Abstract
The efficacy of currently available decontamination strategies for the treatment of indoor furnishings contaminated with bioterrorism agents is poorly understood. Efficacy testing of decontamination products in a controlled environment is needed to ensure that effective methods are used to decontaminate domestic and workplace settings. An experimental room supplied with materials used in office furnishings (i.e., wood laminate, painted metal, and vinyl tile) was used with controlled dry aerosol releases of endospores of Bacillus atrophaeus (“Bacillus subtilis subsp. niger,” also referred to as BG), a Bacillus anthracis surrogate. Studies were performed using two test products, a foam decontaminant and chlorine dioxide gas. Surface samples were collected pre- and posttreatment with three sampling methods and analyzed by culture and quantitative PCR (QPCR). Additional aerosol releases with environmental background present on the surface materials were also conducted to determine if there was any interference with decontamination or sample analysis. Culture results indicated that 105 to 106 CFU per sample were present on surfaces before decontamination. After decontamination with the foam, no culturable B. atrophaeus spores were detected. After decontamination with chlorine dioxide gas, no culturable B. atrophaeus was detected in 24 of 27 samples (89%). However, QPCR analysis showed that B. atrophaeus DNA was still present after decontamination with both methods. Environmental background material had no apparent effect on decontamination, but inhibition of the QPCR assay was observed. These results demonstrate the effectiveness of two decontamination methods and illustrate the utility of surface sampling and QPCR analysis for the evaluation of decontamination strategies.
Deposition of airborne microorganisms on surfaces may result in biocontamination in indoor environments. It has been demonstrated that biocontaminants do not remain permanently settled on surfaces, as they may become reentrained into the indoor air with air currents and human activity (4, 12). Human exposure to bioaerosols can occur by inhalation, dermal contact, and ingestion, but inhalation is the most common route that results in adverse health effects (10). Exposure to airborne microorganisms can result in allergies, hypersensitivity reactions, asthma, infections, irritation of the skin or mucous membranes, and flu-like symptoms. In addition, exposure of the domestic population to the accidental or purposeful release of biological agents in indoor environments can result in fatalities or severe human illness (9).
Decontamination is the process of neutralizing, destroying, or removing infectious agents from a person, object, or space, rendering them safe (8). The efficacy of currently available decontamination strategies for the treatment of materials contaminated with bioterrorism agents is poorly understood. Several agents have been used for this purpose, but most of the data available have been obtained from laboratory experiments and not from contaminated buildings. Bacterial endospores are highly resistant to environmental conditions and are easily dispersed and reentrained in the indoor air, making inactivation and removal a difficult task. The organism specified by the U.S. Department of Defense for use in this study, Bacillus atrophaeus (“Bacillus subtilis subsp. niger,” also sometimes referred to as BG), has historically been used as a Bacillus anthracis surrogate because it is a noninfectious, endospore-forming bacterium that is easily cultured and has a distinct colony morphology.
Chlorine dioxide gas and a foam decontaminant were selected for testing in this study. Chlorine dioxide gas is an agent used to control noxious microorganisms on inanimate objects and surfaces and has been registered as a sterilant (a type of antimicrobial pesticide) with the U.S. Environmental Protection Agency since 1988 (11). The foam decontaminant is supplied as two agents that when mixed together can be used to remove chemical and biological warfare agents on objects and surfaces. The mixed agent is composed of cationic detergents, fatty alcohols, stabilized hydrogen peroxide, water, and inert ingredients.
Enhanced detection methods for biocontaminants on surfaces are needed to assess the level of contamination and to evaluate the efficacy of decontamination procedures used on building material furnishings. However, monitoring is hampered by the lack of methods that provide precise, accurate, and representative exposure estimates for bioaerosols and microbe-contaminated surfaces (1). Traditional microbial monitoring relies on the collection of air and surface samples and analysis by either culture on artificial growth media or microscopy (5). Currently, swab samples are the primary collection method employed in indoor environments suspected of contamination with biological agents (7). However, the sensitivity of detection with this method is low due to the relatively small surface area sampled. Furthermore, swab sampling may produce an excessive number of samples, which can delay the reporting of results, tax laboratory resources and staff, and increase analysis costs. Because environmental and sampling stresses can affect the viability of biocontaminants, culture analysis methods underestimate concentrations due to the enumeration and identification of those cells that are culturable while nonculturable organisms go undetected (6). The inaccuracy of traditional sampling methods and lengthy analysis required to characterize air and surface biocontaminant concentrations underscore the need for developing new sampling and analysis techniques that can provide rapid, reliable data for bioaerosol exposure. Quantitative PCR (QPCR) has been used in the analysis of surface samples for the enhanced detection of bacteria (3) and was used in this research for the rapid detection and measurement of B. atrophaeus on surfaces in an experimental room. Building furnishings were contaminated with B. atrophaeus and then sampled before and after treatment with a decontamination agent. Concentrations of B. atrophaeus were determined by culture and QPCR analyses, with and without environmental background material present. Previously developed protocols for B. atrophaeus DNA extraction and sample processing methods compatible with PCR analysis were utilized (3). In addition to culture and QPCR analyses, an antibody-based assay was also utilized in the chlorine dioxide testing experiments. This assay, referred to as a hand-held assay (HHA), consists of a cardboard ticket containing immobilized antibodies specific to the target organism, B. atrophaeus. Application of a liquid sample initiates a color change reaction when B. atrophaeus is present at a concentration of ≥105 spores. Although this assay is not quantitative, it has the advantage of speed in determining the presence of the target organism.
The purpose of this research was to determine the efficacy of decontamination strategies and evaluate surface sampling methods and QPCR for the enhanced detection of a target biocontaminant present on building material furnishings. These studies involved both laboratory experiments and the use of an experimental room designed for bioaerosol studies. The use of a controlled environment with a variety of contaminated materials provided data to ensure that effective sampling, analysis, and decontamination methods can be employed in domestic and workplace settings. This research established sampling and analysis methods for characterization of a specific microorganism on building material furnishings that are fast, quantitative, sensitive, and useful for efficacy testing of decontamination strategies in indoor environments.
MATERIALS AND METHODS
Test organism and culture media.
Dry endospores of B. atrophaeus were obtained from the U.S. Army Dugway Proving Ground, Utah. Samples containing B. atrophaeus were cultured on tryptic soy agar (pH 7.0; Difco Laboratories, Sparks, Md.) and incubated at 28°C for 2 days. B. atrophaeus was typically the only microorganism cultured from surface samples of clean test materials. For experiments with environmental background material present, B. atrophaeus was readily distinguishable from background microbial contaminants by colony morphology. In addition to decontamination testing using B. atrophaeus as the target organism, commercially available paper strips containing spores of the bacterium Bacillus stearothermophilus (Steris, Erie, Pa.) were placed throughout the experimental room before treatment with chlorine dioxide gas. Six spore strips were placed in the room for each experiment and collected following decontamination. Spore strips were placed in tryptic soy broth (Difco) and incubated at 55°C for 72 h. Growth determination following incubation was ascertained by the presence of turbidity.
Experimental room.
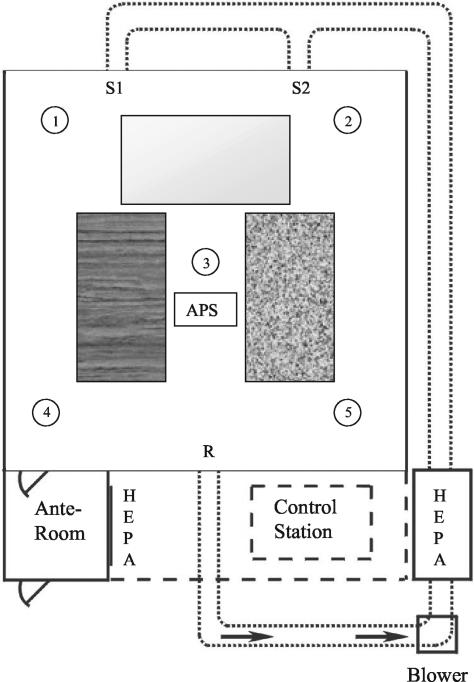
Materials were contaminated by aerosolization and deposition of B. atrophaeus spores in a room-sized experimental chamber that was designed to resemble a residential indoor environment and has been previously used for bioaerosol studies (Fig. 1) (2, 3). The room, which measures 4.0 by 4.0 by 2.2 m high, has a sheet vinyl tile floor. The interior walls, exterior walls, and ceiling are covered with gypsum wallboard and coated with interior latex paint. The room is equipped with a heating, ventilation, and air conditioning (HVAC) system sized to simulate a residential system with rectangular bare metal ductwork. During aerosolization experiments, the HVAC was operated with an airflow of 4.2 m3/min, resulting in approximately 7 room air volume exchanges per h. The room was maintained at a positive static pressure (+0.02 in. of water) during operation to minimize contamination from the surrounding area into the room. An anteroom equipped with a HEPA-filtered air shower attached to the room entrance reduced mixing of air resulting from entering and exiting the room during experiments. Temperature was monitored by 20 type T thermocouples (Thermo Electric Co., Saddle Brook, N.J.), and relative humidity (RH) was monitored by five RH probes (Hy-Cal Engineering, El Monte, Calif.) located within the room. During all activities in the experimental room, technicians wore full-face respirators and nonwoven protective clothing. Upon completion of each series of experimental room trials, contaminated surface materials were removed and the interior surfaces of the room were disinfected with a 0.5% sodium hypochlorite solution.
FIG. 1.
Diagram of the experimental room. HEPA-filtered air is delivered into the room via two supply registers (S1 and S2) and exits through the return register (R). Sampling stands 1 to 5 support temperature and RH probes. Rectangles in the room represent sampling benches with wood laminate, metal, and vinyl tile surfaces.
Test materials.
Surface materials consisted of 929-cm2 (1-ft2) sections of wood laminate, vinyl tile, and painted metal to simulate laminate table and desk surfaces, flooring, and metal file cabinets and bookshelves, respectively. Materials were disinfected with a 0.5% sodium hypochlorite solution, rinsed with sterile water, dried, and placed in the experimental room prior to aerosolization of B. atrophaeus. Any materials damaged during the decontamination processes were retreated or replaced.
B. atrophaeus aerosolization.
Dry B. atrophaeus spores were aerosolized into the experimental room by acoustic vibration using the Pitt 3 dry aerosol generator as previously described (3). Airborne particle concentrations were measured in the room using a model 3320 aerodynamic particle sizer (APS; TSI Inc., St. Paul, Minn.). Particle concentrations in the B. atrophaeus spore size range above background levels were used to determine the B. atrophaeus bioaerosol concentration. Following aerosolization of B. atrophaeus spores, the room HVAC system was turned off and remained off overnight to permit deposition of spores onto surfaces in the room. After settling of the B. atrophaeus aerosol overnight, surface samples were collected (as described below) on two consecutive days for analysis by culture and QPCR.
Environmental background.
To illustrate the effects of the presence of environmental background material on the efficacy of the decontamination agents, a trial was performed with environmental background material for each decontamination method. Environmental background material was obtained from outdoor air filters of commercial buildings from several locations in the United States. The filters were vacuumed using high-efficiency vacuum bags, and the collected material was sieved and pooled. Culture analysis of the environmental background material indicated bacterial concentrations of 106 CFU/g. No B. atrophaeus was cultured from the environmental background material. Approximately 10 g of the environmental background material was aerosolized into the room and allowed to settle on the test materials. The approximate soiling level was 2 mg per 100 cm2 of surface area. This level of contamination was visible and similar in appearance to a moderately dusty surface. B. atrophaeus was then aerosolized into the experimental room as described above.
Decontamination agents.
Two decontamination agents were tested in the experimental room for their efficacy in killing B. atrophaeus spores, a foam (Modec decontamination formulation [MDF]; Modec Inc., Denver, Colo.) and chlorine dioxide gas (CDG Research Corporation, Bethlehem, Pa.). For each decontamination agent, three trials were performed with B. atrophaeus present on clean surface materials and one trial was performed with B. atrophaeus and environmental material applied to the surface materials. For all experiments, surface samples of 929 cm2 (1 ft2) in area were collected using three sampling methods before and after decontamination.
The foam decontaminant is supplied as two agents that when mixed together can be used to remove chemical and biological warfare agents on objects and surfaces. The mixed agent is composed of cationic detergents, fatty alcohols, stabilized hydrogen peroxide, water, and inert ingredients. Decontamination with the foam was performed by mixing the two chemical components (MDF I and II) in equal parts (total volume, 3 liters) in a commercially available liquid sprayer according to the manufacturer's instructions, pressurizing the sprayer by hand pumping, and spraying the decontaminant onto surfaces until they were completely wetted. Using this application technique the product was similar in appearance to a liquid detergent. The entire volume of the foam decontaminant was applied to the test materials, walls, and floor.
Decontamination with chlorine dioxide gas was performed by CDG Research Corporation personnel in the manner used for building decontamination and in accordance with the safety plan prepared by the University of Nevada, Las Vegas and CDG Research Corporation. The experimental room containing B. atrophaeus-contaminated materials was preconditioned by raising the humidity to 85% RH and maintaining that level for approximately 2 h before decontamination. Chlorine dioxide gas was then introduced into the experimental room to a concentration of approximately 1,400 ppm and maintained at that level for 4 h. The concentration of chlorine dioxide gas was monitored by continuous measurement of absorbance of room air using a Hach DR/4000 spectrophotometer (Hach Co., Loveland, Colo.). During decontamination, the RH was maintained at approximately 85% and the temperature was approximately 24°C. Four small portable fans were operated inside the experimental room to provide adequate mixing of the chlorine dioxide gas. The room was operated at a slight negative pressure (−0.03 in. of water) to prevent leakage of chlorine dioxide gas from the room. After 4 h of chlorine dioxide decontamination, the room air was exhausted through a scrubber that reduced chlorine dioxide levels to <2 ppm before venting to the outdoor air. An additional B. atrophaeus release was conducted as a control experiment without the use of chlorine dioxide to determine if physical factors such as disturbance by the operation of the portable fans and evacuation of room air through the scrubber, or environmental factors such as 85% RH, affected the postdecontamination results. During the control experiment, all conditions were identical to decontamination trials with the exception that the room was treated with only the nitrogen carrier gas (no chlorine dioxide) for 4 h.
Surface sampling methods.
Three commercially available products were used for surface sampling, a swipe (Speci-sponge; Nasco, Fort Atkinson, Wis.), a Heavy Wipe (Handy Wipes Heavy Wipe; First Brands Corporation, Danbury, Conn.), and a swab sample processing (SSP) kit (ASD, Ft. Lauderdale, Fla.). One sample was collected from each material with each sampling method before and after decontamination.
Swipe sampling consisted of moistening the sponge in a sterile bag with 30 ml of 0.01 M phosphate buffer with 0.05% Tween (PBT; pH 7.0). The swipe was squeezed to remove the excess buffer and then used to sample surface material sections by wiping the entire area (929 cm2) in a horizontal direction. The swipe was turned over to expose the unused side and used to sample the same surface material section in a vertical direction.
The Heavy Wipe was folded, placed in a sterile bag with 40 ml of PBT, and used to sample in the same manner as for the swipe sampling. The swipe and Heavy Wipe were returned to the sample bags, and the samples were hand mixed for 1 min, followed by squeezing to elute the liquid sample. The squeezed swipe and Heavy Wipe were then discarded.
The SSP kit was utilized as per the manufacturer's protocol, which consisted of moistening the surface material section with 20 drops of the supplied buffer followed by sampling the first half of the surface material area with a foam swab, turning the swab over to expose the unused side, and swabbing the second half of the surface material. The swab was returned to a collection bottle containing 55 drops of buffer and incubated for 15 s, followed by hand mixing for 1 min (manufacturer's protocol; fraction B). The swab was then pressed against the sides of the bottle to elute the liquid sample, and the swab was discarded. Sample volume ranged from approximately 1.0 to 1.5 ml. Laboratory studies indicated that the highest concentration of the target organism was present in fraction B, and further purification of the liquid sample, as described in the manufacturer's protocol, resulted in losses of B. atrophaeus. Therefore, only fraction B was analyzed for all SSP kit samples.
Surface samples were analyzed by culture and QPCR. In addition, HHAs were used for chlorine dioxide experiments.
DNA extraction and purification.
A previously developed DNA extraction and concentration protocol was used for preparation of quantitation standards and for all surface samples (3). Twenty milliliters of the swipe and Heavy Wipe samples was concentrated by filtration for subsequent DNA extraction. Processed samples were filtered through a 0.65-μm-pore-diameter mixed cellulose ester filter membrane (Millipore Corp., Bedford, Mass.), and the filter was resuspended in 0.5 ml of PBT. Some samples (e.g., Heavy Wipe samples and samples containing environmental background) could not be filtered due to blockage of the membrane and required prefiltration through a 0.80-μm mixed cellulose ester membrane. For these samples, both membranes were resuspended in PBT. Due to the small sample volume obtained with the SSP kit samples, these samples were not filter concentrated, and a 0.5-ml aliquot was reserved for DNA extraction. Each sample was pretreated with sodium dodecyl sulfate (0.5% final concentration) and proteinase K (20 μg/ml final concentration), incubated at 50°C for 5 min, and then boiled for 15 min. The sample was chilled on ice for 2 min, and bovine serum albumin (0.05% final concentration) was added, followed by incubation for 5 min at 37°C in a rotary shaker at 230 rpm. Following DNA extraction, samples containing membranes were vortexed briefly and the membranes were removed. The DNA from all samples was purified using the Pellet Paint protocol (Novagen, Madison, Wis.) and resuspended in 50 μl of Tris-EDTA buffer (pH 8.0). PCR quantitation standards were prepared from a purified B. atrophaeus spore suspension enumerated electronically with a Coulter Multisizer II (Beckman Coulter, Inc., Hialeah, Fla.) using the same DNA extraction and purification methods used to process samples.
QPCR.
The ABI Prism 7700 sequence detection system (Applied Biosystems, Foster City, Calif.) was used for QPCR analysis as previously described (3). This involved amplification of a segment of the B. atrophaeus recA gene with primer and probe sequences obtained from the Naval Medical Research Center (Silver Spring, Md.), which produced a 131-bp amplicon. Sequences for the primers were ACCAGACAATGCTCGACGTT (forward) and CCCTCTTGAAATTCCCGAAT (reverse). The TaqMan probe sequence was 6-carboxyfluorescein-5′-ACTGAACAGCTGATCGAGACAGCTGCA-3′-tetramethyl carboxyrhodamine. Primers were obtained from Operon Technologies (Alameda, Calif.), and the probe was obtained from Synthetic Genetics (San Diego, Calif.). The amplification conditions specified by the Naval Medical Research Center for use with Applied Biosystems reagents were as follows: B. atrophaeus DNA template; 1× TaqMan buffer A, 5 mM MgCl2, 0.1 mM dATP, 0.1 mM dCTP, 0.1 mM dGTP, 0.2 mM dUTP, 2.5 U of AmpliTaq Gold, 0.5 U of AmpErase uracyl N-glycosylase, a 0.2 μM concentration of each primer, and 0.2 μM probe, for a total reaction volume of 50 μl. The TaqMan cycling conditions were as follows: 2 min at 50°C, 10 min at 95°C, and 40 cycles of 15 s at 95°C followed by 1 min at 60°C. An internal positive control (IPC) (IPC-VIC probe; Applied Biosystems) was incorporated into the PCR to determine whether the samples contained PCR inhibitors. The IPC kit consisted of control DNA, primers, and a specific probe. This fluorescent probe was labeled with a dye that was different from the target DNA probe to allow for the differentiation of fluorescent signals generated during amplification. A known amount of IPC DNA (0.4 pg per reaction mixture) was amplified with the sample, and inhibition was observed by an increase in the threshold detection cycle (described below) of control DNA.
Quantitation was achieved by amplification of standards containing B. atrophaeus DNA extracted from spore suspensions of known concentration (100 to 105 templates per reaction mixture). Extraction of B. atrophaeus standards was performed in the same manner as that of samples, allowing absolute quantitation of B. atrophaeus templates. Quantitation by this method corrects for losses of target DNA during extraction, assuming losses of sample DNA during extraction are consistent and similar to losses in standard DNA. In addition, this method corrects for the occurrence of external B. atrophaeus DNA present in the samples, assuming that the amount of external DNA present in B. atrophaeus spore populations is consistent. Standards were amplified in duplicate at the same time and under the same conditions as the replicate unknown samples. Once amplification was completed, the data were analyzed using the software provided with the ABI Prism 7700 sequence detection system. Using the concentrations assigned to each standard, the software constructed a standard curve of Ct value versus concentration. Ct refers to the PCR cycle at which fluorescence (i.e., amplification product) is first detected and is inversely proportional to the initial DNA template concentration. Concentration values for the unknown samples were extrapolated from the standard curve by the software and reported as the means of two replicates.
HHAs.
The HHAs (Critical Reagents Program, Aberdeen Proving Ground, Md.) were used in the chlorine dioxide gas decontamination trials. The HHAs consisted of individually wrapped cardboard tickets containing immobilized antibodies specific to the target organism, B. atrophaeus. Application of a liquid sample initiates an immunological reaction when B. atrophaeus is present at a concentration of ≥105 spores. The HHA has a positive control (C) well and test (T) well. If a pink or red line develops in both the C and T wells, the test is considered positive. If a pink or red line develops only in the C well, the test is considered negative. The HHAs were stored at 4°C until ready to use. A volume of 100 μl of each surface sample was dispensed in the HHA sample well labeled S and incubated at room temperature for 15 min.
Data and statistical analyses.
The B. atrophaeus CFU per milliliter of sample values were converted to the number of CFU per sample of surface material. The concentrations of B. atrophaeus CFU per sample determined for the three trials without environmental background were averaged and log10 transformed. The numbers of initial target DNA in PCR amplification reactions were converted to the numbers of templates per sample. The concentrations of B. atrophaeus templates per sample determined for the three trials without environmental background were averaged and log10 transformed. Analysis of variance tests were performed on log-transformed data to compare results. Lower detection limits were determined based on detection of 1 B. atrophaeus CFU per milliliter and 1 B. atrophaeus template per PCR and conversion to number of CFU per sample and number of templates per sample, respectively.
RESULTS
Foam decontaminant.
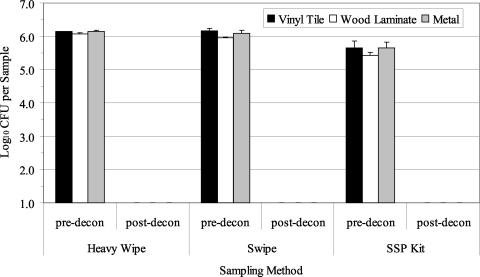
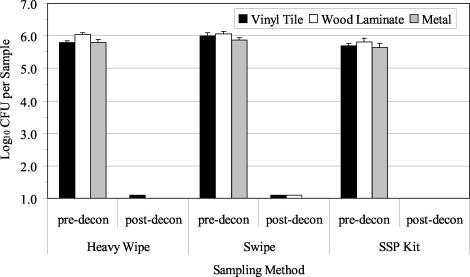
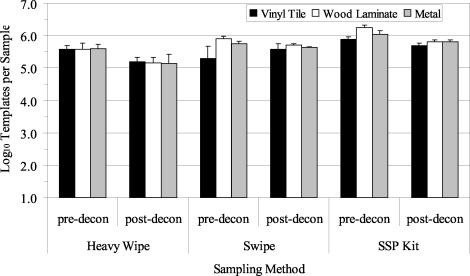
Three decontamination trials with the foam decontaminant were performed with B. atrophaeus spores aerosolized and deposited onto clean surface materials. The average airborne concentration (±1 standard error [SE]) of B. atrophaeus spores as determined by the APS during the three aerosol releases was 1.48 × 106± 8.99 × 104 spores/m3. Culture results from predecontamination samples indicated a contamination level of 105 to 106 B. atrophaeus spores/sample (Fig. 2). The Heavy Wipe and swipe data were significantly greater than those obtained with the SSP kit (P = 0.000). There was no significant difference between results obtained from the three materials sampled (P = 0.455). After foam decontamination, culturable B. atrophaeus spores were not detected (Fig. 2; lower detection limit, approximately 10 to 40 B. atrophaeus CFU/ft2 [929 cm2]). QPCR results from predecontamination samples indicated a contamination level of 105 to 106 B. atrophaeus templates per sample, depending on the sampling method and surface material (Fig. 3). B. atrophaeus spore viability estimated prior to decontamination was 42.6% ± 7.6% (n = 9), based on the ratio of culturable spores to total spores, as measured by QPCR for SSP kit samples. QPCR data showed no significant differences between sampling methods (P = 0.347) or materials (P = 0.182). After foam decontamination, B. atrophaeus DNA was detected with QPCR in all surface samples at a level of 104 to 105 B. atrophaeus templates per sample.
FIG. 2.
Summary of culture results obtained for the detection of B. atrophaeus from surfaces in three experimental room trials before and after use of the foam decontaminant. Materials were contaminated by aerosolization of B. atrophaeus spores into the room, followed by settling of the bioaerosol onto surfaces in the room. Surface samples (929 cm2 [1 ft2]) were collected with Heavy Wipes, swipes, and SSP kits prior to decontamination (pre-decon). Surfaces were treated with the foam and sampled postdecontamination (post-decon). All postdecontamination samples contained no culturable B. atrophaeus (lower detection limit range, 10 to 40 CFU per sample). Bar heights represent the mean (log10) of three samples ± 1 SE.
FIG. 3.
Summary of QPCR results obtained for the detection of B. atrophaeus from surfaces before and after use of a foam decontaminant. Materials were contaminated by aerosolization of B. atrophaeus spores into the room, followed by settling of the bioaerosol onto surfaces. Surface samples (929 cm2 [1 ft2]) were collected with Heavy Wipes, swipes, and SSP kits prior to decontamination (pre-decon). Surfaces were treated with the foam and sampled postdecontamination (post-decon). Bar heights represent the mean (log10) of three samples ± 1 SE.
For the single foam decontamination trial performed with B. atrophaeus and environmental material present on the surface materials, the average airborne concentration of B. atrophaeus spores during the aerosol release was 1.49 × 106 spores/m3. Culture results from surface samples with environmental background material present were comparable to those obtained in previous trials without environmental background, indicating a contamination level of 105 to 106 B. atrophaeus spores per sample before decontamination (data not shown). After decontamination, no culturable B. atrophaeus spores were detected. All QPCR results from samples collected before decontamination showed lower concentrations of B. atrophaeus than culture analysis, indicating that the environmental background material had an inhibitory effect on the assay. The greatest inhibitory effect was observed with swipe samples, where QPCR measurements of total B. atrophaeus were several orders of magnitude lower than culturable B. atrophaeus levels (data not shown). Postdecontamination QPCR results indicated that B. atrophaeus DNA was still present, as shown in trials without environmental background.
Chlorine dioxide gas.
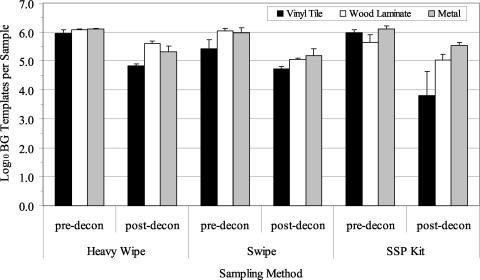
Three decontamination trials were performed using chlorine dioxide gas with B. atrophaeus spores aerosolized and deposited onto clean surface materials. The average airborne concentration of B. atrophaeus spores as determined by the APS during the three aerosol releases was 1.61 × 106± 1.76 × 105 (1 SE) spores/m3. Culture results from predecontamination samples indicated a contamination level of 105 to 106 B. atrophaeus spores/sample (Fig. 4). There was a significant difference observed between the sampling methods (P = 0.009). Culture data were highest for the swipe, followed by the Heavy Wipe and the SSP kit. There was no significant difference between results obtained from the three materials sampled (P = 0.069). After chlorine dioxide decontamination, culturable B. atrophaeus spores were not detected (lower detection limit, approximately 10 to 23 B. atrophaeus CFU/ft2 [929 cm2]) in 24 of 27 samples (Fig. 4). The three positive samples contained culturable B. atrophaeus at the lower detection limit of 1 culturable spore. QPCR results from predecontamination samples indicated a contamination level of 105 to 106 B. atrophaeus templates per sample, depending on the sampling method and surface material (Fig. 5). B. atrophaeus spore viability estimated prior to decontamination was 47.3% ± 5.1% (n = 9), based on the ratio of culturable spores to total spores as measured by QPCR for SSP kit samples. QPCR data showed significant differences between sampling methods (P = 0.006). In contrast with culture results, the SSP kit data were significantly higher than those for the other sampling methods. No significant difference was observed between data obtained from the three materials sampled (P = 0.174). After chlorine dioxide decontamination, B. atrophaeus DNA was detected with QPCR in all surface samples at a level of 105 B. atrophaeus templates per sample.
FIG. 4.
Summary of culture results obtained for the detection of B. atrophaeus from surfaces before and after decontamination with chlorine dioxide gas. Materials were contaminated by aerosolization of B. atrophaeus spores into the room, followed by settling of the bioaerosol onto surfaces. Surface samples (929 cm2 [1 ft2]) were collected with Heavy Wipes, swipes, and SSP kits prior to decontamination (pre-decon). Surfaces were treated with chlorine dioxide gas and sampled postdecontamination (post-decon). Bar heights represent the mean (log10) of three samples ± 1 SE.
FIG. 5.
Summary of QPCR results obtained for the detection of B. atrophaeus from surfaces before and after decontamination with chlorine dioxide gas. Materials were contaminated by aerosolization of B. atrophaeus spores into the room, followed by settling of the bioaerosol onto surfaces. Surface samples (929 cm2 [1 ft2]) were collected with Heavy Wipes, swipes, and SSP kits prior to decontamination (pre-decon). Surfaces were treated with chlorine dioxide gas and sampled postdecontamination (post-decon). Bar heights represent the mean (log10) of three samples ± 1 SE.
A control experiment was conducted to determine if factors related to chlorine dioxide decontamination affected the results, such as disturbance by the operation of the portable fans, 85% RH, or evacuation of room air through the scrubber. This experiment was conducted in an identical manner as the other B. atrophaeus contamination trials (no environmental background material present), with the exception that the chlorine dioxide gas was not released into the room. Surface sample results indicated that no differences in B. atrophaeus contamination levels were caused by disturbances associated with the chlorine dioxide decontamination process (Table 1).
TABLE 1.
Results of a control experiment without chlorine dioxide gas decontaminationa
| Sampling method | Material | Culture analysis (CFU/sample)
|
QPCR analysis (templates/sample)
|
||
|---|---|---|---|---|---|
| Pre | Post | Pre | Post | ||
| Heavy wipe | Vinyl tile | 6.43 × 105 | 5.41 × 105 | 3.37 × 105 | 4.47 × 105 |
| Wood laminate | 7.97 × 105 | 8.43 × 105 | 1.42 × 105 | 4.52 × 105 | |
| Metal cabinet | 5.62 × 105 | 6.77 × 105 | 2.50 × 105 | 4.39 × 105 | |
| Swipe | Vinyl tile | 7.45 × 105 | 7.79 × 105 | 6.15 × 105 | 9.48 × 105 |
| Wood laminate | 1.56 × 106 | 7.85 × 105 | 6.16 × 104 | 7.54 × 105 | |
| Metal cabinet | 7.81 × 105 | ND | 5.95 × 105 | 6.66 × 105 | |
| SSP kit | Vinyl tile | 4.55 × 105 | 5.16 × 105 | 1.04 × 106 | 1.23 × 106 |
| Wood laminate | 9.06 × 105 | 4.02 × 105 | 1.26 × 106 | 1.33 × 106 | |
| Metal cabinet | 5.83 × 105 | 5.20 × 105 | 1.15 × 106 | 1.22 × 106 | |
Data were obtained in a control experiment with no chlorine dioxide gas decontamination to determine if physical factors during treatment affected postdecontamination results. The control experiment was conducted in an identical manner as the other B. atrophaeus decontamination trials, with the exception that chlorine dioxide gas was not released into the room. Surface samples (929 cm2, [1 ft2]; n = 1 for each material) were collected with Heavy Wipes, swipes, and SSP kits prior to treatment (Pre) and posttreatment (Post). Culture and QPCR results were obtained for the detection of B. atrophaeus from surfaces in the experimental room. ND, not determined.
For the single chlorine dioxide decontamination trial performed with B. atrophaeus and environmental material present on the surface materials, the average airborne concentration of B. atrophaeus spores during the aerosol release was 1.84 × 106 spores/m3. Culture results from surface samples with environmental background material present were comparable to those obtained in previous trials without environmental background (data not shown), indicating a contamination level of 105 to 106 B. atrophaeus spores per sample before decontamination. After decontamination, culturable B. atrophaeus spores were detected in only one of nine samples, the wood laminate surface sampled with the swipe, at a level of 213 B. atrophaeus CFU/ft2. QPCR results from samples collected before decontamination indicated that the environmental background material had an inhibitory effect on the assay compared to when no environmental background was present (data not shown). The greatest inhibitory effect was observed with the swipe and SSP kit samples, where B. atrophaeus measurements were below the limits of detection (12 and 29 templates per sample, respectively). Postdecontamination QPCR results indicated that B. atrophaeus DNA was still present, as shown in trials without environmental background.
In addition to decontamination testing with B. atrophaeus as the target organism, commercially available paper strips containing spores of the bacterium B. stearothermophilus were placed throughout the experimental room before treatment with chlorine dioxide gas. Postdecontamination results showed no growth of B. stearothermophilus from any of the test strips, whereas growth was observed for all spore strips in the control experiment without treatment with chlorine dioxide gas.
Positive results were obtained with HHAs for all samples, both before and after chlorine dioxide decontamination (data not shown). The decontamination showed no apparent effect on the HHA signals. The SSP kit samples had a stronger signal than the swipe and Heavy Wipe samples, due to lower sample volumes and therefore greater B. atrophaeus concentrations.
DISCUSSION
The B. atrophaeus dry aerosolization method proved to be an effective and repeatable means of contamination of materials in the experimental room, as evidenced by the uniformity of predecontamination culturable B. atrophaeus levels. The three surface sampling methods tested, the swipe, the Heavy Wipe, and the SSP kit, demonstrated comparable efficiency of recovery of B. atrophaeus spores from surfaces. In addition, similar levels of B. atrophaeus were obtained from the surfaces of the three materials tested.
No culturable B. atrophaeus spores were detected after decontamination with the foam, a reduction of approximately 5 orders of magnitude. However, QPCR results showed that amplifiable B. atrophaeus DNA was still present after decontamination, although at significantly lower levels compared with predecontamination samples. The decrease in the amount of B. atrophaeus DNA measured after decontamination may have been due to interference with the DNA extraction or with the QPCR that was caused by the presence of the foam decontaminant in the samples, or it may have been due to the destruction of B. atrophaeus spores and DNA by the foam. The latter explanation is more likely, as IPC data indicated no apparent PCR inhibition was caused by the foam in any of the postdecontamination QPCR samples. With environmental background material present, culture analysis appeared to be unaffected but QPCR was inhibited in both pre- and postdecontamination samples.
The results showed that the foam was an effective decontamination agent. The advantages of using the foam are that it is a fast and easy process, requiring no special equipment, personnel, or training. The method seems particularly well-suited to environments containing nonporous, washable surfaces. However, several drawbacks to using the foam decontaminant were noted. The foam partially dissolved the floor polish applied to the vinyl tile test sections and the experimental room floor. The foam also caused bubbling of the wood laminate surfaces of tables and stripped the paint from portions of the metal and walls. This caused problems with sample processing and in cleanup of the experimental room. Filter concentration of samples from the vinyl tile sections was difficult due to clogging of the membranes, presumably by partially dissolved floor wax, and additional membranes were required for these samples. This may have contributed to the lower values obtained with QPCR for vinyl tile postdecontamination samples.
The chlorine dioxide gas decontamination method was an effective treatment, reducing the levels of culturable B. atrophaeus by 4 to 5 orders of magnitude. With the gas concentration and physical conditions tested in the experimental room, only 3 of 27 samples (11%) contained culturable B. atrophaeus. Each of the three positive samples was at the lower detection limit of 1 culturable B. atrophaeus spore. QPCR results showed that B. atrophaeus DNA was still present and postdecontamination B. atrophaeus DNA levels were only slightly less than those obtained before decontamination. This indicates that while the chlorine dioxide gas decontamination rendered the majority of B. atrophaeus spores nonculturable, B. atrophaeus DNA remained amplifiable. Positive HHA results confirmed that B. atrophaeus antigens were still present and detectable after chlorine dioxide decontamination. With environmental background material, culture analysis appeared to be unaffected, but QPCR was inhibited in both pre- and postdecontamination samples. It appeared that the presence of environmental material did not have an effect on the efficacy of decontamination with chlorine dioxide, with the exception of the single positive sample (213 B. atrophaeus CFU/ft2 [929 cm2]). As observed with the foam, the effectiveness of the QPCR analysis was inhibited by the presence of environmental background material.
The chlorine dioxide gas decontamination method has several advantages. The use of a gas treatment allows entire rooms to be treated, and their contents are treated in place. Room furnishings were essentially undamaged, with the exception of corrosion to the surface of an aluminum cart. The corrosion occurred over the course of several trials and may have been due to the repeated exposure of the material to chlorine dioxide gas and high humidity conditions. The disadvantages of the method include the requirement for gases and special equipment to be delivered on-site and assembled and operated by trained personnel.
In summary, both of the methods evaluated for the decontamination of indoor environments demonstrated effectiveness against culturable B. atrophaeus spores. The treatments appeared to be unaffected by environmental background material; however, analysis of environmental samples by QPCR was inhibited. Both of the decontamination strategies tested have advantages and disadvantages that should be considered in the design of a decontamination protocol. It is important to note that DNA and antigenic compounds remained after the decontamination process; therefore, additional cleanup steps may be required, depending on the biocontaminant, to ensure a safe environment.
The results of treatment of B. atrophaeus-contaminated surfaces in the experimental room with the two decontamination methods were assessed by culture, QPCR, and HHA analyses. The test organism, B. atrophaeus, is a nonpathogenic surrogate for B. anthracis. Additional work is needed to determine whether these decontamination methods are effective for the remediation of actual biological warfare agents, such as B. anthracis. A particular area of concern that could not be addressed in this study is the infection potential of biological agents following decontamination treatments. Currently, culturability is used to verify the effectiveness of remediation, but the infection potential of nonculturable B. anthracis after remediation of contaminated environments is unknown.
Acknowledgments
The research described in this article was funded by the Technical Support Working Group, Arlington, Va.
We thank CDG Research Corporation personnel for providing their expertise and assistance with the chlorine dioxide gas experiments and Stephanie Piccininni for her technical assistance.
REFERENCES
- 1.American Conference of Governmental Industrial Hygienists. 1999. Bioaerosols: assessment and control. American Conference of Government Industrial Hygienists, Cincinnati, Ohio.
- 2.Buttner, M. P., and L. D. Stetzenbach. 1993. Monitoring of fungal spores in an experimental indoor environment to evaluate sampling methods and the effects of human activity on air sampling. Appl. Environ. Microbiol. 59:219-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buttner, M. P., P. Cruz-Perez, and L. D. Stetzenbach. 2001. Enhanced detection of surface-associated bacteria in indoor environments by quantitative PCR. Appl. Environ. Microbiol. 67:2564-2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buttner, M. P., P. Cruz-Perez, L. D. Stetzenbach, P. J. Garrett, and A. E. Luedtke. 2002. Measurement of airborne fungal spore dispersal from three types of flooring materials. Aerobiologia 18:1-11. [Google Scholar]
- 5.Buttner, M. P., K. Willeke, and S. Grinshpun. 2002. Sampling and analysis of airborne microorganisms, p. 814-826. In C. J. Hurst, G. Knudsen, M. McInerney, M. V. Walter, and L. D. Stetzenbach (ed.), Manual of environmental microbiology, 2nd ed. ASM Press, Washington, D.C.
- 6.Cox, C. S. 1989. Airborne bacteria and viruses. Sci. Prog. 73:469-500. [PubMed] [Google Scholar]
- 7.Higgins, J. A., M. Cooper, L. Schroeder-Tucker, S. Black, D. Miller, J. S. Karns, E. Manthey, R. Breeze, and M. L. Perdue. 2003. A field investigation of Bacillus anthracis contamination of U.S. Department of Agriculture and other Washington, D. C., buildings during the anthrax attack of October 2001. Appl. Environ. Microbiol. 69:593-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morris, C. (ed.). 1992. Dictionary of science and technology. Academic Press, San Diego, Calif.
- 9.Sheeran, T. J. 2002. Bioterrorism, p. 771-782. In G. Bitton (ed.), Encyclopedia of environmental microbiology, vol. 2. John Wiley & Sons, New York, N.Y. [Google Scholar]
- 10.Stetzenbach, L. D. 2002. Introduction to aerobiology, p. 801-813. In C. J. Hurst, R. L. Crawford, G. R. Knudsen, and L. D. Stetzenbach (ed.), Manual of environmental microbiology, 2nd ed. American Society for Microbiology, Washington, D.C.
- 11.U.S. Environmental Protection Agency. 2003. Pesticides: topical and chemical fact sheets. Chlorine dioxide. U.S. Environmental Protection Agency, Washington, D.C. [Online.] http://www.epa.gov/pesticides/factsheets/chemicals/chlorinedioxidefactsheet.htm.
- 12.Weis, C. P., A. J. Intrepido, A. K. Miller, P. G. Cowin, M. A. Durno, J. S. Gebhardt, and R. Bull. 2002. Secondary aerosolization of viable Bacillus anthracis spores in a contaminated U.S. Senate office. JAMA 288:2853-2858. [DOI] [PubMed] [Google Scholar]