), similarly high percentages of [3H]leucine-labeled cells were recorded in the controls and the amended carboys, with an average of 71% positive cells. On day 5 the percentage of cells assimilating [3H]leucine was similar to that of cells assimilating [35S]DMSP in all carboys. On day 8, however, cells incorporating DMSP were significantly less abundant than those incorporating leucine (Fig. 5).
The composition of the heterotrophic bacterial assemblage was analyzed using a variety of molecular techniques that focused on 16S rRNA genes: 16S rRNA clone libraries, denaturing gradient gel electrophoresis, terminal restriction fragment length polymorphism, and FISH. The Roseobacter clade dominated the bacterioplankton community, and representatives of the Cytophaga-Flavobacterium, SAR11, and SAR86 groups were also important (Pinhassi et al., unpublished). In order to test whether Roseobacter was the main bacterial group responsible for DMSP uptake, MicroFISH was performed on day 8 in the amended carboy C using fluorescent probes for eubacteria and for Roseobacter (Fig. 6 and 7). Hybridization with the Non338 probe did not give any positive cells, indicating that nonspecific binding of the probes was not significant. The Roseobacter clade accounted for 23% of total (DAPI-stained) prokaryotes (Fig. 6A). As many as 87% of Roseobacter cells identified by FISH were labeled with 35S (Fig. 6B), whereas 50% of FISH-identified Roseobacter cells were labeled with [3H]leucine (Fig. 6C). Among all the 35S AU-positive cells that occurred in this sample, 43% belonged to Roseobacter (Fig. 6D). In contrast, Roseobacter contributed only 21% of the [3H]leucine-labeled cells (Fig. 6E).
FIG. 6.
Results of MicroFISH in the DIME study (Gulf of Mexico, 8 to 16 June 2000) and in the Blanes Bay time series (NW Mediterranean, 13 January and 4 March 2003). (A) FISH+, cells hybridizing with each probe as a percentage of total DAPI count. (B) DMSP AU+, percentage of cells hybridizing with each probe that was labeled with [35S]DMSP. (C) Leu AU+: percentage of cells hybridizing with each probe that was labeled with [3H]leucine. (D) DMSP AU+ FISH+, percentage of cells labeled with [35S]DMSP that hybridized with each probe. (E) Leu AU+ FISH+, percentage of cells labeled with [3H]leucine that hybridized with each probe. Eub, eubacteria; Alpha, α-proteobacteria; Roseo, Roseobacter; gamma, γ-proteobacteria; CF, Cytophaga-Flavobacter.
FIG. 7.
Result of MicroFISH in the DIME study in the Gulf of Mexico (microcosm NUT2 enriched with nutrients, 3 days after the Chl peak) and in the Blanes Bay time series (NW Mediterranean). Results are shown for [35S]DMSP-labeled and [3H]leucine-labeled cells separately. Values of labeled (AU+) cells appear above the x axis, and those of nonlabeled cells (AU−) appear below it. Two probes were used for FISH: eubacteria (Eub) and Roseobacter (Ros). The percentage of cells labeled with the Ros probe (black portion of bars) is shown as a subset of the percentage of cells labeled with the Eub probe (black plus dashed portion of bars). “Other” indicates DAPI-stained cells that did not hybridize with the Eub probe.
Coastal NW Mediterranean.
Characteristics of the coastal NW Mediterranean seawater at the time of sampling are given in Table 1. Chl a concentrations increased from 1.43 μg liter−1 in January to 2.21 μg liter−1 in March (Table 1), while DMSPp concentrations increased from 8 to 42 nM during this same period. As a consequence, the DMSPp/Chl a ratio, which is a proxy for the relative abundance of DMSP producers among the phytoplankton, was lower in January (6 nmol/μg) than in March (19 nmol/μg).
Microautoradiography showed that the proportion of total DAPI-positive cells labeled with [35S]DMSP and [3H]leucine increased between January and March (Fig. 7). On the two sample dates in January, 5 to 10% of the DAPI-positive prokaryotes incorporated [35S]DMSP, while 21 to 24% took up [3H]leucine. In March, the percentage of cells labeled with [35S]DMSP had increased to 42% and was similar to the fraction of cells labeled with [3H]leucine (40%).
In both January and March samples, the Roseobacter clade accounted for an average of 16% of the DAPI counts according to FISH (Fig. 6A). Based on MicroFISH data, 13 to 19% of the 35S-labeled cells could be identified by the Roseobacter probe (Fig. 6D). The percentage of Roseobacter cells that had assimilated [35S]DMSP increased from 7 to 52% between January and March (Fig. 6B).
The fraction of Roseobacter cells that assimilated [3H]leucine was higher than the fraction incorporating [35S]DMSP in early January, but the two were very similar in March (Fig. 6B and C). The fraction of [3H]leucine-labeled cells that could be identified by the Roseobacter probe decreased from 29 to 17% from January to March as detected by MicroFISH (Fig. 6E). Nevertheless, the fraction of Roseobacter cells that assimilated [3H]leucine remained constant (43 to 48%) (Fig. 6c).
With the March sample we used FISH probes for other groups of bacteria in combination with AU. The different groups tested accounted for similar percentages of the total DAPI-positive prokaryote count (Fig. 6A). The α-proteobacteria and Roseobacter probes produced very similar numbers, suggesting that most of the α-proteobacteria that could be detected by FISH were indeed Roseobacter. As can be seen in Fig. 6B to C, all groups were active in both DMSP and leucine incorporation, suggesting that DMSP use is widespread among different phylogenetic groups of bacterioplankton.
Figure 8 shows the percentage of total DAPI-positive prokaryotic cells that hybridized with each probe and incorporated each substrate for the March sampling. All groups had a significant fraction of cells that assimilated sulfur from [35S]DMSP. Roseobacter and γ-proteobacteria showed a similar percentage of DMSP- and leucine-labeled cells (i.e., they were close to the 1:1 line in the plot of Fig. 8). In the Cytophaga-Flavobacterium group, on the other hand, the percentage of cells that took up S from [35S]DMSP was lower than that of cells incorporating [3H]leucine.
FIG. 8.
Results of MicroFISH in Blanes Bay, NW Mediterranean (4 March). Comparison of [35S]DMSP-labeled cells versus [3H]leucine-labeled cells (as a percentage of total DAPI counts) for each phylogenetic group tested. The diagonal indicates a 1:1 relationship, i.e., equivalent numbers of cells assimilating both substrates. Eub, eubacteria; Ros, Roseobacter; gamma, γ-proteobacteria; alpha, α-proteobacteria; CF, Cytophaga-Flavobacter.
DISCUSSION
DMSP and overall bacterial activity.
Microautoradiography with [35S]DMSP has provided a direct measure of the proportion of bacteria involved in utilization of this organic sulfur molecule. The percentage of bacteria that incorporate DMSP varies with sample characteristics but can be very high (Table 1; Fig. 5 and 7); under certain conditions, it can be similar to that of cells incorporating leucine. Leucine is widely accepted as a universal substrate for bacteria, to the extent that it is commonly used to provide estimates of bacterial heterotrophic production (21). We thus take the number of cells assimilating [3H]leucine as a measure of the fraction of active heterotrophic cells. In our samples, when the percentage of active cells was higher (>40% of cells were 3H positive in either DIME or NW Mediterranean blooms), the incorporation of DMSP was widespread (≥40% of cells were 35S positive). When the bacterial assemblage was less active, such as in the January samples from Blanes Bay, the fraction of cells assimilating DMSP in that sample was also low (<10%) (Table 1; Fig. 5 and 7).
There are limitations of the AU method that call for caution in interpreting results. Exposed silver grains indicate the presence of a labeled atom or moiety but not necessarily the original substrate molecule. In the case of [35S]DMSP, which is a very labile substance that usually turns over in less than 1 day (12, 18; Pinhassi et al., unpublished), degradation products (i.e., [35S]DMS or [35S]MeSH [17]) may be released into seawater and assimilated by other bacteria during the incubation period of 10 to 14 h. Hence, counts of 35S AU-positive cells assess direct and indirect incorporation of sulfur from DMSP and may therefore be an overestimate of cells actually incorporating DMSP. In any case, the conclusion that DMSP provided sulfur for a wide variety of marine bacteria is not altered. This indirect uptake of the isotope is unlikely to have occurred in the case of the [3H]leucine, for which incubations were significantly shorter (5 h), and because leucine is incorporated directly into proteins with little degradation (22).
The low specific activity of the biologically synthesized [35S]DMSP and the fast decay rates of 35S necessitated the use of a DMSP concentration in the DIME study (ca. 60 nM) that was well above the endogenous DMSPd concentration. Therefore, AU-positive cells may represent the number of cells capable of incorporating DMSP, rather than those actively incorporating DMSP at in situ concentrations. In the NW Mediterranean, chemically synthesized [35S]DMSP of higher specific activity was added at levels below the endogenous DMSPd concentrations (<0.3 nM). [3H]leucine amendments in DIME (2.5 nM) were likewise above estimated endogenous concentrations (<1 nM) (5), while the added leucine levels in the NW Mediterranean (0.5 nM) were likely below endogenous levels. Therefore, since we applied the same criterion for the additions of DMSP and leucine, the comparisons of the relative extent of the incorporation of the two compounds by heterotrophic bacterioplankton are not affected.
Comparison of AU results obtained with DMSP to those obtained with leucine show that on some occasions DMSP assimilation is as widespread as leucine assimilation, but on other occasions DMSP is assimilated by significantly fewer cells (Table 1; Fig. 5 to 8). Leucine is directly incorporated into bacterial proteins, which is the basis for its use as a measure of bacterial protein synthesis (22). Malmstrom et al. (27) recently proposed that since DMSP assimilation appears to satisfy most of the bacterial sulfur demand in different oceanic environments (e.g., see references 16 and 35) and all bacteria synthesizing protein need sulfur, then all bacteria synthesizing protein should assimilate DMSP. Our results indicate that natural assemblages of bacterioplankton may indeed rely heavily on DMSP as a sulfur source when it is available, but certainly not all bacteria synthesizing protein assimilate DMSP (Fig. 7 and 8).
Exploring the links between Roseobacter and DMSP.
In recent years, significant advances have been made in determining the identity of prokaryotic plankton in the ocean (e.g., see reference 8). A major remaining challenge is to connect microbial phylogeny with the role that microbes play in the biogeochemical cycles of key elements. The involvement of the Roseobacter clade in bacterial metabolism of dimethylated sulfur in marine environments represents a clear example of how such a connection can be approached (28). By drawing jointly on the expertise of molecular microbial ecologists and biogeochemists, and by combining laboratory and field work, research on the linkage between DMSP cycling and the Roseobacter clade is as firmly established as for any other organic substrate and bacterioplankton taxon.
Previous studies have provided strong, yet circumstantial, evidence for a prominent role of Roseobacter in DMSP degradation. Every one of 15 cultured members of this group was able to cleave DMSP into DMS and acrylate, despite the fact that only half were isolated on DMSP-containing media (9). Five of the 15 isolates expressed the demethylation/demethiolation route, and all of these were also capable of incorporating DMSP sulfur into proteins (19). In the open ocean, Roseobacter was found to be the most abundant group in the bacterial assemblage associated with blooms of high-DMSP-producing phytoplankton (10, 45). González et al. (10) reported that Roseobacter DNA abundance was positively correlated with both Chl and DMSP concentrations, while Zubkov et al. (45) found that one species-level Roseobacter taxon was positively correlated with bacterial production and DMSP loss (45).
By means of MicroFISH with a Roseobacter probe, we observed that this group contributed to 35S assimilation in all samples studied. In the sample for which the MicroFISH was done with the set of five phylogenetic probes (NW Mediterranean, March), Roseobacter and γ-proteobacteria were the two largest contributors to 35S-positive cells (Fig. 6 and 8). In agreement with the conclusion of Malmstrom et al. (27), these results indicate that Roseobacter is a prominent group among DMSP-utilizing bacterioplankton, yet by no means the only group involved.
Implications for elucidating the DMS-DMSP cycle.
The demethylation/demethiolation pathway, and the subsequent utilization of the methanethiol moiety as an S source, has been found in previous studies to dominate the microbial degradation of DMSP in seawater over the DMS production pathway (18, 20, 42). Single-cell resolution of DMSP uptake shows that the capability for incorporating S from DMSP is widespread among marine bacteria (Fig. 6B and D). The question of how widespread is the occurrence of the DMS-producing, DMSP lyase pathway among bacterioplankton remains yet to be answered.
Bacterial degradation by either of the two pathways is the fate of only a portion of algal DMSP in the ocean. Other fates include cleavage by algal DMSP lyases during oxidative stress, grazing, or autolysis and assimilation by herbivores (35, 38, 41, 42). Moreover, bacteria are involved not only in the degradation of DMSP but also in the consumption of the evolved DMS and MeSH (13, 19). In particular, microbial consumption is one of the major mechanisms for DMS loss from the surface ocean. As such, it is one of the major factors controlling DMS emission to the atmosphere (34), and knowledge of the phylogeny and dynamics of the bacteria involved is crucial for understanding the marine DMS(P) cycle. Previous studies have suggested that DMS is a minor sulfur source, mainly used as a supplementary carbon source (45) by methylotrophic bacteria (11). The MicroFISH protocols developed in the present study, if applied with [35S]DMS, could provide new insights into the microbial cycling of DMS.
Implications for the ecology and biogeochemistry of marine bacterioplankton.
As stated by Giovannoni and Rappé (8), the dominance of certain groups of heterotrophic prokaryotes in the surface ocean likely results from their competence in using labile dissolved organic matter derived from the primary producers. Several recent studies have shown that algae-derived DMSP, although occurring at nanomolar concentrations in seawater, turns over on the order of a few hours (18). With techniques such as AU and MicroFISH, we have been able to examine coincidence of DMSP incorporation and identity in single cells. All the bacterioplankton groups examined had cells that exhibited the capability to assimilate sulfur from DMSP. In future studies, the comparison of MicroFISH with DMSP relative to other substrates (such as glucose and amino acids) together with rate measurements of substrate utilization by bacteria should provide information on the importance of a rather small set of ubiquitous molecules released by phytoplankton for satisfying bacterioplankton C and energy demands.
The observation that Roseobacter and γ-proteobacteria had a greater affinity for DMSP than other phylogroups (Fig. 8) is complementary to the findings of Malmstrom et al. (27) in a MicroFISH study of the NW Atlantic. These authors observed larger silver-grain surface areas around Roseobacter cells relative to other phylogenetic groups, which led them to suggest that Roseobacter was more capable of utilizing DMSP on a per-cell basis. If further studies confirm that affinity for DMSP varies among bacterial phylotypes, then we can speculate that conditions with a higher contribution of DMSP to the C and S pools and fluxes might favor bacterioplankton assemblages with a higher proportion of specialized DMSP utilizers. In other words, we can speculate that succession of phytoplankton towards higher DMSP producers will be followed by succession of bacterioplankton towards better DMSP consumers. Obviously, addressing such a hypothesis will require a comprehensive application of the tools developed in the present study to natural communities, along with the determination of DMSP production, occurrence, and transformation fluxes under changing environmental conditions, e.g., throughout time series or spatial gradients of trophic status.
Acknowledgments
This work was supported by project 2000/20185 from the U.S.-Spain Joint Commission for Scientific and Technological Cooperation as well as by EU project BASICS (EVK3-CT2002-00078, to J. M. Gasol), Spanish MCyT projects MicroDiFF (REN2001-2120/MAR, to J. M. Gasol) and REN2000-2457-E (to R. Simó), and U.S. NSF grants OCE-9907471 to R. P. Kiene and MCB-0084164 to M. A. Moran.
The tireless efforts of Laura Linn during DIME are gratefully acknowledged. R. Massana and J. M. Gasol provided valuable comments, L. Alonso provided some bacterial heterotrophic production data, A. Riera and E. Blanc assisted with cell counts, and E. Flo, R. Scharek, and M. Latasa provided some Chl a concentrations.
REFERENCES
- 1.Andreae, M. O., and P. J. Crutzen. 1997. Atmospheric aerosols: biogeochemical sources and role in atmospheric chemistry. Science 276:1052-1058. [Google Scholar]
- 2.Bates, T. S., R. P. Kiene, G. V. Wolfe, P. A. Matrai, F. P. Chavez, K. R. Buck, B. W. Blomquist, and R. L. Cuhel. 1994. The cycling of sulfur in surface seawater of the northeast Pacific. J. Geophys. Res. 99:7835-7843. [Google Scholar]
- 3.Brock, M. L., and T. D. Brock. 1968. The application of microautoradiographic techniques to ecological studies. Mitt. Int. Ver.. Theor. Angew. Limnol. 15:1-29. [Google Scholar]
- 4.Burkill, P. H., S. D. Archer, C. Robinson, P. D. Nightingale, S. B. Groom, G. A. Tarran, and M. V. Zubkov. 2002. Dimethyl sulphide biogeochemistry within a coccolithophore bloom (DISCO): an overview. Deep-Sea Res. II 49:2863-2885. [Google Scholar]
- 5.Carlucci, A. F., D. B. Craven, and S. M. Henrichs. 1984. Diel production and microheterotrophic utilization of dissolved free amino acids in waters off Southern California. Appl. Environ. Microbiol. 48:165-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cottrell, M. T., and D. L. Kirchman. 2000. Natural assemblages of marine proteobacteria and members of the Cytophaga-Flavobacter cluster consuming low- and high-molecular-weight dissolved organic matter. Appl. Environ. Microbiol. 66:1692-1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fuhrman, J. A., K. McCallum, and A. A. Davis. 1993. Phylogenetic diversity of subsurface marine microbial communities from the Atlantic and Pacific Oceans. Appl. Environ. Microbiol. 59:1294-1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giovannoni, S., and M. Rappé. 2000. Evolution, diversity, and molecular ecology of marine prokaryotes, p. 47-84. In D. L. Kirchman (ed.), Microbial ecology of the oceans. Wiley-Liss, New York, N.Y.
- 9.González, J. M., Kiene, R. P., and M. A. Moran. 1999. Transformation of sulfur compounds by an abundant lineage of marine bacteria in the α-subclass of the class Proteobacteria. Appl. Environ. Microbiol. 65:3810-3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.González, J. M., R. Simó, R. Massana, J. S. Covert, E. O. Casamayor, C. Pedrós-Alió, and M. A. Moran. 2000. Bacterial community structure associated with a dimethylsulfonopropionate-producing North Atlantic algal bloom. Appl. Environ. Microbiol. 66:4237-4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kiene, R.P. 1993. Microbial sources and sinks for methylated sulfur compounds in the marine environment, p. 15-33. In J. C. Murrell and D. P. Kelly (ed.), Microbial growth on C1 compounds. Intercept, Andover, United Kingdom.
- 12.Kiene, R. P. 1996. Production of methane thiol from dimethylsulfoniopropionate in marine surface waters. Mar. Chem. 54:69-83. [Google Scholar]
- 13.Kiene, R. P., and T. S. Bates. 1990. Biological removal of dimethyl sulphide from seawater. Nature 345:702-705. [Google Scholar]
- 14.Kiene, R. P., and G. Gerard. 1994. Determination of trace levels of dimethylsulfoxide (DMSO) in seawater and rainwater. Mar. Chem. 47:1-12. [Google Scholar]
- 15.Kiene, R. P., L. P. Hoffmann Williams, and J. E. Walker. 1998. Seawater microorganisms have a high affinity glycine betaine uptake system which also recognizes dimethylsulfoniopropionate. Aquat. Microb. Ecol. 15:39-51. [Google Scholar]
- 16.Kiene, R. P., and L. J. Linn. 2000. Distribution and turnover of dissolved DMSP and its relationship with bacterial production and dimethylsulfide in the Gulf of Mexico. Limnol. Oceanogr. 45:849-861. [Google Scholar]
- 17.Kiene, R. P., and L. J. Linn. 2000b. The fate of dissolved dimethylsulfoniopropionate (DMSP) in seawater: tracer studies using 35S-DMSP. Geochim. Cosmochim. Acta 64:2797-2810. [Google Scholar]
- 18.Kiene, R. P., L. J. Linn, and J. A. Bruton. 2000. New and important roles for DMSP in marine microbial communities. J. Sea Res. 43:209-224. [Google Scholar]
- 19.Kiene, R. P., L. Linn, J. Gonzalez, M. A. Moran, and J. Bruton. 1999. Dimethylsulfoniopropionate and methanethiol are important precursors of methionine and protein-sulfur in marine bacterioplankton. Appl. Environ. Microbiol. 65:4549-4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kiene, R. P., P. T. Visscher, M. D. Keller, and G. O. Kirst (ed.). 1996. Biological and environmental chemistry of DMSP and related sulfonium compounds. Plenum, New York, N.Y.
- 21.Kirchman, D. L. 2001. Measuring bacterial biomass production and growth rates from leucine incorporation in natural aquatic environments, p. 227-237. In J. Paul(ed.), Methods in microbiology. Academic Press, New York, N.Y.
- 22.Kirchman, D. L., E. K'nees, and R. E. Hodson. 1985. Leucine incorporation and its potential as a measure of protein synthesis by bacteria in natural aquatic systems. Appl. Environ. Microbiol. 49:599-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ledyard, K. M., and J. W. H. Dacey. 1994. Dimethylsulfide production from dimethylsulfoniopropionate by a marine bacterium. Mar. Ecol. Prog. Ser. 110:95-103. [Google Scholar]
- 24.Ledyard, K. M., and J. W. H. Dacey. 1996. Microbial cycling of DMSP and DMS in coastal and oligotrophic seawater. Limnol. Oceanogr. 41:33-40. [Google Scholar]
- 25.Lee, N., P. H. Nielsen, H. Andreasen, S. Juretschko, J. L. Nielsen, K.-H. Schleifer, and M. Wagner. 1999. Combination of fluorescent in situ hybridization and microautoradiography—a new tool for structure-function analyses in microbial ecology. Appl. Environ. Microbiol. 65:1289-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Malin, G., and G. O. Kirst. 1997. Algal production of dimethyl sulfide and its atmospheric role. J. Phycol. 33:889-896. [Google Scholar]
- 27.Malmstrom, R. R., R. P. Kiene, and D. L. Kirchman. 2004. Identification and enumeration of bacteria assimilating dimethylsulfoniopropionate (DMSP) in the North Atlantic and Gulf of Mexico. Limnol. Oceanogr. 49:597-606. [Google Scholar]
- 28.Moran, M. A., J. M. González, and R. P. Kiene. 2003. Linking a bacterial taxon to sulfur cycling in the sea: studies of the marine Roseobacter group. Geomicrobiol. J. 20:1-14. [Google Scholar]
- 29.Ouverney, C. C., and J. A. Fuhrman. 1999. Combined microautoradiography-16S rRNA probe technique for determination of radioisotope uptake by specific microbial cell types in situ. Appl. Environ. Microbiol. 65:1746-1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parsons, T. R., Y. Maita, and C. M. Lalli. 1984. A manual for chemical and biological methods for seawater analysis. Pergamon, New York, N.Y.
- 31.Pedrós-Alió, C., and S. Y. Newell. 1989. Microautoradiography study of thymidine uptake in brackish waters around Sapelo island, Georgia, USA. Mar. Ecol. Prog. Ser. 55:83-94. [Google Scholar]
- 32.Pernthaler, J., F. O. Glöckner, W. Schönhuber, and R. Amann. 2001. Fluorescence in situ hybridization (FISH) with rRNA-targeted oligonucleotide probes, p. 207-226. In J. H. Paul (ed.), Marine microbiology. Academic Press, London, United Kingdom.
- 33.Porter, K. G., and Y. S. Feig. 1980. The use of DAPI for identifying and counting the aquatic microflora. Limnol. Oceanogr. 25:943-948. [Google Scholar]
- 34.Simó, R. 2001. Production of atmospheric sulfur by oceanic plankton: biogeochemical, ecological and evolutionary links. Trends Ecol. Evol. 16:287-294. [DOI] [PubMed] [Google Scholar]
- 35.Simó, R., S. D. Archer, C. Pedrós-Alió, L. Gilpin, and C. E. Stelfox-Widdicombe. 2002. Coupled dynamics of dimethylsulfoniopropionate and dimethylsulfide cycling and the microbial food web in surface waters of the North Atlantic. Limnol. Oceanogr. 47:53-61. [Google Scholar]
- 36.Simó, R., J. O. Grimalt, and J. Albaigés. 1996. Sequential method for the field determination of nanomolar concentrations of dimethyl sulfoxide in natural waters. Anal. Chem. 68:1493-1498. [DOI] [PubMed] [Google Scholar]
- 37.Simó, R., and C. Pedrós-Alió. 1999. Short-term variability in the open ocean cycle of dimethylsulfide. Global Biogeochem. Cycles 13:1173-1181. [Google Scholar]
- 38.Simó, R., and C. Pedrós-Alió. 1999b. Role of vertical mixing in controlling the oceanic production of dimethyl sulphide. Nature 402:396-399. [Google Scholar]
- 39.Smith, D. C., and F. Azam. 1992. A simple, economical method for measuring bacterial protein synthesis rates in seawater using 3H-leucine. Mar. Microb. Food Webs 6:107-114. [Google Scholar]
- 40.Stefels, J. 2000. Physiological aspects of the production and conversion of DMSP in marine algae and higher plants. J. Sea Res. 43:183-197. [Google Scholar]
- 41.Sunda, W., D. J. Kieber, R. P. Kiene, and S. Huntsman. 2002. An antioxidant function for DMSP and DMS in marine algae. Nature 418:317-320. [DOI] [PubMed] [Google Scholar]
- 42.Tang, K. W., and R. Simó. 2003. Trophic uptake and transfer of DMSP in simple planktonic food chains. Aquat. Microb. Ecol. 31:193-202. [Google Scholar]
- 43.Yoch, D. C. 2002. Dimethylsulfoniopropionate: its sources, role in the marine food web, and biological degradation to dimethylsulfide. Appl. Environ. Microbiol. 68:5804-5815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yoch, D. C., J. H. Ansede, and K. S. Rabinowitz. 1997. Evidence for intracellular and extracellular dimethylsulfoniopropionate (DMSP) lyases and DMSP uptake site in two species of marine bacteria. Appl. Environ. Microbiol. 63:3182-3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zubkov, M. V., B. M. Fuchs, S. D. Archer, R. P. Kiene, R. Amann, and P. H. Burkill. 2001. Linking the composition of bacterioplankton to rapid turnover of dissolved dimethylsulphoniopropionate in an algal bloom in the North Sea. Environ. Microbiol. 3:304-311. [DOI] [PubMed] [Google Scholar]