Abstract
The role of immune response is emerging as a key factor in the complex multistep process of cancer. Tumor microenvironment contains different types of immune cells, which contribute to regulate the fine balance between anti and protumor signals. In this context, mechanisms of crosstalk between cancer and immune cells remain to be extensively elucidated. Interestingly, microRNAs (miRNAs) have been demonstrated to function as crucial regulators of immune response in both physiological and pathological conditions. Specifically, different miRNAs have been reported to have a role in controlling the development and the functions of tumor-associated immune cells. This review aims to describe the most important miRNAs acting as critical modulators of immune response in the context of different solid tumors. In particular, we discuss recent studies that have demonstrated the existence of miRNA-mediated mechanisms regulating the recruitment and the activation status of specific tumor-associated immune cells in the tumor microenvironment. Moreover, various miRNAs have been found to target key cancer-related immune pathways, which concur to mediate the secretion of immunosuppressive or immunostimulating factors by cancer or immune cells. Modalities of miRNA exchange and miRNA-based delivery strategies are also discussed. Based on these findings, the modulation of individual or multiple miRNAs has the potential to enhance or inhibit specific immune subpopulations supporting antitumor immune responses, thus contributing to negatively affect tumorigenesis. New miRNA-based strategies can be developed for more effective immunotherapeutic interventions in cancer.
Keywords: MicroRNAs, Cancer, Immune System, Immune-related MicroRNAs, Innate Immunity, Adaptive Immunity, Cancer-Related Immune Response, Anticancer Immunotherapy
Background
Local immune response has emerged in the last decade as a key element in the modulation of the multistep process of cancer development [1]. The connection between tumor onset and inflammation has been envisaged after the demonstration that some tumors arise from sites of chronic inflammation. Moreover, not only some tumors are infiltrated by both the innate and adaptive arms of the immune system, but these cells are present even within tumor microenvironment [2]. Immunity has been reported to act both as a pro- or anti-tumorigenic factor depending on the fine-tuned equilibrium between innate and adaptive immune system [3]. In this context, the intercellular communication between cancer and infiltrating immune cells has the main role to modulate this immune response, thus positively influencing tumor development [4]. Among the different molecular players in the field, microRNAs (miRNAs) have been described as small non–coding RNA molecules regulating different physiological and pathological processes, including inflammation and cancer [5–7]. The multifaceted role of miRNAs derives from their mechanism of action, based on post-transcriptional modulation of multiple genes by base-pairing to target messenger RNAs (mRNAs) [8]. Mature miRNAs are 18-24 nucleotides long, and their biogenesis process has been widely studied in the past years. Briefly, primary miRNA (pri-miRNA) transcripts are produced in the nucleus by RNA polymerases II or III and subsequently processed into precursor-miRNAs (pre-miRNAs) by the RNase III domain of endonuclease Drosha, complexed with DCGR8 [8]. Pre-miRNAs are then actively translocated into the cytoplasm by the exportin 5 nuclear protein (EXP5) in complex with Ran-GTP unit. The RNase III endoribonuclease Dicer converts pre-miRNAs into RNA duplexes about 22 nucleotides long, containing one mature miRNA molecule [9]. This strand is finally incorporated into the RNA-induced silencing complex (RISC), which is the functional unit of Argonaute-mediated miRNA/mRNA binding and regulation [10]. Target sequences are usually located in the 3’untranslated region (UTR) of mRNAs and the grade of complementarity determines whether it will be targeted for degradation (total complementarity) or translational repression (partial complementarity) [11].
The growing body of literature demonstrating the importance of miRNAs in tumor onset, progression and response to therapy has defined these molecules as potential cancer biomarkers [12–14]. Different delivery strategies have been developed suggesting novel promising miRNA-based therapeutic approaches in order to overcome limits of current treatment of cancer [15]. Recently, due to the improvements in delivery systems, miRNA-targeting drugs entered into human clinical trials and the first results about the efficacy of this therapy is expected to be presented soon [16, 17]. In the past decades, the importance of miRNAs in the modulation of normal and pathological immune function has been shown in various studies in which deregulation of miRNAs was demonstrated to characterize diseases associated with excessive or uncontrolled inflammation [18]. There is an increasing number of studies directed to investigate miRNA-immunity-cancer connection and all these new data need to be comprehensively understood for novel therapy applications [19, 20]. Specifically, the mechanisms of miRNA exchange between cancer and immune cells have to be further explored and clarified in a tumor context-dependent manner. Exosomes have been demonstrated to represent vehicles of this miRNA transport within tumor microenvironment and recently also in circulation of cancer patients [21]. This review aims to summarize the most important miRNAs as regulators of immune cells (immune-related miRNAs) and to deeply discuss their role in modulating crucial checkpoints of cancer-related immune response in different types of solid tumors. An extensive understanding of effects of these immune-related miRNAs in cancer will very likely allow to identify specific miRNAs as potential targets for cancer immunotherapy.
The role of immune response in cancer
The human immune system has developed as a complex network of pathways regulating the response to pathogens. Innate immunity provides the initial defense against pathogenic infections, while its propagation leads to the activation of adaptive immune response. At this stage, adaptive immune cells sustain an extremely versatile mechanism of host organism defense in response to the exposure to a known antigen and/or to the reinfection with the same pathogen. In the context of cancer, immunity has clearly emerged as crucial biological event that contributes to the complex process of tumor development [22]. The pathogenesis of ∼ 15–20 % of human tumors is linked to infection-driven inflammation and interestingly, the presence of an inflammatory component characterizes the microenvironment of some tumors that are not causally linked to pathogens [23]. Two general molecular and cellular pathways have been proposed to describe the interaction between inflammation and cancer: the intrinsic pathway and the extrinsic one [23]. The intrinsic pathway which consists in a series of genetic events (e.g. activation of oncogenes, inactivation of tumor suppressor genes, different genetic aberrations) causing neoplastic transformation, initiates the induction of inflammation-related programs which influence the development of an inflammatory microenvironment (e.g. papillary thyroid carcinomas, breast cancers). On the other hand, the extrinsic pathway is driven by inflammatory leukocytes and soluble mediators that sustain inflammatory conditions increasing cancer risk (e.g. colon, prostate, pancreas cancers). All these pathways converge in the first activation of pro-inflammatory transcription factors in cancer cells and in their consequential production of inflammatory mediators (cytokines, chemokines, cyclooxygenase-2, and prostaglandins) [24]. Immune cells of innate system, such as macrophages, myeloid-derived suppressor cells, mast cells, eosinophils and neutrophils, are the first one to be recruited upon inflammation, and their activation contributes to reinforce the pro-inflammatory milieu [25]. The additional secretion of inflammatory mediators leads to the re-induction of the same pro-inflammatory pathways in cancer cells [23]. Independently of the cause that triggers inflammation, these immune signals present in the tumor microenvironment play a crucial role in all stages of cancer evolution, from initiation to metastasis. Indeed, an effective immune response ensues from the harmonious collaboration between innate and adaptive immune system, with the negative feedback of immune checkpoints and immunosuppressive mechanisms. On the other hand, immune response may exert either pro- or anti-tumorigenic effects on tumor microenvironment. A robust body of literature demonstrates that both innate and adaptive immunity are able to execute their role to achieve immunosurveillance, eliminating nascent tumors through the recognition of tumor neo-antigens as non-self [26]. On the contrary, following changes occurring in malignant cell populations or in host immune response, the immune system can fail to eliminate all cancer cells and tumors with reduced immunogenicity may escape the immune attack [27]. This dynamic balance between host-protective and tumor-promoting functions of inflammatory/immune cells have led to hypothesize the concept of cancer immunoediting, the process responsible for both eliminating tumors and modulating the immunogenic phenotypes of tumors that arise in immunocompetent hosts [26]. In conditions of equilibrium, immune system can induce a state of functional dormancy in cells, which can evade from this form of immune-mediated latent tumor through different strategies that allow them to proliferate [26, 28–33]. In established tumors, the escape from immunosurveillance occurs through different mechanisms, either at cancer (e.g. antigen loss, immunogenic tolerance) or immune cell level (e.g. T-cell activation inhibition) [34, 35]. In this second case, the cancer cells immersed in a local immunosuppressed microenvironment are required to mediate the inhibition of effector immune cells, such as T cells, natural killer cells (NK) or dendritic cells and the recruitment of immunosuppressive cells (regulatory T cells and myeloid-derived suppressor cells). Therefore, the heterogeneous composition of tumor microenvironment in terms of immune cell type has the potential to define a pro or an antitumor milieu [36]. Indeed, the presence of tumor-infiltrating lymphocytes (TILs) in human tumors has been described as a prognostic factor, supporting the evidence of immunoediting in this context [37]. Higher frequency of T cells, NK cells and natural killer T (NKT) cells have been correlated with better prognosis in patients with different types of cancer types [38–43]. Specifically, significant association between different subsets of T cells and clinical response have been found in cancer patients. In particular, a positive effect has been ascribed to CD8+ T cytotoxic (CTL), CD4+ T helper 1 (TH1) and T follicular helper (TFH) cells, as opposed to the negative role assigned to CD4+ regulatory T (TREG), T helper 2 (TH2) and T helper 17 (TH17) cells [44]. In particular, high ratios of CD4+/CD8+ and TH2/TH1 lymphocyte markers have been associated with poor prognosis specifically in breast cancer [45]. Among immune cells of myeloid origin, tumor-associated macrophages (TAMs) represent the most abundant cell component of tumor microenvironment, playing an important role in cancer development [46]. Two main different subsets of TAMs have been distinctively described according to their gene expression profiles and pattern of secreted molecules [47]. The natural plasticity of TAMs allows them to easily alter their phenotype during tumor development, converting from a pro-inflammatory (M1-like) form at early stages of tumor to a pro-angiogenic/immunosuppressive (M2-like) form during later phases of tumor progression (angiogenesis, invasion and metastasis) [48]. Several human tumors are mostly characterized by the presence of TAMs with M2-like phenotype [49, 50], while the presence of high levels of either M2- or M1- like TAMs has been identified as a poor prognostic factor in diverse solid tumors [51, 52]. However, understanding of the exact role of each TAM subset in cancer needs further investigation. Similarly, neutrophils have been demonstrated to exert both antitumor and protumor activities, including the sustainment of T cell responses and the promotion of angiogenesis and metastasis [46]. In addition, myeloid-derived suppressor cells (MDSCs) represent another component of TILs with multiple immunosuppressive functions on both innate and adaptive cells [53]. On the contrary, dendritic cells (DCs) play an important role as antigen presenting cells (APCs), which contribute to mount antitumor CTL immune response [54, 55]. Therefore, the immune-related nature of tumor microenvironment is very complex and depends on finely regulated interactions between cells [56]. This continuous crosstalk between tumor-infiltrating immune cells and cancer cells need to be extensively studied in order to define mechanisms underlying immunosurveillance and tumor immune escape.
MicroRNAs as regulators of cancer-related immunity in solid tumors
It has been widely demonstrated that differential expression patterns of miRNAs are associated with several human pathologies, including cancer in all its stages [57–59]. To date, miRNAs have been classified either as oncogenic (e.g. miR-155, miR-17-5p or miR-21), or having a tumor suppressor role (e.g. miR-34, miR-15a, let-7) [60–64]. Deregulation of a single miRNA or distinctive miRNA profiles have been correlated with survival, clinical outcome and response to therapy in various solid tumors [65–69]. Interestingly, recent studies have also correlated aberrant expression of crucial proteins related to miRNA biogenesis, with poor outcome [70]. Moreover, miRNAs are important regulators of both innate and adaptive immunity, controlling the maintenance and the development of immune progenitors as well as the differentiation and the functions of mature immune cell (Table 1) [71–74]. Therefore, the complexity of mechanisms underlying the connection between cancer and immunity has led to investigate miRNAs as additional key molecular players. Since specific miRNAs are essential for proper immune cell functioning, it is not surprising that aberrations in expression of immune-related miRNAs can lead to an altered antitumor immune response and contribute to cancer development [75, 76]. Indeed, some miRNAs with validated oncogenic or antitumor properties have shown the potential to exert a modulation of immune cell activity in the tumor microenvironment [20]. A large body of literature has reported different mechanisms by which single immune-related miRNAs have a double role in cancer development and immunity by modulating both immune and non-immune targets (Table 2). In the next subsections we are going to investigate the most relevant miRNAs with immunomodulatory effects, which can function as a bridge between immune response and cancer.
Table 1.
MicroRNAs involved in Innate and Adaptive Immune System Functions
| Cell lineage | Cellular process | MicroRNAs | |
|---|---|---|---|
| Immune cell progenitors | |||
| Hematopoietic stem cells | Cell maintenance | let-7ea, miR-29a, miR-99ba, miR-125a, miR-126, miR-212/132 cluster | |
| Multipotent progenitors | Cell development | miR-10 family, miR-126, miR-196b, miR-221/222 | |
| Common myeloid progenitors | Cell development | miR-17, miR-24, miR-126, miR-128, miR-155, miR-181a | |
| Common lymphoid progenitors | Cell development | miR-126, miR-128, miR-146, miR-181a | |
| Granulocyte–macrophage progenitors | Cell development | miR-16, miR-103, miR-107 | |
| Macrophage progenitors | Cell development | miR-17-5p, miR-20a, miR-106a | |
| Granulocyte progenitors | Cell development | miR-223 | |
| Erythroid precursors | Cell development | miR-155, miR-221/222 | |
| Megakaryocyte precursors | Cell development | miR-10a/b, miR-17, miR-20, miR-126 | |
| Innate immunity | |||
| Monocytes | Cell differentiation | miR-17-5p, miR-20a, miR-21, miR-106a, miR-155, miR-196b, miR-223, miR-338, miR-342, miR-424 | |
| Cell activation | miR-155, miR-424 | ||
| Dendritic cells | Cell differentiation | miR-21, miR-34a | |
| Cell function | miR-10a, miR-148/152, miR-155, miR-223 | ||
| Macrophages | Cell differentiation | miR-15a, miR-16, miR-19a-3p, miR-21, miR-107, miR-146a, miR-424 | |
| Cell function | Let-7, miR-9, miR-21, miR-101, miR-125b, miR-146a, miR-147, miR-155, miR-187, miR-212/132 cluster, miR-378, miR-487b, miR-1224 | ||
| Cell polarization | let-7c, let-7f, miR-9, miR-21, miR-33, miR-101, miR-124, miR-125, miR-146, miR-147, miR-155, miR-187, miR-223, miR-342, miR-378, miR-511 | ||
| Granulocytes | Cell differentiation | miR-15a, miR-21, miR-27, miR-196b, miR-223 | |
| Cell function | miR-223 | ||
| Neutrophils | Cell function | miR-223 | |
| MDSCs | Cell function | miR-494, miR-17-5p/20a | |
| Megakaryocytes | Cell differentiation | miR-10a, miR-130a, miR-146a, miR-150, miR-155, miR-223 | |
| Erythrocytes | Cell differentiation | miR-15a, miR-16, miR-24, miR-144, miR-150, miR-155, miR-221/222 cluster, miR-223, miR-451 | |
| Natural killer cells | Cell differentiation | miR-150, miR-181a/b | |
| Cell function | miR-15/16, miR-27a, miR-29, miR-30c-1, miR-30e, miR-155, miR-223, miR-378 | ||
| Adaptive immunity | |||
| B cells | Cell differentiation | miR-17/92 cluster, miR-23a, miR-34a, miR-142, miR-150, miR-155, miR-181 family, miR-212/132 cluster | |
| Cell activation | miR-9, miR-17/92 cluster, miR-30, miR-125b, miR-155, miR-181b, miR-223 | ||
| Plasma cells | Cell differentiation | miR-148a | |
| T cells | Cell differentiation | miR-17/92 cluster, miR-21, miR-142-3p, miR-150, miR-181a, miR-223 | |
| Cell activation | miR-155, miR-181a, miR-182, miR-214 | ||
| T helper cells | Cell differentiation | miR-125b, miR-150 | |
| Cell function | miR-182, miR-214, miR-297, miR-669c | ||
| T helper 1 cells | Cell differentiation | miR-17/92 cluster, miR-29, miR-146a, miR-148a, miR-155, miR-210, miR-326 | |
| T helper 2 cells | Cell differentiation | miR-21, miR-27, miR-28 | |
| Cell function | miR-155 | ||
| T cytotoxic cells | Cell differentiation | Let-7f, miR-15b, miR-16, miR-17/92 cluster, miR-21, miR-139, miR-142, miR-150, miR-155, miR-342 | |
| Cell function | miR-17/92 cluster, miR-21, miR-29, miR-23a, miR-24, miR-27a, miR-30b, miR-130/301, miR-139, miR-146a, miR-150, miR-155, miR-214 | ||
| T regulatory cells | Cell differentiation | miR-17/92 cluster, miR-10, miR-99a/miR-150, miR-155 | |
| Cell function | miR-142-3p, miR-146a, miR-155 | ||
| T helper 17 cells | Cell differentiation | miR-10a, miR-19b, miR-17, miR-155, miR-210, miR-212/132 cluster, miR-301, miR-326 | |
| T follicular helper cells | Cell differentiation | miR-10a, miR-17/92 cluster |
The most relevant miRNAs are in bold. MDSCs, Myeloid-Derived Suppressor Cells
aFurther investigations are required
Table 2.
Main deregulated microRNAs/targets, and the biological roles in immune- and cancer-related pathways in solid tumors
| Cancer type | miRNA | Expression status and cell localizationb | Target | Immune-related role | Cancer-related role | Refc |
|---|---|---|---|---|---|---|
| Breast | ↑miR-10b | Cancer cells | ↓MICB | Suppression of NK-mediated killing of tumor cells | Metastasis developmentd | [107] |
| ↑miR-19a-3p | M2 Macrophages | ↓FRA-1 | Macrophage polarization | Inhibition of cancer progression and metastasis development | [95] | |
| ↑miR-21 | Cancer cells | ↓PIAS3 | Reduced chemokine production and lymphocyte migration, immunoresistance to cancer immunotherapyd | Cancer cell survival, cell proliferation | [137] | |
| ↓miR-23a/ 27a/24-2 |
Macrophages | ↑A20 ↑JAK1 ↑STAT6 |
M2 Macrophage polarization | Xenograft tumor growth | [97] | |
| ↓miR-126/126a | Cancer cells | ↑SDF-1α | Downregulation of Ccl2 expression, Suppression of Inflammatory monocyte recruitment | Repression of MSC recruitment, lung metastasis promotion | [100] | |
| ↓miR-146a | Cancer cells | ↑IRAK1 ↑TRAF6 | Modulation of inflammationc | Cell Invasion and Migration impairment (NF-kB signaling block) | [92] | |
| ↑miR-155 | Cancer cells | ↓SOCS1 | STAT3 signaling activation | Cancer cell proliferation, colony formation, and xenograft tumor growth | [141] | |
| Myeloid cells | ↓SHIP1 | Tumor-infiltrating innate immune cell recruitment | Antitumor activity | [98] | ||
| ↑miR-223a | M2 macrophages and cancer cells | ↓MEF2Cd | Macrophage differentiationd | Promotion of cancer cell invasion | [163] | |
| ↑miR-494 | MDSCs | ↓PTEN | Accumulation of MDSCs | Tumor cell invasion and metastasis development | [117] | |
| Gastric | ↓miR-146a | Cancer cells | ↑IRAK1 ↑TRAF6 ↓IL8 |
Modulation of inflammationd | Antitumor activity | [146] |
| Ovarian | ↑miR-20a | Cancer cells | ↓MICA/B | Suppression of NK-mediated killing of tumor cells | Long-term cellular proliferation, invasion capabilities | [104] |
| ↓miR-199a | Cancer cells | ↑IKKβ | Cytokine production | Tumor progression, chemosensitivity (NF-kB signaling modulation) | [148] | |
| ↑miR-424 | Cancer cells | ↓PDL1 ↓CD80 |
T cell activation | Chemosensitivity | [151] | |
| Colorectal | ↓miR-17-5p/miR-20a/miR-124 | MDSCs | ↑STAT3 | Inhibition of immunosuppressive potential of MDSCs | Tumor growth | [142–144] |
| ↑miR-21/miR-29ba | Cancer cells and immune cells | ↑IL-6 (Indirectly) | Activation of pro inflammatory immune cellsd | Promotion of cancer cell invasion, tumor progressiond | [164] | |
| Hepatocellular | ↑miR-20a, miR-96, miR-106b | Cancer cells | ↓MICA | Suppression of NK-mediated killing of tumor cells | Long-term cellular proliferation, invasion capabilitiesd | [108] |
| HBV+/Hepatocellular | ↓miR-34a | Cancer cells | ↑CCL2 | Regulation of Treg recruitment | Suppression of tumor growth/metastasis development | [113] |
| Melanoma | ↓miR-34a/c | Cancer cells | ↑ULPB2 | Suppression of NK-mediated killing of tumor cells | Cell cycle arrest, senescence, apoptosis | [111–112] |
| ↓miR-17 | T cells | ↑STAT3 | Impairment of T cell responsed | Tumor growthd | [142] | |
| Melanoma and Lewis lung cancer | ↓miR-155 | Immune cells | ↑HIF1a | Recruitment of MDSC cells to tumor microenvironment | Promotion of tumor growthd | [118] |
| Lung | ↑miR-23a | T cells | ↓BLIMP1 | Suppression of CD8+ T cell functiond | Tumor progression, TGF-β-mediated immune evasiond | [145] |
| Glioma | ↓miR-124 | T cells | ↑STAT3 | Impairment of T cell responses | Tumor growth | [143] |
| Various solid tumors | ↓miR-29 | Cancer cells | ↑B7-H3 | Inhibition of NK and T cell functiond | Protumor activityd | [114–115] |
| ↓miR-214a | Cancer cells and CD4+CD25+ T cells | ↓PTEN | Expansion of Treg cells | Promotion of tumor growth | [166] |
a Detailed mechanism involving microvesicles-cell interactions (see also subsection “MicroRNAs and cell-to-cell communication”)
b These data are referred to studies in either tissue samples or in vitro/in vivo models
c Reference number listed in bibliography
d Further investigation is needed
Upregulation of miRNA or miRNA target. Downregulation of miRNA or miRNA target
HBV, Hepatitis B Virus; MDSCs, Myeloid-Derived Suppressor Cells; NKs, Natural Killer cells; TAMs, Tumor Associated Macrophages
MicroRNAs and tumor-associated immune cells
The differentiation and the activation of different tumor-associated immune cells have been described to be dependent on expression of specific miRNAs (Fig. 1) [77, 78]. In particular, deregulation of various miRNAs has been identified to affect monocyte-macrophage lineage maturation [79]. Members of miR-17/92 and miR-106a/92 clusters (miR-17-5p, miR-20a, and miR-106a) have been demonstrated to negatively regulate monocyte commitment by targeting Runt-related transcription factor 1 (RUNX1) [80]. Interestingly, modulation of miR-155, miR-125a/b, miR-146a, miR-21, and let-7e have been shown to be crucial for macrophage differentiation and activation into different phenotypes [81]. Specifically, the upregulation of these miRNAs have been observed to occur in macrophages after activation of Toll-like receptor (TLR) signaling and to sustain (i.e. miR-155, miR-125a/b) or repress (i.e. miR-146a, let-7e) pro-inflammatory M1- like TAM activation [79]. In the context of macrophage cell functions, miR-146a and miR-155 are also the two most well characterized miRNAs regulating immune response mediated by these cells [76]. These two miRNAs act as mediators of inflammatory stimuli with opposite effects on inflammatory response, miR-146a as negative and miR-155 as positive regulators of immune response through the direct targeting of IRAK1 and TRAF6, and SOCS1 and BCL6, respectively [82, 83]. However, this gene network is a part of a bidirectional mechanism in which inflammatory pathways are able to modulate miR-146a and miR-155 expression, and in turn innate immune response [83–86]. Along with TAMs, NK cells are another key component of the innate immune response and their differentiation has reported to be determined by miR-150 and miR-181a/b expression level [87]. In addition to tumor-associated innate immune cells, miRNAs have been found to regulate cell differentiation and functions of different T cell subsets (Fig. 1). Specifically, T helper effector differentiation has been found to be regulated by miR-155 expression in favor of TH1 phenotype, by miR-326 in promoting Th17 differentiation, and by miR-10a and miR-17-92 cluster regulating T follicular helper maturation [88–90]. In the lineage of CTL cells, the maturation and the activation of cells into effector or memory cell subsets have been demonstrated to be promoted by different miRNAs, including miR-17/92, miR-21, miR-30b and miR-155 [91]. On the contrary, miR-130/301 and miR-146a have displayed inhibiting effects on CTL immune responses [91]. miRNA roles have been also elucidated in Treg cell biology, with particular attention to miR-142-3p, miR-146a and miR-155 for cell function [92, 93].
Fig. 1.
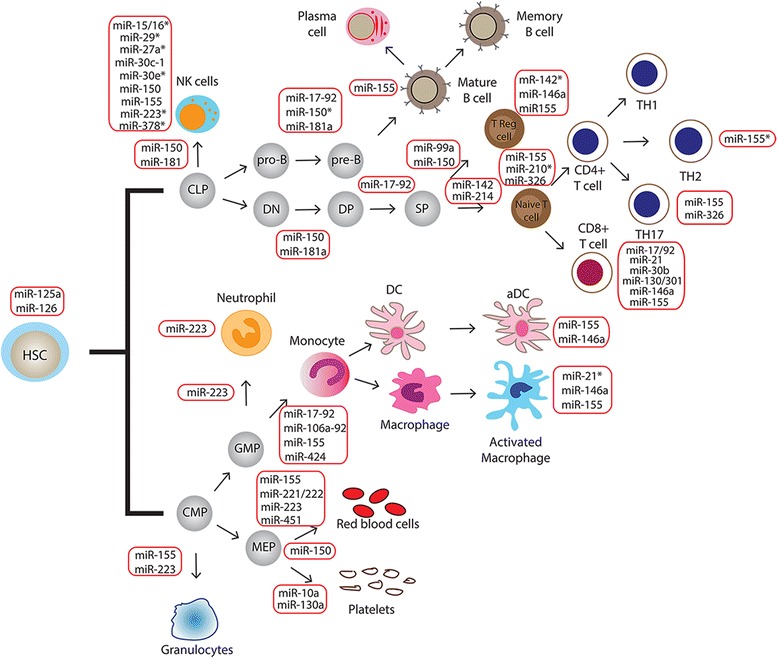
Schematic representation of microRNA regulation in immune cell development and activity. MiRNAs have been demonstrated to function as important regulators of both innate and adaptive immunity, including differentiation and functions of different immune cell subsets. The most relevant miRNAs are included in the figure. The miRNAs in red boxes are involved in regulating the developmental transition indicated by the arrows, as well as the activity of cells in the immune system (full explanation in text). *MicroRNAs involved in negative regulation of immune cell development or function. HSC: Hematopoietic Stem Cell; CMP: Common Myeloid Progenitor; GMP: Granulocyte–Monocyte Progenitor; MEP: Megakaryocyte-Erythrocyte Progenitor; CLP: Common Lymphoid Progenitor; DC: Dendritic cell; aDC: activated Dendritic Cell; DN: Double Negative; DP: Double Positive; SP: Single Positive
All these infiltrating inflammatory cells are recruited to the tumor microenvironment and their activation can be modulated by molecular signals produced by stromal and malignant cells [94]. In this context, distinctive miRNA-mediated mechanisms have been identified in different models of cancer (Table 2). In breast cancer miR-19a-3p has been reported to regulate the switch of TAMs from a M2-like phenotype into M1-like macrophages by targeting the Fra-1 proto-oncogene and other genes of its downstream signaling pathway (VEGF, STAT3 and pSTAT3), and to contribute to the inhibition of metastasis development [95]. In particular, Fra-1 has been already demonstrated to have a key role in the polarization of TAMs from the M1- to the M2-like phenotype [96]. Specifically, in the Balb/c mouse model, in vivo miR-19a-3p intratumoral injection has been found to both decrease the population of M2-like TAMs and inhibit lung metastasis of 4 T1 breast cancer cell-derived tumors [95]. Similarly, the miR-23a/27a/24-2 cluster has been demonstrated to mediate macrophage polarization and to contribute to tumor progression in breast cancer [97]. These studies support the concept that the modulation of the expression of single miRNAs (miR-19a-3p or miR-23a/27a/24-2 cluster downregulation) can promote the activation of specific signaling pathways, and the differentiation of a specific immune cell type (M2 phenotype of TAMs) in the tumor microenvironment.
Interestingly, miR-155 has been also reported to mediate the antitumor potential of distinctive immune cell subsets in breast cancer. In particular, miR-155 upregulation has been recently demonstrated to be required in the myeloid cell compartment for the promotion of antitumor immunity in early stages of breast cancer carcinogenesis [98]. In a spontaneous breast cancer model, specific miR-155 knock down in myeloid cells is able to induce faster tumor growth, reduction of M1-like TAMs and enrichment of protumor cytokines within tumor milieu, all concurring to create an immunosuppressive microenvironment [98]. In particular, the proposed mechanism involves the regulation of SHIP1, which is the main negative regulator of the pro-inflammatory PI3K/AKT pathway. The inhibition of this pathway was demonstrated to revert the common pro-inflammatory and protumor events mediated by AKT activation [99].
In the same direction, miR-126/126* pair has been shown to have an antitumor role by inhibiting breast cancer cell invasion and metastasis [100], either through the direct targeting of stromal cell-derived factor-1 alpha, SDF-1α, and with the indirect suppression of chemokine (C-C motif) ligand 2, CCL2, in cancer cells. These two chemokines mediate the sequential recruitment of two different non malignant cell types to primary tumor site: SDF-1α is responsible for attraction of mesenchymal stem cells (MSCs), while the second for inflammatory monocytes. MSCs are supposed to create a paracrine loop with cancer cells to induce cell invasion and migration, meanwhile monocytes act to promote the extravasation of tumor cells [101, 102]. Therefore, miR-126/126* pair is able to modulate the composition of the microenvironment of primary tumors in order to contrast breast cancer metastasis. These findings are perfectly in line with discoveries correlating reduced expression of miR-126 to poor metastasis-free survival of breast cancer patients [103].
As previously described, the complexity of tumor microenvironment includes innate immune components recruited to eradicate latent cancer cells. Among them, NK cells are a subset of lymphocytes that can rapidly respond to the presence of tumor cells and initiate an antitumor immune response. NK cells express receptors through which they are capable to detect their targets on cancer cells. MiR-20 has been demonstrated to regulate NK cytotoxicity in ovarian cancer through the targeting of MICA/B, a MHC class I chain-related molecules widely expressed on epithelial tumor cells [104]. This protein is recognized by NK cells through the NK group 2 member D receptor (NKG2D), whose pathway is critical for direct recognition of malignant cells by immune surveillance system [105]. In vitro and in vivo studies have shown that miR-20-mediated downregulation of MICA/B induced the reduction of NKG2D recognition resulting in the diminished killing of malignant cells by NK compartment, thus leading to enhanced tumor cell survival in vivo [106]. The same mechanism has been demonstrated for miR-10b/MICB pair in murine breast cancer model, and for miR-20a, miR-93, miR-106b/MICB pair in hepatocellular cell lines [107, 108]. These data propose a miRNA-based immune escape mechanism for tumor cells, which can partially explain the correlation between overexpression of these miRNAs and poor prognosis in cancer patients.
Similarly to human ovarian cancer cells, human melanoma cells have been reported to express NKG2D receptor ligands such as MICA and ULBP2 [109]. Recently, Heinemann A. et al. have identified serum ULBP2 overexpression as a strong independent predictor of poor prognosis in melanoma patients [110]. The same group also demonstrated that the tumor suppressor miR-34a/c repressed ULBP2 expression by directly binding to the 3’-UTR [111]. Together with the fact that miR-34a/c expression is frequently lost in cancer, these miRNAs might be crucial for tumor immune surveillance [112]. This immuno-suppressive mechanism is also suggested to predispose HBV-Positive hepatocellular carcinoma (HBV-HCC) patients to the development of intrahepatic venous metastasis. Specifically, miR-34a deregulation has been linked to immune escape mechanism in HBV-HCC, whose development is supposed to be associated with HBV and HCV virus infection [113]. In particular, TGF-β/miR-34a/CCL22 axis induced the recruitment of Treg cells that are known to have an inhibitory role in the immune system and, ultimately, to participate to escape immune surveillance helping tumor cells [113]. Interestingly, TGF-β signaling activation induces the suppression of miR-34a, CCL22 expression and in turn recruitment of Treg cells in liver microenvironment.
In the context of immune cell function regulation, miR-29 has been identified as a negative regulator of B7-H3 protein, which is a surface immunomodulatory glycoprotein inhibiting NK and T cell functions [114]. Specifically, mir-29 and B7-H3 expression levels have been found inversely correlated in both solid tumors and cancer cell line experiments [115]. The downregulation of miR-29 family members has been reported in many cancers, where they influence cell proliferation, apoptosis and metastasis development through the modulation of different targets [116]. Additional in vivo and in vitro studies have to be performed in order to validate the role of miR-29 in the promotion of antitumor immunity mediated by NK and T cell. Along with NK and T cells, the expansion and function of MDSCs depend on soluble factors released by tumor and stromal compartments and by activated immune cells [116]. Recently, it has been demonstrated that two different miRNAs, miR-494 and miR-155, are fundamental for the recruitment of MDSCs to the tumor site, contributing to the modulation of their immunosuppressive function and to tumor growth in breast cancer and glioma models, respectively [117, 118].
MicroRNAs and cancer-related immune pathways in solid tumors
Cancer and immune cells produce various growth or angiogenic factors, proteinases, chemokines and cytokines, which contribute to removal of tumor cell or to the formation of an immunosuppressive microenvironment [119, 120]. In particular, the activation of specific transcription factors and the presence of primary inflammatory cytokines represent key connection elements between immune and cancer cells [121, 122]. Among them, nuclear factor kappa-B (NF-kB), transducers activator of transcription 3 (STAT3), tumor necrosis factor (TNF) and transforming growth factor β (TGF-β), have been demonstrated to mediate the activation of numerous oncogenic pathways [122–124]. NF-kB is a major regulator of inflammation and innate immunity, and its aberrant regulation has been described in many human tumors [124]. Different upstream stimuli have been identified to trigger NF-kB pathway, including pro-inflammatory cytokines (e.g. TNF-α, IL-1β) and microorganism infections (bacterial and viral infections), which both determine the activation of the IkB kinase (IKK) complex. This protein unit phosphorylates inhibitor of NF-kB and allows the translocation of NF-kB into nucleus [125]. The downstream effects of NF-kB are cell-type dependent, consisting in induction of gene expression related to pro-inflammatory signals in inflammatory cells and of anti-apoptotic genes in tumor cells, favoring tumor development [125]. One of the most important activators of NF-kB signaling is TNF-α, which binds to its specific receptor expressed by immune or cancer cells [126]. This pro-inflammatory cytokine has been also demonstrated to exert tumor–promoting activities, including promotion of angiogenesis and metastasis [127]. Similar to NF-kB, STAT3 is constitutively activated in both tumor and immune cells increasing tumor cell proliferation, survival and invasion and is activated by NFkB-induced genes such as Interleukin 6 (IL6) [128–129]. On the contrary, STAT3 signaling is required for immunosuppressive and pro-tumorigenic functions of immune cells of both innate (MDSCs, TAMs) and adaptive system (Treg, Th17), opposing the role of NF-kB in favoring antitumor immune response [130, 131]. Immunosuppressive and anti-inflammatory signals derive also from the action of TGF-β, which attenuates the production of pro-inflammatory cytokines [132]. Furthermore, TGF-β is known to be a key cytokine during carcinogenesis, being secreted and upregulated in a wide range of tumors [133–136]. Therefore, different signaling represent crucial pathways in the model of origin of cancer-related inflammation and large body of literature supports the central role of miRNAs as both down- or up-stream modulators of the activation of these factors (Table 2).
Among these miRNAs, miR-21 and miR-155 have been found to be part of the complex immune regulatory network in the context of breast cancer [137–141]. These two oncomiRNAs are linked in different ways to the constitutive activation of STAT3 pathway, promoting the expression of immunosuppressive factors and thus having immune suppressive effects. Inhibition of miR-21 expression levels in MCF7 cell line has recently been reported to increased chemokine and lymphocyte migration, which is paralleled to an increase in levels of PIAS3, an inhibitor of activated STAT3, and of STAT3 phosphorylation, making miR-21 a good therapeutic target [137]. In particular, this mechanism is explained by the activity of miR-21, which inhibits the release of RANTES and IP-10, T cell chemoattractants, through the targeting of PIAS3 [137]. These data have demonstrated a positive relationship between miR-21 and STAT3 in a tumor cell line context, as previously reported [138, 139]. In addition to the protumor activities affecting cell survival and proliferation, miR-21/STAT3 network could contribute to create an immune suppressive tumor milieu, thus supporting the miRNA role as regulator of drug resistance [140]. Similarly to this miRNA, miR-155 exerts its oncogenic role by negatively regulating the tumor suppressor gene of cytokine signaling 1 (Socs1) and consequently promoting cell proliferation, colony formation and xenograft tumor growth in breast cancer model [141]. The immunomodulating property is represented by the constitutive activation of STAT3 signaling as tumor promoting inflammatory mechanism.
Other miRNAs have been linked to the regulation of STAT3 signaling. As previously described, effective adaptive immune responses have a key role in contrasting tumor progression and different immune cell subsets contribute to maintain this potential. In this context, STAT3 mRNA has been demonstrated to be directly targeted by miR-17-5p, miR-20a and miR-124 in different cancer models where forced miRNA upregulation has been showed to sustain T cell response in favor of an antitumor activity [142–144]. Conversely, miR-23a contributes in suppressing CTL function, at least in a murine model of lung cancer [145]. Specifically, the TGF-β-mediated upregulation of miR-23a in CTL cells induced a tumor immune-evasion mechanism by targeting the transcription factor BLIMP-1, in turn promoting tumor progression [145].
As previously mentioned, miR-146a is a miRNA involved in the control of both inflammatory response to infection and of innate immune system. In particular it has been described as a NF-kB-dependent gene, which in turn downregulates the expression of immune target genes, such as IRAK1 and TRAF6 (two adaptor molecules downstream of toll-like and cytokine receptors), and represses NF-kB signaling in LPS-stimulated monocytes [83]. MiR-146a axis has been investigated also in gastric cancer cells, where a link between miR-146a and the inflammatory response to Helicobacter pylori has been envisaged [146]. Similarly, forced ectopic expression of miR-146a in MDA-MB-231 breast cancer cell line inhibits endogenous NF-kB expression and activity through IRAK1 and TRAF6 targeting, reducing their metastatic potential [147]. Further investigations are needed in order to fully elucidate how miR-146a contribute to tumor development.
A study by Chen R. et al. has described a similar miR-199a/IKKβ/NF-kB axis also in epithelial ovarian cancer cells (EOCs) [148]. MiR-199a has been identified as a negative regulator of IKKβ mRNA, which encodes for the β subunit of IKK, the direct upstream activator of NF-kB pathway [149]. In EOCs, IKKβ contributes to a pro-inflammatory environment by functionally inducing NF-kB pathway through the activation of Toll-like receptor 4 (TLR4)-MyD88 signaling sustaining tissue repair processes and the secretion of pro-inflammatory signals [150]. Consequently, EOC cells are induced to secrete pro-inflammatory/protumor cytokines, including IL-6, IL-8, MCP-1, MIP-1α, RANTES, GRO-α, GM-CSF and MIF. MiR-199a targets IKKβ resulting in the inhibition of NF-kB signaling. Therefore, the loss of this anti-inflammatory miRNA may be an important step that contributes to tumor progression. These data may have important implications in tissue repair, tumor progression and chemoresistance. Interestingly, chemosensitivity of EOCs has been shown to be restored by miR-424 overexpression, which activates T cell immune response through direct targeting of critical immune checkpoints, such as the programmed death-ligand 1 (PD-L1) and CD80 [151].
MicroRNAs and cell-to-cell communication
miRNAs have emerged as crucial mediators of intercellular communication occurring between immune and tumor cells within the tumor microenvironment [152]. The vast majority of studies described the mechanisms of deregulation of endogenous miRNAs in immune or tumor cells, which can modulate the cancer-related immune response, thus also affecting tumor progression. More recently, a mechanism of indirect cell-to-cell communication has been described, showing that exogenous miRNAs can be transferred from a donor to a recipient cell in order to modulate gene expression [21]. This process is based on cell-derived extracellular vesicles (EVs) containing both proteins and RNAs, including miRNAs and mRNAs. Generally EVs are subdivided into three major classes of particles according to size, ectosomes or shedding vesicles (200–1000 nm) and exosomes (30–200 nm), which differ from apoptotic bodies (0.5–3 μm) that derive from cells in apoptosis or under stress [153]. In particular, exosomes have been identified as the most important carriers of functional miRNAs [21]. Exosomes are small membrane-derived vesicles of endocytic origin, which can be released by different types of cell, including immune and tumor cells, under both normal and pathological conditions [154]. The mechanism of horizontal exchange of exosome containing miRNAs between cells is described in Fig. 2. Interestingly, the composition of RNA molecules in exosomes is different from the content of cell of origin suggesting the existence of not well defined active mechanisms for sorting specific RNAs into exosomes [155–157]. In particular, the ESCRT (endosomal sorting complex required for transport) protein complex is likely to be involved in some of these processes [157–159]. However, the exact nature of these mechanisms remain to be extensively characterized. The transfer of this genetic information can alter gene expression in neighboring and even distant cells. Recent data have demonstrated involvement of exosome at different levels, including immune responses and cancer [160–162]. These findings suggested that exosome-mediated miRNA transfer between immune and tumor cells could be crucial to identify new miRNAs as modulators of tumor microenvironment and potential target for cancer immunotherapy. Several recent studies provided evidences of this hypothesis (Table 2). A study by Yang M. et al. firstly demonstrated that after transfection into IL-4 activated M2 macrophages, the exogenous miR-223 can shuttle into co-cultivated breast cancer cells from M2-derived exosomes containing miRNA. Interestingly, exosome–transferred miR-223 stimulates the invasive behavior of breast cancer cells by targeting of Mef2c/β-catenin pathway, thus leading to increase cell migration [163]. A more complex regulatory loop between cancer and immune cells has been described in in vitro co-culture model of colorectal cancer: the secretion of IL-6 from immune cells promotes invasiveness of cancer cells, which in turn induces immune-related IL-6 production and miR-21 release through the tumor-derived secretion of miR-21 and miR-29b in the tumor microenvironment [164]. As result, this mechanism mediated by exogenous miRNAs is suggested to be able to support the maintenance of a pro-tumorigenic inflammatory environment. Moreover, the two tumor-secreted miR-21 and miR-29b have been reported to act as ligands binding to receptors of TLR family of macrophages, producing pro-inflammatory signals in the tumor microenvironment and pro-metastatic potential in in vivo models [165]. Similarly, miR-214 has been identified as another tumor-secreted miRNA capable to support simultaneously host immune suppression and tumor growth in in vivo mice models [166]. Specifically, tumor-derived miR-214 was delivered into peripheral CD4+ T cells and found to induce Treg expansion by targeting phosphatase and tensin homolog (PTEN) protein. The secretion of higher levels of IL-10 by miR-214-induced Treg has been demonstrated to promote tumor growth in nude mice [167]. The dual role of tumor-derived exosomes in the modulation of immune responses and in the mediation of tumor progression has been also investigated at circulating level in cancer patients [167–170]. In particular, higher level of circulating exosomes have been found in breast or ovarian cancer patients compared to healthy individuals, and serum exosomes derived from patients with oral or ovarian cancer have been characterized as inhibitors of T cell functions [167–170]. In this context, interestingly circulating tumor-derived exosomes isolated from serum of nasopharyngeal carcinoma patients have been characterized as miRNA-enriched vesicles able to impair in vitro T cell function interfering with ERK and STAT signaling pathways in T cells [171]. In addition to the exosome mediation, miRNA transfer has been demonstrated to happen directly via intercellular contact through gap junctions. Specifically, human macrophages have been reported to be able to secrete miR-142 and miR-223, which are transferred into hepatocarcinoma cells (HCCs) [172]. These miRNAs affect posttranscriptional regulation of target proteins in HCC cells, in particular inducing decreased expression levels of stathmin-1 (STMN1) and insulin-like growth factor-1 receptor (IGF-R1), and, importantly, inhibiting proliferation of cancerous cells in in vitro experiments [172]. In opposition to its antitumor role in HCC model, M2-derived miR-223 has been previously shown to stimulate proliferation in breast cancer cells because of the targeting of Mef2c/β-catenin pathway related to the inhibition of cell migration [163, 173, 174].
Fig. 2.
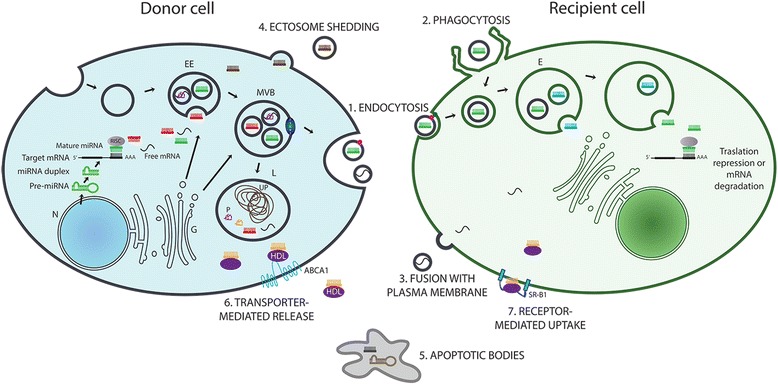
Mechanisms of RNA transfer in cell-to-cell communication. Mechanisms underlying the transfer of RNA molecules between cells are mainly based on two systems, vesicle- and protein-mediated transport. (1,2,3) After exosome release from donor cell, RNA content is delivered into recipient cell by (1) the fusion of the exosome with the recipient cell membrane, by (2) phagocytosis- or (3) endocytosis-like internalization of the exosome. RNA molecules can be exported and transported out of the cells by microvesicles also as (4) shedding ectosomes or (5) apoptotic bodies. (6,7) Different protein complexes (violet boxes) including Argonaute, NPM1 and HDL proteins, bind miRNAs and are transferred out of the cell through (6) transporter-mediated release (ABCA1) and are translocated to target cell by (7) receptor-mediated uptake (SR-B1). All these pathways result in the delivery of microRNA or mRNA molecules to the cytosol of the recipient cell where they may contribute to post translational gene regulation. E: Endosome; EE: Early Endosome; G: Golgi; L: Lysosome; MVB: Multivesicular Bodies; N: Nucleus; P: Protein; UP: Undigested Protein
More data regarding different mechanisms of miRNA transfer from non-immune stromal to tumor cells have been published [175]. All these findings highlight the potential role of shuttled miRNAs in mechanisms used by cancer cells to evade immunosurveillance and to sustain tumor progression.
MicroRNA-based immune response as potential target for anticancer immunotherapies
The potential of miRNAs in regulating different cellular pathways and as mediators of interactions between cells make them ideal drug targets. Indeed, different local and systemic delivery strategies are already under investigation [176]. Their ultimate aim is either enhancing or inhibiting the expression of specific miRNAs acting as tumor suppressor genes or oncogenes, respectively [176]. These effects can be achieved by targeting miRNAs at different levels of their biogenesis and activity (Fig. 3). In the context of cancer cell targeting, promising results have been obtained with the use of miRNA antagonists and miRNA mimics in preclinical studies [177, 178]. miRNA antagonists are single-stranded oligonucleotides complementary to miRNA sequences, designed to target and functionally reduce miRNA activity [179]. On the other hand, restoration of tumor suppressor miRNA level is obtained by the delivery of chemically synthesized short double-stranded oligonucleotides, which functionally mimic pre-miRNA duplexes [180]. To date, these two strategies are under investigation in clinical trials based on targeting of miR-122 and miR-34a for the treatment of hepatitis C virus and advanced HCC, respectively [181]. Due to the recent advances in the understanding of specific miRNA modulatory mechanisms that influence the immune system and tumor-mediated immunity, researchers are now developing novel miRNA-based interventions for cancer immunotherapy. Currently, different immunotherapies have been approved aimed to activate antitumor immunity, including passive immunization with monoclonal antibodies, systemic delivery of cytokines and addition of immune adjuvants into the tumor microenvironment [182]. Among them, the use of tumor targeting monoclonal antibodies seems to be the most promising approach for some hematologic and solid tumors [183]. Novel antibody-based approaches are under development in order to block immunosuppressive networks or stimulate antitumor cytotoxicity [184–188]. In this context, the regulation of cancer-related immune responses through the fine tuning of immune-related miRNA expression could contribute to enhance antitumor immunity and at the same time inhibit tumor development. Interestingly, miRNAs can be targeted not only in cancer cells but also in stromal cells, such as tumor-associated fibroblasts and lymphocytes, which are essential for tumor formation, progression and metastasis. In this case, successful delivery directed to specific tissue and cell, represents a big challenge for in vivo miRNA-mediated immunomodulation. Different types of vehicles have been synthesized as biodegradable and biocompatible carriers of miRNA mimics and miRNA antagonists, including liposomes, polymers, nanoparticles and viral agents [182]. The versatility of liposomal carriers have made them suitable elements for designing of co-delivery system of miRNAs and small-molecule drugs which concurrently are able to target the same cancer cell resulting in an effective synergic antitumor effect. Firstly employed for small conventional drugs and siRNA delivery in clinical trials, liposomal formulation of miR-34a mimic is currently used in a Phase I clinical trial of patients with advanced HCC [17]. Polymeric micelles, polymeric nanoparticles and carbon nanomaterials have already been employed in co-delivery system of different miRNAs and chemotherapeutics in different tumor models [189–192]. Interestingly, systematic intensive infiltration of myeloid leukocytes in solid tumors and their enhanced endocytic activity make these cells the ideal targets for nanocomplex-mediated delivery. Accordingly, a recent work has obtained the induction of miR-155 activity selectively in DCs in ovarian cancer microenvironment using a non-viral approach, resulting in the promotion of T-cell mediated protective immunity and therefore antitumor responses [193]. In the same context, downregulation of miR-31/miR-214 and upregulation of miR-155 are capable to selectively reprogram normal human fibroblasts into tumor-promoting cancer-associated fibroblasts (CAFs) [194]. Furthermore, downregulation of miR-214 has also been demonstrated to directly influence chemokine (C-C motif) ligand 5 CCL5 production, whose increased levels lead to enhanced tumor growth, by stimulating OvCa cells invasion [195]. Therefore, miR-31/miR-214 restoration and inhibition of miR-155 in protumor CAFs could be considered as potential immunotherapeutic options for ovarian cancer. Similarly, miR-21 has been identified as a potential immunotherapeutic target for its ability to positively regulate STAT3 signaling, which is a prerequisite for effective T cell therapy. In addition, miR-21 can suppress T cell priming and impair responses triggered by anti-mycobacterial vaccination [196].
Fig. 3.
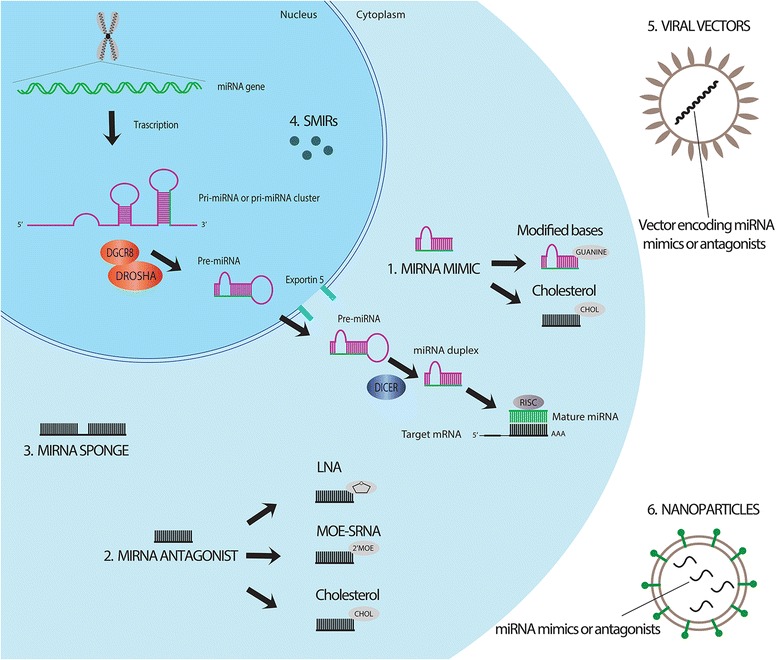
MicroRNA-based strategies for anti-cancer therapy. The main strategies for the modulation of miRNA activity are basically based on enhancing or inhibiting the expression of specific miRNAs with miRNA mimics (1) or miRNA antagonists (2), respectively. Modified miRNA molecules have been developed to increase the stability of miRNA mimics and miRNA antagonists, including miRNA mimics containing modified cyclopentyl-guanine based, cholesterol-conjugated 2′-O methyl-modified miRNA mimics/anti-miRs, locked nucleic acid (LNA)-modified anti-miRs and 2′-O-methoxyethyll-4′-thioRNA (MOE-SRNA). A different approach consists in miRNA sponges (3), which are complex constructs able to interfere with miRNA/mRNA interaction. Interference at miRNA biogenesis level is obtained with small-molecule inhibitors of miRNAs, SMIRs (4). Modified or unmodified miRNA modulators can be delivered to target cells by using viral (5) or non-viral vectors consisting in different types of biocompatible and biodegradable nanoparticles (6)
Opposed to previous described non-viral delivery approach, viral vectors have been investigated as potential carriers of miRNAs in order to improve the efficiency and the specificity of the systemic delivery. Different examples of application of virus can be mentioned, starting from lentiviral vectors containing miR-494 antagonists with the potential of reducing tumor-infiltrating MDSCs and their protumor activity in in vivo breast cancer model [117]. Lentiviruses are able to integrate their reversed DNA into human cells, implicating the potential risk of different oncogenic pathways’ activation. Adenovirus and adeno-associated virus are more suitable for therapeutic purposes, due to their non-integrative activity [197]. However, the general increased immunogenic microenvironment and the limits in their large-scale production make viral vectors a less safe delivery system than non-viral approach.
In the context of extracellular vesicle-based therapy, the identification of exosomes as miRNA carriers and mediators of communication between cells of tumor microenvironment constitutes an opportunity to study new targeted approaches [198]. Similar applications have been already investigated using tumor-derived EVs to deliver a therapeutic miRNA to breast cancer cells with specific phenotypes [199]. In particular, the tumor suppressor miRNA let-7a has been efficiently delivered to epidermal growth factor receptor (EGFR)-expressing breast cancer cell, exerting the in vivo inhibition of tumor development. The specific targeting of receptor-expressing cells was obtained by using exosomes derived from engineered donor cells with EGFR ligand on plasma membrane. As regards the immune system, EVs derived from immune and non-immune cells are able to positively and negatively regulate the immune response [200]. Thus, cell-derived exosomes containing immune-related miRNAs could have the potential to be used as therapeutic agents to enhance immunostimulatory signaling pathways and antitumor immunity. In this context, tumor-derived exosomes has been primarily investigated, showing their potential to concur to immune evasion [201, 202]. Recently, miRNAs have been reported to be associated with the RISC-Loading Complex in breast cancer-derived exosomes [203]. These entities display cell-independent miRNA biogenesis able to sustain Dicer-dependent mRNA targeting in recipient cells. Specifically, cancer exosomes can induce malignant transformation of normal epithelial cells in a Dicer-dependent manner. This work contributes to propose miRNA-containing exosomes as active players of cancer progression. However, the use of exosomes as carriers of miRNAs in cancer therapies is only at the beginning and needs to be further investigated.
Conclusions
The importance of specific miRNAs for immune cell development and function has been widely demonstrated, and their association with different human diseases is a matter of fact. Alteration in levels of these immune-related miRNAs could determine the modulation of immune pathways and the crosstalk between cells in the tumor microenvironment, thus resulting in a non-effective cancer-related immune response. Altogether, these findings suggest the development of novel miRNA-based approaches directed to target immunomodulatory mechanisms. Reported data have also highlighted that miRNAs can exert their function in a cell type context-dependent manner. Thus, the design of more effective in vivo strategies of miRNA delivery needs to be improved, in particular focusing on enhancing tissue and cell specificity of miRNA vectors. Accordingly, exosome- and immune cell-based delivery represent two interesting potential strategies for miRNA-based cancer immunotherapy. In addition, the surface of carrier vesicles can be specifically modified with ligands or antibodies which are able to bind to the endogenous receptors of tumor or stromal cells. Therefore, the characterization of the tumor microenvironment in terms of miRNA/mRNA expression and their localization at cellular level is crucial. In addition, the mechanisms of miRNA-based modulation of immune responses need to be investigated in relation to different immunomodulatory therapies. In this context, the combination of miRNA-related immunotherapy with conventional cytotoxic drug agents or targeted therapy could represent a valuable opportunity for effective therapeutic interventions in human cancer.
Abbreviations
CAF, cancer-associated fibroblast; CCL2, chemokine (C-C motif) ligand 2; CCL5, chemokine (C-C motif) ligand 5; CTL, CD8+ T cytotoxic cell; EGFR, epidermal growth factor receptor; ESCRT, endosomal sorting complex required for transport; EV, extracellular vesicle; EXP5, exportin 5; HCC, hepatocellular carcinoma; IKK, IkB kinase; IL6, interleukin 6; mRNA, messenger RNA; miRNA, microRNA; NFkB, nuclear factor kappa-B; NK, natural killer; NKG2D, NK group 2 member D receptor; NKT, natural killer T cells; pre-miRNA, precursor-miRNA; pri-miRNA, primary-miRNA; RISC, RNA-induced silencing complex; RUNX1, Runt-related transcription factor 1; STAT3, transducers activator of transcription 3; TAM, tumor-associated macrophage; TIL, tumor-infiltrating lymphocytes; TFH, T follicular helper cell; TH1, T helper 1 cells; TH2, T helper 2 cells; TH17, T helper 17 cells; TLR, Toll-like Receptor; TREG, regulatory T cells; UTR, untranslated region
Acknowledgements
The authors are grateful to the Associazione Italiana Ricerca sul Cancro (AIRC) for funds support (Grant 6251 to L.S.) and Fondazione Italiana Ricerca sul Cancro (FIRC) (Grant 18328 to G.B.). GAC is The Alan M. Gewirtz Leukemia & Lymphoma Society Scholar. Work in GAC’s laboratory is supported in part by the National Institutes of Health (NIH)/National Cancer Institute (NCI) grants 1UH2TR00943-01 and 1 R01 CA1829-01, 05, the University of Texas MD Anderson Cancer Center SPORE in Melanoma grant from NCI P50 CA093459, Aim at Melanoma Foundation and the Miriam and Jim Mulva research funds, the Brain SPORE grant 2P50CA127001, the Center for Radiation Oncology Research Project, the Center for Cancer Epigenetics Pilot Project, a 2014 Knowledge Global Academic Programs (GAP) MD Anderson Cancer Center grant, a Chronic Lymphocytic Leukemia (CLL) Moonshot pilot project, the University of Texas MD Anderson Cancer Center Duncan Family Institute for Cancer Prevention and Risk Assessment, a Sister Institution Network Fund (SINF) grant for colon cancer research, the Laura and John Arnold Foundation, the RGK Foundation and the Estate of C. G. Johnson, Jr.
Authors’ contributions
All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Contributor Information
George A. Calin, Phone: +1 (713) 792-5461, FAX: +1 (713) 745-4528, Email: gcalin@mdanderson.org
Libero Santarpia, Phone: +39 02 8224-5173, FAX: +39 02 8224-5191, Email: Libero.Santarpia@humanitasresearch.it, Email: liberosantarpia@yahoo.it.
References
- 1.Kitamura T, Qian BZ, Pollard JW. Immune cell promotion of metastasis. Nat Rev Immunol. 2015;15:73–86. doi: 10.1038/nri3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balkwill F, Mantovani A. Inflammation and cancer: back to Virchow? Lancet. 2001;357:539–45. doi: 10.1016/S0140-6736(00)04046-0. [DOI] [PubMed] [Google Scholar]
- 3.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883–99. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Nardo DG, Coussens LM. Inflammation and breast cancer. Balancing immune response: crosstalk between adaptive and innate immune cells during breast cancer progression. Breast Cancer Res. 2007;9:212. doi: 10.1186/bcr1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ebert MS, Sharp PA. Roles for microRNAs in conferring robustness to biological processes. Cell. 2012;149:515–24. doi: 10.1016/j.cell.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raisch J, Darfeuille-Michaud A, Nguyen HT. Role of microRNAs in the immune system, inflammation and cancer. World J Gastroenterol. 2013;19:2985–96. doi: 10.3748/wjg.v19.i20.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dai R, Ahmed SA. MicroRNA, a new paradigm for understanding immunoregulation, inflammation, and autoimmune diseases. Transl Res. 2011;157:163–79. doi: 10.1016/j.trsl.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 9.Lund E, Güttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–8. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 10.Mendell JT, Olson EN. MicroRNAs in stress signaling and human disease. Cell. 2004;148:1172–87. doi: 10.1016/j.cell.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11:228–24. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 12.Santarpia L, Calin GA, Adam L, Ye L, Fusco A, Giunti S, et al. A miRNA signature associated with human metastatic medullary thyroid carcinoma. Endocr Relat Cancer. 2013;20:809–23. doi: 10.1530/ERC-13-0357. [DOI] [PubMed] [Google Scholar]
- 13.Bottai G, Pasculli B, Calin GA, Santarpia L. Targeting the microRNA-regulating DNA damage/repair pathways in cancer. Expert Opin Biol Ther. 2014;14:1667–83. doi: 10.1517/14712598.2014.950650. [DOI] [PubMed] [Google Scholar]
- 14.De Mattos-Arruda L, Bottai G, Nuciforo PG, Di Tommaso L, Giovannetti E, Peg V, et al. MicroRNA-21 links epithelial-to-mesenchymal transition and inflammatory signals to confer resistance to neoadjuvant trastuzumab and chemotherapy in HER2-positive breast cancer patients. Oncotarget. 2015;6:37269–80. doi: 10.18632/oncotarget.5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sotillo E, Thomas-Tikhonenko A. Shielding the messenger (RNA): microRNA-based anticancer therapies. Pharmacol Ther. 2011;131:18–32. doi: 10.1016/j.pharmthera.2011.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, Munk ME, et al. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bader G. miR-34 — a microRNA replacement therapy is headed to the clinic. Front Genet. 2012;3:120. doi: 10.3389/fgene.2012.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu G, Abraham E. MicroRNAs in immune response and macrophage polarization. Arterioscler Thromb Vasc Biol. 2013;33:170–7. doi: 10.1161/ATVBAHA.112.300068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tili E, Michaille JJ, Wernicke D, Alder H, Costinean S, Volinia S, et al. Mutator activity induced by microRNA-155 (miR-155) links inflammation and cancer. Proc Natl Acad Sci U S A. 2011;108:4908–13. doi: 10.1073/pnas.1101795108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmitt MJ, Margue C, Behrmann I, Kreis S. MiRNA-29: a microRNA family with tumor-suppressing and immune-modulating properties. Curr Mol Med. 2011;13:572–85. doi: 10.2174/1566524011313040009. [DOI] [PubMed] [Google Scholar]
- 21.Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–69. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 22.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 23.Crusz SM, Balkwill FR. Inflammation and cancer: advances and new agents. Nat Rev Clin Oncol. 2015;12:584–96. doi: 10.1038/nrclinonc.2015.105. [DOI] [PubMed] [Google Scholar]
- 24.Del Prete A, Allavena P, Santoro G, Fumarulo R, Corsi MM, Mantovani A. Molecular pathways in cancer-related inflammation. Biochem Med. 2011;21:264–75. doi: 10.11613/BM.2011.036. [DOI] [PubMed] [Google Scholar]
- 25.de Visser KE, Eichten A, Coussens LM. Paradoxical roles of the immune system during cancer development. Nat Rev Cancer. 2006;6:24–37. doi: 10.1038/nrc1782. [DOI] [PubMed] [Google Scholar]
- 26.Dunn GP, Bruce AT, Ikeda H, Old LJ, Schreiber RD. Cancer immunoediting: from immunosurveillance to tumor escape. Nat Immunol. 2002;3:991–8. doi: 10.1038/ni1102-991. [DOI] [PubMed] [Google Scholar]
- 27.Vesely MD, Kershaw MH, Schreiber RD, Smyth MJ. Natural innate and adaptive immunity to cancer. Annu Rev Immunol. 2011;29:235–71. doi: 10.1146/annurev-immunol-031210-101324. [DOI] [PubMed] [Google Scholar]
- 28.Aguirre-Ghiso JA. Models, mechanisms and clinical evidence for cancer dormancy. Nat Rev Cancer. 2007;7:834–46. doi: 10.1038/nrc2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zitvogel L, Tesniere A, Kroemer G. Cancer despite immunosurveillance: immunoselection and immunosubversion. Nat Rev Immunol. 2006;6:715–27. doi: 10.1038/nri1936. [DOI] [PubMed] [Google Scholar]
- 30.Dunn GP, Old LJ, Schreiber RD. The immunobiology of cancer immunosurveillance and immunoediting. Immunity. 2004;21:137–48. doi: 10.1016/j.immuni.2004.07.017. [DOI] [PubMed] [Google Scholar]
- 31.Dunn GP, Old LJ, Schreiber RD. The three Es of cancer immunoediting. Annu Rev Immunol. 2004;22:329–60. doi: 10.1146/annurev.immunol.22.012703.104803. [DOI] [PubMed] [Google Scholar]
- 32.Smyth MJ, Dunn GP, Schreiber RD. Cancer immunosurveillance and immunoediting: the roles of immunity in suppressing tumor development and shaping tumor immunogenicity. Adv Immunol. 2006;90:1–50. doi: 10.1016/S0065-2776(06)90001-7. [DOI] [PubMed] [Google Scholar]
- 33.Khong HT, Restifo NP. Natural selection of tumor variants in the generation of “tumor escape” phenotypes. Nat Immunol. 2002;3:999–1005. doi: 10.1038/ni1102-999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Willimsky G, Blankenstein T. Sporadic immunogenic tumours avoid destruction by inducing T-cell tolerance. Nature. 2005;437:141–6. doi: 10.1038/nature03954. [DOI] [PubMed] [Google Scholar]
- 35.Schell TD, Knowles BB, Tevethia SS. Sequential loss of cytotoxic T lymphocyte responses to simian virus 40 large T antigen epitopes in T antigen transgenic mice developing osteosarcomas. Cancer Res. 2000;60:3002–12. [PubMed] [Google Scholar]
- 36.Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med. 2013;19:1423–37. doi: 10.1038/nm.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gooden MJ, de Bock GH, Leffers N, Daemen T, Nijman HW. The prognostic influence of tumour-infiltrating lymphocytes in cancer: a systematic review with meta-analysis. Br J Cancer. 2011;105:93–103. doi: 10.1038/bjc.2011.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clemente CG, Mihm MC, Jr, Bufalino R, Zurrida S, Collini P, Cascinelli N. Prognostic value of tumor infiltrating lymphocytes in the vertical growth phase of primary cutaneous melanoma. Cancer. 1996;77:1303–10. doi: 10.1002/(SICI)1097-0142(19960401)77:7<1303::AID-CNCR12>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 39.Schumacher K, Haensch W, Roefzaad C, Schlag PM. Prognostic significance of activated CD8(+) T cell infiltrations within esophageal carcinomas. Cancer Res. 2001;61:3932–6. [PubMed] [Google Scholar]
- 40.Zhang L, Conejo-Garcia JR, Katsaros D, Gimotty PA, Massobrio M, Regnani G, et al. Intratumoral T cells, recurrence, and survival in epithelial ovarian cancer. N Engl J Med. 2003;348:203–13. doi: 10.1056/NEJMoa020177. [DOI] [PubMed] [Google Scholar]
- 41.Sato E, Olson SH, Ahn J, Bundy B, Nishikawa H, Qian F, et al. Intraepithelial CD8+ tumor-infiltrating lymphocytes and a high CD8+/regulatory T cell ratio are associated with favorable prognosis in ovarian cancer. Proc Natl Acad Sci U S A. 2005;102:18538–43. doi: 10.1073/pnas.0509182102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Galon J, Costes A, Sanchez-Cabo F, Kirilovsky A, Mlecnik B, Lagorce-Pages C, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313:1960–4. doi: 10.1126/science.1129139. [DOI] [PubMed] [Google Scholar]
- 43.Liu S, Foulkes WD, Leung S, Gao D, Lau S, Kos Z, et al. Prognostic significance of FOXP3+ tumor-infiltrating lymphocytes in breast cancer depends on estrogen receptor and human epidermal growth factor receptor-2 expression status and concurrent cytotoxic T-cell infiltration. Breast Cancer Res. 2014;16:432. doi: 10.1186/s13058-014-0432-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shalapour S, Karin M. Immunity, inflammation, and cancer: an eternal fight between good and evil. J Clin Invest. 2015;125:3347–55. doi: 10.1172/JCI80007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dushyanthen S, Beavis PA, Savas P, Teo ZL, Zhou C, Mansour M, et al. Relevance of tumor-infiltrating lymphocytes in breast cancer. BMC Med. 2015;13:202. doi: 10.1186/s12916-015-0431-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Galdiero MR, Bonavita E, Barajon I, Garlanda C, Mantovani A, Jaillon S. Tumor associated macrophages and neutrophils in cancer. Immunobiology. 2013;218:1402–10. doi: 10.1016/j.imbio.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 47.Murray PJ, Wynn TA. Protective and pathogenic functions of macrophage subsets. Nat Rev Immunol. 2011;11:723–37. doi: 10.1038/nri3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ruffell B, Coussens LM. Macrophages and therapeutic resistance in cancer. Cancer Cell. 2015;27:462–72. doi: 10.1016/j.ccell.2015.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lewis CE, Pollard JW. Distinct role of macrophages in different tumor microenvironments. Cancer Res. 2006;66:605–12. doi: 10.1158/0008-5472.CAN-05-4005. [DOI] [PubMed] [Google Scholar]
- 50.Biswas SK, Mantovani A. Macrophage plasticity and interaction with lymphocyte subsets: cancer as a paradigm. Nat Immunol. 2010;11:889–96. doi: 10.1038/ni.1937. [DOI] [PubMed] [Google Scholar]
- 51.Komohara Y, Jinushi M, Takeya M. Clinical significance of macrophage heterogeneity in human malignant tumors. Cancer Sci. 2014;105:1–8. doi: 10.1111/cas.12314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Asgharzadeh S, Salo JA, Ji L, Oberthuer A, Fischer M, Berthold F, et al. Clinical significance of tumor-associated inflammatory cells in metastatic neuroblastoma. J Clin Oncol. 2012;30:3525–32. doi: 10.1200/JCO.2011.40.9169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gabrilovich DI, Nagaraj S. Myeloid-derived suppressor cells as regulators of the immune system. Nat Rev Immunol. 2009;9:162–74. doi: 10.1038/nri2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fuertes MB, Kacha AK, Kline J, Woo SR, Kranz DM, Murphy KM, et al. Host type I IFN signals are required for antitumor CD8+ T cell responses through CD8{alpha} + dendritic cells. J Exp Med. 2011;208:2005–16. doi: 10.1084/jem.20101159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Broz ML, Binnewies M, Boldajipour B, Nelson AE, Pollack JL, Erle DJ, et al. Dissecting the tumor myeloid compartment reveals rare activating antigen-presenting cells critical for T cell immunity. Cancer Cell. 2014;26:638–52. doi: 10.1016/j.ccell.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Joice JA, Pollard JW. Microenvironmental regulation of metastasis. Nat Rev Cancer. 2009;9:239–52. doi: 10.1038/nrc2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Olson P, Lu J, Zhang H, Shai A, Chun MG, Wang Y, et al. MicroRNA dynamics in the stages of tumorigenesis correlate with hallmark capabilities of cancer. Genes Dev. 2009;23:2152–65. doi: 10.1101/gad.1820109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–61. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Farazi TA, Hoell JI, Morozov P, Tuschl T. MicroRNAs in human cancer. Adv Exp Med Biol. 2013;774:1–20. doi: 10.1007/978-94-007-5590-1_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mattiske S, Suetani RJ, Neilsen PM, Callen DF. The oncogenic role of miR-155 in breast cancer. Cancer Epidemiol Biomarkers Prev. 2012;21:1236–43. doi: 10.1158/1055-9965.EPI-12-0173. [DOI] [PubMed] [Google Scholar]
- 61.Li H, Bian C, Liao L, Li J, Zhao RC. miR-17-5p promotes human breast cancer cell migration and invasion through suppression of HBP1. Breast Cancer Res Treat. 2011;126:565–75. doi: 10.1007/s10549-010-0954-4. [DOI] [PubMed] [Google Scholar]
- 62.Han M, Liu M, Wang Y, Chen X, Hu J, Sun Y, et al. Antagonism of miR-21 reverses epithelial-mesenchymal transition and cancer stem cell phenotype through AKT/ERK1/2 inactivation by targeting PTEN. PLoS One. 2012;7:e39520. doi: 10.1371/journal.pone.0039520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Di Leva G, Garofalo M, Croce CM. MicroRNAs in cancer. Annu Rev Pathol. 2014;9:287–314. doi: 10.1146/annurev-pathol-012513-104715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lujambio A, Lowe SW. The microcosmos of cancer. Nature. 2012;482:347–55. doi: 10.1038/nature10888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gasparini P, Cascione L, Fassan M, Lovat F, Guler G, Balci S, et al. microRNA expression profiling identifies a four microRNA signature as a novel diagnostic and prognostic biomarker in triple negative breast cancers. Oncotarget. 2014;5:1174–84. doi: 10.18632/oncotarget.1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kleivi Sahlberg K, Bottai G, Naume B, Burwinkel B, Calin GA, Børresen-Dale AL, et al. A serum microRNA signature predicts tumor relapse and survival in triple-negative breast cancer patients. Clin Cancer Res. 2015;21:1207–14. doi: 10.1158/1078-0432.CCR-14-2011. [DOI] [PubMed] [Google Scholar]
- 67.Lyng MB, Lænkholm AV, Søkilde R, Gravgaard KH, Litman T, Ditzel HJ. Global microRNA expression profiling of high-risk ER+ breast cancers from patients receiving adjuvant tamoxifen mono-therapy: a DBCG study. PLoS One. 2012;7:e36170. doi: 10.1371/journal.pone.0036170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li R, Gao K, Luo H, Wang X, Shi Y, Dong Q, et al. Identification of intrinsic subtype-specific prognostic microRNAs in primary glioblastoma. J Exp Clin Cancer Res. 2014;33:9. doi: 10.1186/1756-9966-33-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhou X, Wang X, Huang Z, Xu L, Zhu W, Liu P. An ER-associated miRNA signature predicts prognosis in ER-positive breast cancer. J Exp Clin Cancer Res. 2014;33:94. doi: 10.1186/s13046-014-0094-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gurtner A, Falcone E, Garibaldi F, Piaggio G. Dysregulation of microRNA biogenesis in cancer: the impact of mutant p53 on Drosha complex activity. J Exp Clin Cancer Res. 2016;35:45. doi: 10.1186/s13046-016-0319-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee HM, Nguyen DT, Lu LF. Progress and challenge of microRNA research in immunity. Front Genet. 2014;5:178. doi: 10.3389/fgene.2014.00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Merkerova M, Belickova M, Bruchova H. Differential expression of microRNAs in hematopoietic cell lineages. Eur J Haematol. 2008;81:304–10. doi: 10.1111/j.1600-0609.2008.01111.x. [DOI] [PubMed] [Google Scholar]
- 73.Kluiver J, Kroesen BJ, Poppema S, van den Berg A. The role of microRNAs in normal hematopoiesis and hematopoietic malignancies. Leukemia. 2006;20:1931–6. doi: 10.1038/sj.leu.2404387. [DOI] [PubMed] [Google Scholar]
- 74.Davidson-Moncada J, Papavasiliou FN, Tam W. MicroRNAs of the immune system: roles in inflammation and cancer. Ann N Y Acad Sci. 2010;1183:183–94. doi: 10.1111/j.1749-6632.2009.05121.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.O’Connell RM, Rao DS, Chaudhuri AA, Baltimore D. Physiological and pathological roles for microRNAs in the immune system. Nat Rev Immunol. 2010;10:111–22. doi: 10.1038/nri2708. [DOI] [PubMed] [Google Scholar]
- 76.O’Connell RM, Zhao JL, Rao DS. MicroRNA function in myeloid biology. Blood. 2011;118:2960–9. doi: 10.1182/blood-2011-03-291971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tili E, Michaille JJ, Croce CM. MicroRNAs play a central role in molecular dysfunctions linking inflammation with cancer. Immunol Rev. 2013;253:167–84. doi: 10.1111/imr.12050. [DOI] [PubMed] [Google Scholar]
- 78.Curtale G, Citarella F. Dynamic nature of noncoding RNA regulation of adaptive immune response. Int J Mol Sci. 2013;14:17347–77. doi: 10.3390/ijms140917347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Squadrito ML, Etzrodt M, De Palma M, Pittet MJ. MicroRNA-mediated control of macrophages and its implications for cancer. Trends Immunol. 2013;34:350–9. doi: 10.1016/j.it.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fontana L, Pelosi E, Greco P, Racanicchi S, Testa U, Liuzzi F, et al. MicroRNAs 17-5p-20a-106a control monocytopoiesis through AML1 targeting and M-CSF receptor upregulation. Nat Cell Biol. 2007;9:775–87. doi: 10.1038/ncb1613. [DOI] [PubMed] [Google Scholar]
- 81.Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–80. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 82.O’Connell RM, Chaudhuri AA, Rao DS, Baltimore D. Inositol phosphatase SHIP1 is a primary target of miR-155. Proc Natl Acad Sci U S A. 2009;106:7113–8. doi: 10.1073/pnas.0902636106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NFkappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A. 2006;103:12481–6. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sheedy FJ, Palsson-McDermott E, Hennessy EJ, Martin C, O'Leary JJ, Ruan Q, et al. Negative regulation of TLR4 via targeting of the proinflammatory tumor suppressor PDCD4 by the MicroRNA miR-21. Nat Immunol. 2010;11:141–7. doi: 10.1038/ni.1828. [DOI] [PubMed] [Google Scholar]
- 85.Androulidaki A, Iliopoulos D, Arranz A, Doxaki C, Schworer S, Zacharioudaki V, et al. The kinase Akt1 controls macrophage response to lipopolysaccharide by regulating microRNAs. Immunity. 2009;31:220–31. doi: 10.1016/j.immuni.2009.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.O’Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc Natl Acad Sci U S A. 2007;104:1604–9. doi: 10.1073/pnas.0610731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Beaulieu AM, Bezman NA, Lee JE, Matloubian M, Sun JC, Lanier LL. MicroRNA function in NK-cell biology. Immunol Rev. 2013;253:40–52. doi: 10.1111/imr.12045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR, et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–11. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Du C, Liu C, Kang J, Zhao G, Ye Z, Huang S, et al. MicroRNA miR-326 regulates TH-17 differentiation and is associated with the pathogenesis of multiple sclerosis. Nat Immunol. 2009;10:1252–9. doi: 10.1038/ni.1798. [DOI] [PubMed] [Google Scholar]
- 90.Baumjohann D, Kageyama R, Clingan JM, Morar MM, Patel S, de Kouchkovsky D, et al. The microRNA cluster miR-17 approximately 92 promotes TFH cell differentiation and represses subset-inappropriate gene expression. Nat Immunol. 2013;14:840–8. doi: 10.1038/ni.2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Liang Y, Pan HF, Ye DQ. microRNAs function in CD8 + T cell biology. J Leukoc Biol. 2015;97:487–97. doi: 10.1189/jlb.1RU0814-369R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lu LF, Boldin MP, Chaudhry A, Lin LL, Taganov KD, Hanada T, et al. Function of miR-146a in controlling Treg cell-mediated regulation of Th1 responses. Cell. 2010;142:914–29. doi: 10.1016/j.cell.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lu LF, Thai TH, Calado DP, Chaudhry A, Kubo M, Tanaka K, et al. Foxp3-dependent microRNA155 confers competitive fitness to regulatory T cells by targeting SOCS1 protein. Immunity. 2009;30:80–91. doi: 10.1016/j.immuni.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fridman WH, Pagès F, Sautès-Fridman C, Galon J. The immune contexture in humantumours: impact on clinical outcome. Nat Rev Cancer. 2012;12:298–306. doi: 10.1038/nrc3245. [DOI] [PubMed] [Google Scholar]
- 95.Yang J, Zhang Z, Chen C, Liu Y, Si Q, Chuang TH, et al. MicroRNA-19a-3p inhibits breast cancer progression and metastasis by inducing macrophage polarization through downregulated expression of Fra-1 proto-oncogene. Oncogene. 2014;33:3014–23. doi: 10.1038/onc.2013.258. [DOI] [PubMed] [Google Scholar]
- 96.Wang Q, Ni H, Lan L, Wei X, Xiang R, Wang Y. Fra-1 protooncogene regulates IL-6 expression in macrophages and promotes the generation of M2d macrophages. Cell Res. 2010;20:701–12. doi: 10.1038/cr.2010.52. [DOI] [PubMed] [Google Scholar]
- 97.Ma S, Liu M, Xu Z, Li Y, Guo H, Ge Y, et al. A double feedback loop mediated by microRNA-23a/27a/24-2 regulates M1 versus M2 macrophage polarization and thus regulates cancer progression. Oncotarget. 2015;7:13502–19. doi: 10.18632/oncotarget.6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zonari E, Pucci F, Saini M, Mazzieri R, Politi LS, Gentner B, et al. A role for miR-155 in enabling tumor-infiltrating innate immune cells to mount effective antitumor responses in mice. Blood. 2013;122:243–522. doi: 10.1182/blood-2012-08-449306. [DOI] [PubMed] [Google Scholar]
- 99.Mondal S, Subramanian KK, Sakai J, Bajrami B, Luo HR. Phosphoinositide lipid phosphatase SHIP1 and PTEN coordinate to regulate cell migration and adhesion. Mol Biol Cell. 2012;23:1219–30. doi: 10.1091/mbc.E11-10-0889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhang Y, Yang P, Sun T, Li D, Xu X, Rui Y, et al. miR-126 and miR-126* repress recruitment of mesenchymal stem cells and inflammatory monocytes to inhibit breast cancer metastasis. Nat Cell Biol. 2013;15:284–94. doi: 10.1038/ncb2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Karnoub AE, Dash AB, Vo AP, Sullivan A, Brooks MW, Bell GW, et al. Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature. 2007;449:557–63. doi: 10.1038/nature06188. [DOI] [PubMed] [Google Scholar]
- 102.Qian BZ, Li J, Zhang H, Kitamura T, Zhang J, Campion LR, et al. CCL2 recruits inflammatory monocytes to facilitate breast-tumour metastasis. Nature. 2011;475:222–5. doi: 10.1038/nature10138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tavazoie SF, Alarcón C, Oskarsson T, Padua D, Wang Q, Bos PD, et al. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147–3. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Xie J, Liu M, Li Y, Nie Y, Mi Q, Zhao S. Ovarian tumor-associated microRNA-20a decreases natural killer cell cytotoxicity by downregulating MICA/B expression. Cell Mol Biol. 2014;11:495–502. doi: 10.1038/cmi.2014.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Oppenheim DE, Roberts SJ, Clarke SL, Filler R, Lewis JM, Tigelaar RE, et al. Sustained localized expression of ligand for the activating NKG2D receptor impairs natural cytotoxicity in vivo and reduces tumor immunosurveillance. Nat Immunol. 2005;6:928–37. doi: 10.1038/ni1239. [DOI] [PubMed] [Google Scholar]
- 106.Wyman SK, Parkin RK, Mitchell PS, Fritz BR, O’Briant K, Godwin AK, et al. Repertoire of microRNAs in epithelial ovarian cancer as determined by next generation sequencing of small RNA cDNA libraries. PLoS ONE. 2009;4:e5311. doi: 10.1371/journal.pone.0005311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tsukerman P, Stern-Ginossar N, Gur C, Glasner A, Nachmani D, Bauman Y, et al. MiR-10b downregulates the stress-induced cell surface molecule MICB, a critical ligand for cancer cell recognition by natural killer cells. Cancer Res. 2012;72:5463–72. doi: 10.1158/0008-5472.CAN-11-2671. [DOI] [PubMed] [Google Scholar]
- 108.Yang H, Lan P, Hou Z, Guan Y, Zhang J, Xu W, et al. Histone deacetylase inhibitor SAHA epigenetically regulates miR-17-92 cluster and MCM7 to upregulate MICA expression in hepatoma. Br J Cancer. 2015;112:112–21. doi: 10.1038/bjc.2014.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Vetter CS, Groh V, Thor Straten P, Spies T, Brocker EB, Becker JC. Expression of stress-induced MHC class I related chain molecules on human melanoma. J Invest Dermatol. 2002;118:600–5. doi: 10.1046/j.1523-1747.2002.01700.x. [DOI] [PubMed] [Google Scholar]
- 110.Paschen A, Sucker A, Hill B, Moll I, Zapatka M, Nguyen XD, et al. Differential clinical significance of individual NKG2D ligands in melanoma: soluble ULBP2 as an indicator of poor prognosis superior to S100B. Clin Cancer Res. 2009;15:5208–15. doi: 10.1158/1078-0432.CCR-09-0886. [DOI] [PubMed] [Google Scholar]
- 111.Heinemann A, Zhao F, Pechlivanis S, Eberle J, Steinle A, Diederichs S, et al. Cell Receptor NKG2D Expression of ULBP2, a Stress-Induced Ligand of the Natural Killer Tumor Suppressive MicroRNAs miR-34a/c. Cancer Res. 2012;72:460–71. doi: 10.1158/0008-5472.CAN-11-1977. [DOI] [PubMed] [Google Scholar]
- 112.Lodygin D, Tarasov V, Epanchintsev A, Berking C, Knyazeva T, Körner H, et al. Inactivation of miR-34a by aberrant CpG methylation in multiple types of cancer. Cell Cycle. 2008;7:2591–600. doi: 10.4161/cc.7.16.6533. [DOI] [PubMed] [Google Scholar]
- 113.Yang P, Li QJ, Feng Y, Zhang Y, Markowitz GJ, Ning S, et al. TGF-b-miR-34a-CCL22 signaling-induced Treg cell recruitment promotes venous metastases of HBV-positive hepatocellular carcinoma. Cancer Cell. 2012;22:291–303. doi: 10.1016/j.ccr.2012.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Xu H, Cheung IY, Guo HF, Cheung NK. MicroRNA miR-29 modulates expression of immunoinhibitory molecule B7-H3: potential implications for immune based therapy of human solid tumors. Cancer Res. 2009;69:6275–81. doi: 10.1158/0008-5472.CAN-08-4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wang Y, Zhang X, Li H, Yu J, Ren X. The role of miRNA-29 family in cancer. Eur J Cell Biol. 2013;92:123–8. doi: 10.1016/j.ejcb.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 116.Filipazzi P, Huber V, Rivoltini L. Phenotype, function and clinical implications of myeloid-derived suppressor cells in cancer patients. Cancer Immunol Immunother. 2012;61:255–63. doi: 10.1007/s00262-011-1161-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Liu Y, Lai L, Chen Q, Song Y, Xu S, Ma F, et al. MicroRNA-494 is required for the accumulation and functions of tumor-expanded myeloid-derived suppressor cells via targeting of PTEN. J Immunol. 2012;188:5500–10. doi: 10.4049/jimmunol.1103505. [DOI] [PubMed] [Google Scholar]
- 118.Wang J, Yu F, Jia X, Iwanowycz S, Wang Y, Huang S, et al. MicroRNA-155 deficiency enhances the recruitment and functions of myeloid-derived suppressor cells in tumor microenvironment and promotes solid tumor growth. Int J Cancer. 2015;136:E602–13. doi: 10.1002/ijc.29151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zamarron BF, Chen W. Dual roles of immune cells and their factors in cancer development and progression. Int J Biol Sci. 2011;7:651–8. doi: 10.7150/ijbs.7.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Whiteside TL. The tumor microenvironment and its role in promoting tumor growth. Oncogene. 2008;27:5904–12. doi: 10.1038/onc.2008.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Rius J, Guma M, Schachtrup C, Akassoglou K, Zinkernagel AS, Nizet V, et al. NF-kappaB links innate immunity to the hypoxic response through transcriptional regulation of HIF-1alpha. Nature. 2008;453:807–11. doi: 10.1038/nature06905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Balkwill F. Tumour necrosis factor and cancer. Nat Rev Cancer. 2008;9:361–71. doi: 10.1038/nrc2628. [DOI] [PubMed] [Google Scholar]
- 123.Massagué J. TGFbeta in Cancer. Cell. 2008;134:215–30. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Karin M. Nuclear factor-kappaB in cancer development and progression. Nature. 2006;441:431–6. doi: 10.1038/nature04870. [DOI] [PubMed] [Google Scholar]
- 125.Pikarsky E, Porat RM, Stein I, Abramovitch R, Amit S, Kasem S, et al. NF-kB functions as a tumour promoter in inflammation-associated cancer. Nature. 2004;431:461–4. doi: 10.1038/nature02924. [DOI] [PubMed] [Google Scholar]
- 126.Bezbradica JS, Medzhitov R. Integration of cytokine and heterologous receptor signaling pathways. Nat Immunol. 2009;10:333–9. doi: 10.1038/ni.1713. [DOI] [PubMed] [Google Scholar]
- 127.Mocellin S, Rossi CR, Pilati P, Nitti D. Tumor necrosis factor, cancer and anticancer therapy. Cytokine Growth Factor Rev. 2005;16:35–53. doi: 10.1016/j.cytogfr.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 128.Kortylewski M, Kujawski M, Wang T, Wei S, Zhang S, Pilon-Thomas S, et al. Inhibiting Stat3 signaling in the hematopoietic system elicits multicomponent antitumor immunity. Nat Med. 2005;11:1314–21. doi: 10.1038/nm1325. [DOI] [PubMed] [Google Scholar]
- 129.Xie TX, Huang FJ, Aldape KD, Kang SH, Liu M, Gershenwald JE, et al. Activation of stat3 in human melanoma promotes brain metastasis. Cancer Res. 2006;66:3188–96. doi: 10.1158/0008-5472.CAN-05-2674. [DOI] [PubMed] [Google Scholar]
- 130.Yu H, Yu H, Kortylewski M, Pardoll D. Crosstalk between cancer and immune cells: role of STAT3 in the tumour microenvironment. Nat Rev Immunol. 2007;7:41–51. doi: 10.1038/nri1995. [DOI] [PubMed] [Google Scholar]
- 131.Burdelya L, Shain K, Zhang S, Bhattacharya R, Gabrilovich D, Heller R, et al. Regulation of the innate and adaptive immune responses by Stat-3 signaling in tumor cells. Nat Med. 2004;10:48–54. doi: 10.1038/nm976. [DOI] [PubMed] [Google Scholar]
- 132.Massagué J, Wotton D. Transcriptional control by the TGF-beta/Smad signaling system. EMBO J. 2000;19:1745–54. doi: 10.1093/emboj/19.8.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Derynck R, Jarrett JA, Chen EY, Eaton DH, Bell JR, Assoian RK, et al. Human transforming growth factor-β complementary DNA sequence and expression in normal and transformed cells. Nature. 1985;316:701–5. doi: 10.1038/316701a0. [DOI] [PubMed] [Google Scholar]
- 134.Bennicelli JL, Guerry D. Production of multiple cytokines by cultured human melanomas. Exp Dermatol. 1993;2:186–90. doi: 10.1111/j.1600-0625.1993.tb00031.x. [DOI] [PubMed] [Google Scholar]
- 135.De Jaeger K, Seppenwoolde Y, Kampinga HH, Boersma LJ, Belderbos JS, Lebesque JV. Significance of plasma transforming growth factor-beta levels in radiotherapy for non-small-cell lung cancer. Int J Radiat Oncol Biol Phys. 2004;58:1378–87. doi: 10.1016/j.ijrobp.2003.09.078. [DOI] [PubMed] [Google Scholar]
- 136.Polak ME, Borthwick NJ, Gabriel FG, Johnson P, Higgins B, Hurren J, et al. Mechanisms of local immunosuppression in cutaneous melanoma. Br J Cancer. 2007;96:1879–87. doi: 10.1038/sj.bjc.6603763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wang Z, Han J, Cui Y, Zhou X, Fan K. miRNA-21 inhibition enhances RANTES and IP-10 release in MCF-7 via PIAS3 and STAT3 signalling and causes increased lymphocyte migration. Biochem Biophys Res Commun. 2013;439:384–9. doi: 10.1016/j.bbrc.2013.08.072. [DOI] [PubMed] [Google Scholar]
- 138.Xiong Q, Zhong Q, Zhang J, Yang M, Li C, Zheng P, et al. Identification of novel miR-21 target proteins in multiple myeloma cells by quantitative proteomics. J Proteome Res. 2012;11:2078–90. doi: 10.1021/pr201079y. [DOI] [PubMed] [Google Scholar]
- 139.Yang CH, Yue J, Fan M, Pfeffer LM. IFN induces miR-21 through a signal transducer and activator transcription 3-dependent pathway as a suppressive negative feedback on IFN-induced apoptosis. Cancer Res. 2010;70:8108–16. doi: 10.1158/0008-5472.CAN-10-2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kumarswamy R, Volkmann I, Thum T. Regulation and function of miRNA-21 in health and disease. RNA Biol. 2011;8:706–13. doi: 10.4161/rna.8.5.16154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Jiang S, Zhang HW, Lu MH, He XH, Li Y, Gu H, et al. MicroRNA-155 functions as an OncomiR in breast cancer by targeting the suppressor of cytokine signaling 1 gene. Cancer Res. 2010;70:3119–27. doi: 10.1158/0008-5472.CAN-09-4250. [DOI] [PubMed] [Google Scholar]
- 142.Li H, Gupta S, Du WW, Yang BB. MicroRNA-17 inhibits tumor growth by stimulating T cell mediated host immune response. Oncoscience. 2014;1:531–9. doi: 10.18632/oncoscience.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Wei J, Wang F, Kong LY, Xu S, Doucette T, Ferguson SD, et al. miR-124 inhibits STAT3 signaling to enhance T cell-mediated immune clearance of glioma. Cancer Res. 2013;73:3913–26. doi: 10.1158/0008-5472.CAN-12-4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Zhang M, Liu Q, Mi S, Liang X, Zhang Z, Su X, et al. Both miR-17-5p and miR-20a alleviate suppressive potential of myeloid-derived suppressor cells by modulating STAT3 expression. J Immunol. 2011;186:4716–24. doi: 10.4049/jimmunol.1002989. [DOI] [PubMed] [Google Scholar]
- 145.Lin R, Chen L, Chen G, Hu C, Jiang S, Sevilla J, et al. Targeting miR-23a in CD8+ cytotoxic T lymphocytes prevents tumor-dependent immunosuppression. J Clin Invest. 2014;124:5352–67. doi: 10.1172/JCI76561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Li N, Xu X, Xiao B, Zhu ED, Li BS, Liu Z, et al. H. pylori related proinflammatory cytokines contribute to the induction of miR-146a in human gastric epithelial cells. Mol Biol Rep. 2011;39:4655–661. doi: 10.1007/s11033-011-1257-5. [DOI] [PubMed] [Google Scholar]
- 147.Bhaumik D, Scott GK, Schokrpur S, Patil CK, Campisi J, Benz CC. Expression of micro-RNA 146 suppresses NF-kB activity with reduction of metastatic potential in breast cancer cells. Oncogene. 2008;27:5643–7. doi: 10.1038/onc.2008.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Chen R, Alvero AB, Silasi DA, Kelly MG, Fest S, Visintin I, et al. Regulation of IKKbeta by miR-199a affects NF-kB activity in ovarian cancer cells. Oncogene. 2008;27:4712–23. doi: 10.1038/onc.2008.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Greten FR, Karin M. The IKK/NF-kappaB activation pathway-a target for prevention and treatment of cancer. Cancer Lett. 2004;206:193–9. doi: 10.1016/j.canlet.2003.08.029. [DOI] [PubMed] [Google Scholar]
- 150.Fukata M, Michelsen KS, Eri R, Thomas LS, Hu B, Lukasek K, et al. Toll-like receptor-4 is required for intestinal response to epithelial injury and limiting bacterial translocation in a murine model of acute colitis. Am J Physiol Gastrointest Liver Physiol. 2005;288:G1055–65. doi: 10.1152/ajpgi.00328.2004. [DOI] [PubMed] [Google Scholar]
- 151.Xu S, Tao Z, Hai B, Liang H, Shi Y, Wang T, et al. miR-424(322) reverses chemoresistance via T-cell immune response activation by blocking the PD-L1 immune checkpoint. Nat Commun. 2016;7:11406. doi: 10.1038/ncomms11406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Soon P, Kiaris H. MicroRNAs in the tumour microenvironment: big role for small players. Endocr Relat Cancer. 2013;20:R257–67. doi: 10.1530/ERC-13-0119. [DOI] [PubMed] [Google Scholar]
- 153.Mittelbrunn M, Sánchez-Madrid F. Intercellular communication: diverse structures for exchange of genetic information. Nat Rev Mol Cell Biol. 2012;13:328–35. doi: 10.1038/nrm3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Gutiérrez-Vázquez C, Villarroya-Beltri C, Mittelbrunn M, Sánchez-Madrid F. Transfer of extracellular vesicles during immune cell-cell interactions. Immunol Rev. 2013;251:125–42. doi: 10.1111/imr.12013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Skog J, Würdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470–6. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Mittelbrunn M, Gutiérrez-Vázquez C, Villarroya-Beltri C, González S, Sánchez-Cabo F, González MÁ, et al. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat Commun. 2011;2:282. doi: 10.1038/ncomms1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Montecalvo A, Larregina AT, Shufesky WJ, Stolz DB, Sullivan ML, Karlsson JM, et al. Mechanism of transfer of functional microRNAs between mouse dendritic cells via exosomes. Blood. 2012;119:756–66. doi: 10.1182/blood-2011-02-338004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Gibbings DJ, Ciaudo C, Erhardt M, Voinnet O. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat Cell Biol. 2009;11:1143–9. doi: 10.1038/ncb1929. [DOI] [PubMed] [Google Scholar]
- 159.Lee YS, Pressman S, Andress AP, Kim K, White JL, Cassidy JJ, et al. Silencing by small RNAs is linked to endosomal trafficking. Nat Cell Biol. 2009;11:1150–6. doi: 10.1038/ncb1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Bobrie A, Colombo M, Raposo G, Théry C. Exosome secretion: molecular mechanisms and roles in immune responses. Traffic. 2011;12:1659–68. doi: 10.1111/j.1600-0854.2011.01225.x. [DOI] [PubMed] [Google Scholar]
- 161.Bhatnagar S, Schorey JS. Exosomes released from infected macrophages contain Mycobacterium avium glycopeptidolipids and are proinflammatory. J Biol Chem. 2007;282:25779–89. doi: 10.1074/jbc.M702277200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Redis RS, Calin S, Yang Y, You MJ, Calin GA. Cell-to-cell miRNA transfer: From body homeostasis to therapy Roxana. Pharmacol Ther. 2012;136:169–74. doi: 10.1016/j.pharmthera.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Yang M, Chen J, Su F, Yu B, Su F, Lin L, et al. Microvesicles secreted by macrophages shuttle invasion-potentiating microRNAs into breast cancer cells. Mol Cancer. 2011;10:117. doi: 10.1186/1476-4598-10-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Patel SA, Gooderham NJ. IL6 mediates immune and colorectal cancer cell crosstalk via miR-21 and miR-29b. Mol Cancer Res. 2015;3:1502–8. doi: 10.1158/1541-7786.MCR-15-0147. [DOI] [PubMed] [Google Scholar]
- 165.Fabbri M, Paone A, Calore F, Galli R, Gaudio E, Santhanam R, et al. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci U S A. 2012;109:E2110–16. doi: 10.1073/pnas.1209414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Yin Y, Cai X, Chen X, Liang H, Zhang Y, Li J, et al. Tumor-secreted miR-214 induces regulatory T cells: a major link between immune evasion and tumor growth. Cell Res. 2014;24:1164–80. doi: 10.1038/cr.2014.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol. 2008;110:13–21. doi: 10.1016/j.ygyno.2008.04.033. [DOI] [PubMed] [Google Scholar]
- 168.Toth B, Nieuwland R, Liebhardt S, Ditsch N, Steinig K, Stieber P, et al. Circulating microparticles in breast cancer patients: a comparative analysis with established biomarkers. Anticancer Res. 2008;28:1107–12. [PubMed] [Google Scholar]
- 169.Taylor DD, Gercel-Taylor C, Lyons KS, Stanson J, Whiteside TL. T-cell apoptosis and suppression of T-Cell receptor/CD3-zeta by Fas ligand-containing membrane vesicles shed from ovarian tumors. Clin Cancer Res. 2003;9:5113–9. [PubMed] [Google Scholar]
- 170.Kim JW, Wieckowski E, Taylor DD, Reichert TE, Watkins S, Whiteside TL. Fas ligand-positive membranous vesicles isolated from sera of patients with oral cancer induce apoptosis of activated T lymphocytes. Clin Cancer Res. 2005;11:1010–20. [PubMed] [Google Scholar]
- 171.Ye SB, Li ZL, Luo DH, Huang BJ, Chen YS, Zhang XS, et al. Tumor-derived exosomes promote tumor progression and T-cell dysfunction through the regulation of enriched exosomal microRNAs in human nasopharyngeal carcinoma. Oncotarget. 2014;5:5439–52. doi: 10.18632/oncotarget.2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Aucher A, Rudnicka D, Davis DM. MicroRNAs transfer from human macrophages to hepatocarcinoma cells and inhibit proliferation. J Immunol. 2013;191:6250–60. doi: 10.4049/jimmunol.1301728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Vanpoucke G, Goossens S, De Craene B, Gilbert B, van Roy F, Berx G. GATA-4 and MEF2C transcription factors control the tissue-specific expression of the alphaT-catenin gene CTNNA3. Nucleic Acids Res. 2004;32:4155–65. doi: 10.1093/nar/gkh727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Janssens B, Goossens S, Staes K, Gilbert B, van Hengel J, Colpaert C, et al. AlphaT-catenin: a novel tissue-specific beta-catenin-binding protein mediating strong cell-cell adhesion. J Cell Sci. 2001;114:3177–88. doi: 10.1242/jcs.114.17.3177. [DOI] [PubMed] [Google Scholar]
- 175.Thayanithy V, Dickson EL, Steer C, Subramanian S, Lou E. Tumor-stromal cross talk: direct cell-to-cell transfer of oncogenic microRNAs via tunneling nanotubes. Transl Res. 2014;164:359–65. doi: 10.1016/j.trsl.2014.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Baumann V, Winkler J. miRNA-based therapies: strategies and delivery platforms for oligonucleotide and non-oligonucleotide agents. Future Med Chem. 2014;6:1967–84. doi: 10.4155/fmc.14.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Esquela-Kerscher A, Trang P, Wiggins JF, Patrawala L, Cheng A, Ford L. The let‐7 microRNA reduces tumor growth in mouse models of lung cancer. Cell Cycle. 2008;7:759–64. doi: 10.4161/cc.7.6.5834. [DOI] [PubMed] [Google Scholar]
- 178.Obad S, dos Santos CO, Petri A, Heidenblad M, Broom O, Ruse C, et al. Silencing of microRNA families by seed-targeting tiny LNAs. Nat Gen. 2011;43:371–8. doi: 10.1038/ng.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Ma L, Young J, Prabhala H, Pan E, Mestdagh P, Muth D, et al. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat Cell Biol. 2010;12:247–56. doi: 10.1038/ncb2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Pereira DM, Rodrigues PM, Borralho PM, Rodrigues CM. Delivering the promise of miRNA cancer therapeutics. Drug Discov Today. 2013;18:282–9. doi: 10.1016/j.drudis.2012.10.002. [DOI] [PubMed] [Google Scholar]
- 181.Wahid F, Shehzad A, Khan T, Kim YY. MicroRNAs: synthesis, mechanism, function, and recent clinical trials. Biochim Biophys Acta. 1803;2010:1231–43. doi: 10.1016/j.bbamcr.2010.06.013. [DOI] [PubMed] [Google Scholar]
- 182.Dougan M, Dranoff G. Immune therapy for cancer. Annu Rev Immunol. 2009;27:83–117. doi: 10.1146/annurev.immunol.021908.132544. [DOI] [PubMed] [Google Scholar]
- 183.Weiner LM, Surana R, Wang S. Monoclonal antibodies: versatile platforms for cancer immunotherapy. Nat Rev Immunol. 2010;10:317–27. doi: 10.1038/nri2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.van Elsas A, Hurwitz AA, Allison JP. Combination immunotherapy of B16 melanoma using anti-cytotoxic T lymphocyte-associated antigen 4 (CTLA-4) and granulocyte/macrophage colony-stimulating factor (GM-CSF)-producing vaccines induces rejection of subcutaneous and metastatic tumors accompanied by autoimmune depigmentation. J Exp Med. 1999;190:355–66. doi: 10.1084/jem.190.3.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Korman AJ, Peggs KS, Allison JP. Checkpoint blockade in cancer immunotherapy. Adv Immunol. 2006;90:297–339. doi: 10.1016/S0065-2776(06)90008-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.O’Day SJ, Hamid O, Urba WJ. Targeting cytotoxic T-lymphocyte antigen-4 (CTLA-4): a novel strategy for the treatment of melanoma and other malignancies. Cancer. 2007;110:2614–27. doi: 10.1002/cncr.23086. [DOI] [PubMed] [Google Scholar]
- 187.Wong RM, Scotland RR, Lau RL, Wang C, Korman AJ, Kast WM, et al. Programmed death-1 blockade enhances expansion and functional capacity of human melanoma antigen-specific CTLs. Int Immunol. 2007;19:1223–34. doi: 10.1093/intimm/dxm091. [DOI] [PubMed] [Google Scholar]
- 188.Berger R, Rotem-Yehudar R, Slama G, Landes S, Kneller A, Leiba M, et al. Phase I safety and pharmacokinetic study of CT-011, a humanized antibody interacting with PD-1, in patients with advanced hematologic malignancies. Clin Cancer Res. 2008;14:3044–51. doi: 10.1158/1078-0432.CCR-07-4079. [DOI] [PubMed] [Google Scholar]
- 189.Deng X, Cao M, Zhang J, Hu K, Yin Z, Zhou Z, et al. Hyaluronic acid–chitosan nanoparticles for co-delivery of MiR-34a and doxorubicin in therapy against triple negative breast cancer. Biomaterials. 2014;35:4333–44. doi: 10.1016/j.biomaterials.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 190.Choi KY, Silvestre OF, Huang X, Min KH, Howard GP, Hida N, et al. A versatile RNA-interference nanoplatform for systemic delivery of RNAs. ACS Nano. 2014;8:4559–70. doi: 10.1021/nn500085k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Zhi F, Dong H, Jia X, Guo W, Lu H, Yang Y, et al. Functionalized graphene oxide mediated adriamycin delivery and miR-21 gene silencing to overcome tumor multidrug resistance in vitro. PLoS One. 2013;8:e60034. doi: 10.1371/journal.pone.0060034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Qian X, Long L, Shi Z, Liu C, Qiu M, Sheng J, et al. Star-branched amphiphilic PLA-b-PDMAEMA copolymers for co-delivery of miR-21 inhibitor and doxorubicin to treat glioma. Biomaterials. 2014;35:2322–35. doi: 10.1016/j.biomaterials.2013.11.039. [DOI] [PubMed] [Google Scholar]
- 193.Cubillos-Ruiz JR, Baird JR, Tesone AJ, Rutkowski MR, Scarlett UK, Camposeco-Jacobs AL, et al. Reprogramming tumor-associated dendritic cells in vivo using microRNA mimetics triggers protective immunity against ovarian cancer. Cancer Res. 2012;72:1683–93. doi: 10.1158/0008-5472.CAN-11-3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Chou J, Werb Z. MicroRNAs play a big role in regulating ovarian cancer-associated fibroblasts and the tumor microenvironment. Cancer Discov. 2012;2:1078–80. doi: 10.1158/2159-8290.CD-12-0465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Mitra AK, Zillhardt M, Hua Y, Tiwari P, Murmann AE, Peter ME, et al. MicroRNAs reprogram normal fibroblasts into cancer-associated fibroblasts in ovarian cancer. Cancer Discov. 2012;2:1100–8. doi: 10.1158/2159-8290.CD-12-0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Wu Z, Lu H, Sheng J, Li L. Inductive microRNA-21 impairs anti-mycobacterial responses by targeting IL-12 and Bcl-2. FEBS Lett. 2012;586:2459–67. doi: 10.1016/j.febslet.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 197.Verma IM, Weitzman MD. Gene therapy: twenty-first century medicine. Annu Rev Biochem. 2005;74:711–38. doi: 10.1146/annurev.biochem.74.050304.091637. [DOI] [PubMed] [Google Scholar]
- 198.György B, Hung ME, Breakefield XO, Leonard JN. Therapeutic applications of extracellular vesicles: clinical promise and open questions. Annu Rev Pharmacol Toxicol. 2015;55:439–64. doi: 10.1146/annurev-pharmtox-010814-124630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Ohno S, Takanashi M, Sudo K, Ueda S, Ishikawa A, Matsuyama N, et al. Systemically injected exosomes targeted to EGFR deliver antitumor microRNA to breast cancer cells. Mol Ther. 2013;21:185–911. doi: 10.1038/mt.2012.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Robbins PD, Morelli AE. Regulation of immune responses by extracellular vesicles. Nat Rev Immunol. 2014;14:195–208. doi: 10.1038/nri3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Taylor DD, Gercel-Taylor C. Tumour-derived exosomes and their role in cancer-associated T-cell signalling defects. Br J Cancer. 2005;92:305–11. doi: 10.1038/sj.bjc.6602316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Filipazzi P, Burdek M, Villa A, Rivoltini L, Huber V. Recent advances on the role of tumor exosomes in immunosuppression and disease progression. Semin Cancer Biol. 2012;22:342–9. doi: 10.1016/j.semcancer.2012.02.005. [DOI] [PubMed] [Google Scholar]
- 203.Melo SA, Sugimoto H, O'Connell JT, Kato N, Villanueva A, Vidal A. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell. 2014;26:707–21. doi: 10.1016/j.ccell.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]


