Abstract
Enhanced biodegradation in the rhizosphere has been reported for many organic xenobiotic compounds, although the mechanisms are not fully understood. The purpose of this study was to discover whether rhizosphere-enhanced biodegradation is due to selective enrichment of degraders through growth on compounds produced by rhizodeposition. We monitored the mineralization of [U-14C]2,4-dichlorophenoxyacetic acid (2,4-D) in rhizosphere soil with no history of herbicide application collected over a period of 0 to 116 days after sowing of Lolium perenne and Trifolium pratense. The relationships between the mineralization kinetics, the number of 2,4-D degraders, and the diversity of genes encoding 2,4-D/α-ketoglutarate dioxygenase (tfdA) were investigated. The rhizosphere effect on [14C]2,4-D mineralization (50 μg g−1) was shown to be plant species and plant age specific. In comparison with nonplanted soil, there were significant (P < 0.05) reductions in the lag phase and enhancements of the maximum mineralization rate for 25- and 60-day T. pratense soil but not for 116-day T. pratense rhizosphere soil or for L. perenne rhizosphere soil of any age. Numbers of 2,4-D degraders in planted and nonplanted soil were low (most probable number, <100 g−1) and were not related to plant species or age. Single-strand conformational polymorphism analysis showed that plant species had no impact on the diversity of α-Proteobacteria tfdA-like genes, although an impact of 2,4-D application was recorded. Our results indicate that enhanced mineralization in T. pratense rhizosphere soil is not due to enrichment of 2,4-D-degrading microorganisms by rhizodeposits. We suggest an alternative mechanism in which one or more components of the rhizodeposits induce the 2,4-D pathway.
The rhizosphere is the zone of soil directly influenced by the presence of plant roots. It receives inputs of an array of low (e.g., sugars, organic and amino acids, phenolics, and other secondary metabolites)- and high (e.g., cellulose, lignin, mucilage, proteins)-molecular-mass compounds as a result of rhizodeposition and is a zone of complex plant-microbe interactions (56). Seminal work by Hsu and Bartha (23) demonstrated that mineralization of organophosphate pesticides in the rhizosphere is increased relative to that in nonplanted soils. Since this study, numerous reports of rhizosphere-enhanced biodegradation have been published for several classes of organic pollutants, including polyaromatic (2, 35, 48) and aliphatic (5, 38, 48) hydrocarbons, chlorophenols (3, 13), chlorophenoxyacetic acids (3), chlorobenzoates (46), s-triazines (32), and surfactants (29).
One suggested reason for rhizosphere-enhanced biodegradation is the possible presence, as a result of rhizodeposition, of high densities of active and diverse heterotrophic microorganisms on root surfaces, a situation which will facilitate complex degradative pathways through activities of consortia and conjugative horizontal transfer of catabolic genes carried on mobile genetic elements (6). Another suggested reason for rhizosphere-enhanced xenobiotic biodegradation is that components of the rhizodeposits (exudates, root materials, or their decomposition products) may be structural analogs for the pollutant in question (6, 20). It follows that the presence of a pollutant analog in the rhizosphere may select for pollutant-degrading microorganisms capable of directly using the analog for growth, act as a cooxidized substrate during cometabolism, or serve as an effector molecule in the induction of xenobiotic catabolic pathways (43). Furthermore, some investigators (45, 47, 57) hold the view that the rhizodeposition of compounds stimulating xenobiotic biodegradation may not be merely a passive process that fortuitously results in enhanced biodegradation. On the contrary, it is suggested that the plant, on sensing the presence of a xenobiotic in its rhizosphere, actively alters the composition of its rhizodeposits to stimulate xenobiotic degradation and that this response is an extension of the plant's defenses against natural allelopathic chemicals. However, despite much speculation regarding the causes, most experimental studies are descriptive and solely report the phenomenon of rhizosphere-enhanced biodegradation. Thus, empirical studies are required in order to dissect the mechanisms of enhanced biodegradation in the rhizosphere. Only if we gain a better understanding of plant-soil-microorganism-pollutant interactions can we rationally exploit the system for effective rhizoremediation of contaminated land.
One xenobiotic for which rhizosphere-enhanced biodegradation has been reported is 2,4-dichlorophenoxyacetic acid (2,4-D) (3), a herbicide widely used for the control of broad-leaved weeds in both agriculture and domestic applications. Many 2,4-D-degrading bacteria have been isolated both from 2,4-D-exposed (9, 26) and from nonexposed (pristine) (24, 27) soils. Of these, the best studied is the β-proteobacterium Ralstonia eutropha JMP134(pJP4), originally isolated from 2,4-D-exposed soil (9). The initial step in 2,4-D breakdown, as described for R. eutropha JMP134(pJP4), is mediated by an α-ketoglutarate-dependent dioxygenanse, encoded by the tfdA gene, which cleaves the acetate side chain to produce 2,4-dichlorophenol (15, 16, 49). tfdA shows only low homology to one other previously characterized gene (tauD of Escherichia coli [53]). Other 2,4-D-degrading isolates are distributed among the α, β, and γ subdivisions of the class Proteobacteria (24, 27, 34). Isolates in the β- and γ-Proteobacteria possess tfdA-like genes with more than 76% sequence similarity to the canonical tfdA genes in R. eutropha JMP134(pJP4) (34). Until recently it was thought that isolates belonging to the α-Proteobacteria did not possess tfdA-like genes (27, 28, 34), and Kitagawa et al. (28) have characterized a novel gene cluster, cadRABKC, that is responsible for 2,4-D degradation in Bradyrhizobium sp. strain HW13 but shows no similarity to tfdA genes. cadA homologs have been detected in other bradyrhizobia and in α-proteobacterial degraders belonging to the genus Sphingomonas (28). However, more recent research has reported that slow-growing 2,4-D-degrading bacteria isolated from pristine soils and belonging to the Bradyrhizobium-Agromonas-Nitrobacter-Afipia (BANA) cluster within the α-Proteobacteria possess tfdA-like genes with approximately 60% sequence identity to the canonical tfdA gene (25).
The high diversity uncovered in tfdA-like genes suggests that the selection for genotypes may have been operating before the first use of 2,4-D in the early 1940s, and several workers have postulated the existence of an ancestral tfdA gene encoding a degradative enzyme with the highest affinity for a naturally occurring analog of 2,4-D (11, 17, 22, 25). Indeed, the protein encoded by the tfdA-like gene in BANA cluster isolates from pristine soil has a higher affinity for nonchlorinated phenoxyacids than for 2,4-D (25). The facts that pristine soils can mineralize 2,4-D (17) and that 2,4-D-degrading bacteria can be isolated from them (24, 27) suggest that tfdA-like genes are maintained for a function other than 2,4-D degradation. Dunning Hotopp and Hausinger (11) tested a range of alternative substrates for canonical TfdA and concluded that the original substrate may have been a plant-derived compound, such as a cinnamic acid derivative. Given the diversity of compounds produced in the rhizosphere, it is reasonable to hypothesize that other natural substrates for TfdA may be found in the rhizodeposits and that this may explain rhizosphere-enhanced mineralization of 2,4-D. In this study we monitored 2,4-D mineralization kinetics in pristine soils collected from the rhizospheres of Lolium perenne and Trifolium pratense and related them to the number of 2,4-D degraders and the diversity of tfdA-like genes. We present evidence to suggest that enhanced mineralization in the T. pratense rhizosphere is not due to selection of 2,4-D-degrading microorganisms in the rhizosphere by rhizodeposit analogs of 2,4-D but may be due to induction of the 2,4-D pathway by one or more components of the rhizodeposits.
MATERIALS AND METHODS
Soil.
The brown forest soil used in this study was collected from the Macaulay Land Use Research Institute experimental site at Rigg Foot (Sourhope Research Station, Cheviots, Scotland). The Rigg Foot site is upland grassland, dominated by Agrostis capillaris, and has not been exposed to 2,4-D. Detailed vegetation and soil data can be obtained from the Natural Environmental Research Council (United Kingdom) Soil Biodiversity website (http://mwnta.nmw.ac.uk/soilbio/baseline98A.htm). Soil samples were collected from a depth of 10 to 40 cm, sealed in sterile polyethylene bags, and transported to the laboratory, where they were sieved (mesh size, <2.8 mm) and stored at 4°C until use. Some properties of the soil are shown in Table 1.
TABLE 1.
Chemical properties of Sourhope soila
| Soil | pHb | Total organic carbon (mg g−1)c | Freundlich parameters for adsorption of 2,4-dichlorophenold
|
||
|---|---|---|---|---|---|
| Kf | n | KOC | |||
| Nonplanted | 4.03 (0.007) | 24.61 (0.80) | 5.75 (1.02) | 0.83 (0.006) | 233.6 |
| L. perenne rhizosphere | 4.02 (0.003) | 21.03 (0.51) | 5.60 (1.02) | 0.82 (0.007) | 266.3 |
| T. pratense rhizosphere | 4.04 (0.008) | 24.52 (0.71) | 5.47 (1.01) | 0.83 (0.005) | 223.1 |
Soil was sampled from the rhizospheres of L. perenne and T. pratense after 60 days of plant growth. Values are means (standard errors), n = 4.
Determined in CaCl2 after removal of root debris.
Determined by wet oxidation after removal of root debris.
The concentration dependence of 2,4-dichlorophenol adsorption to L. perenne, T. pratense (with incorporated root debris), and nonplanted soil was investigated after 1 h (25°C) over triplicate initial aqueous concentrations of 0.2, 2, 10, 50, and 250 μg of [14C]2,4-dichlorophenol ml−1. Data are expressed as the linear form of the Freundlich equation (r2 > 0.99 in all cases), logS = logKf + nlogA, where S is the adsorbed concentration (in micrograms per gram), A is the aqueous concentration (in micrograms per milliliter), logKf is the intercept, and n is the elasticity of sorption. Koc is calculated as Kf/percent total organic carbon × 100.
Experimental design.
Plants were grown in boiling tubes containing field-moist (0.36 g of H2O g of dry soil−1; 62.3% maximum holding capacity) soil (equivalent dry weight, 12 g). Replicate tubes were planted with 20 seeds of either L. perenne or T. pratense (Herbiseed, Twyford, England) per tube. As controls, additional tubes were left nonplanted but were otherwise treated identically to the planted tubes. Tubes were closed with polyurethane foam bungs and incubated at 20°C with a light-dark cycle of 16 h of light (4,200 lx) and 8 h of dark. The water content was gravimetrically adjusted to the initial field value with sterile distilled water every 2 days. Toward the end of the time course, plants began to show signs of nutrient stress. Therefore, at 87 and 106 days after planting, tubes for each planting treatment received a solution of NH4NO3 and KH2PO4 as part of the 2-day watering to give 35, 25, and 30 μg of N, P and K, respectively, g of soil−1. Additional tubes received no fertilizer. Four replicate tubes per plant treatment were destructively sampled 0, 25, and 60 days after planting, and three replicates were sampled for each plant × fertilizer treatment on day 116: shoots were excised and discarded, and the root was chopped finely with a sterile scalpel and homogenized with the soil (i.e., the entire below-soil-surface contents of the tube were defined as the rhizosphere). Plants within replicate treatments grew equally well (coefficient of variation for dry weight of shoots, <25%). Subsamples were taken for determination of 2,4-D mineralization potential and microbiological and molecular analysis.
Soil biochemical and microbiological analysis.
Soil dehydrogenase activity was determined by the iodonitrotetrazolium chloride (INT; Sigma-Aldrich Co. Ltd., Gillingham, Dorset, United Kingdom) method (51, 55). An aqueous 0.2% (wt/vol) solution of INT was added to the soil sample (dry weight, 0.5 g). Following incubation in the dark (25°C, 48 h), the reaction was terminated by addition of 10 ml of N,N-dimethyl formamide-ethanol (1:1, vol/vol), soil was removed by centrifugation, and the absorbance of the supernatant was determined at 464 nm. The amount of INT formezan (INTF) produced was calculated by reference to an INTF calibration curve.
Soil subsamples (dry weight, 0.5 g) were used as the basis for a 10-fold dilution series for the determination of culturable bacterial numbers and most probable numbers of 2,4-D degraders (MPN2,4-D). For culturable numbers, dilutions (50 μl) were spread onto nutrient agar (Oxoid), and plates were incubated at 25°C and counted after 72 h. For MPN2,4-D, ignition tubes (five replicates for each dilution) containing 1.8 ml of basal medium supplemented with l-[U-14C]2,4-D (0.050 g liter−1; 28.7 kBq liter−1; Sigma-Aldrich Co., Ltd.) were inoculated with dilutions (200 μl). The basal medium (modified from reference 10) was composed of the following (measured in grams per liter unless otherwise stated): Na2HPO4 (2.78), KH2PO4 (1.0), CaNO3 · 4H2O (0.05), (NH4)2SO4 (1.0), MgSO4 · 7H2O (0.2), Casamino Acids (0.005), and trace element solution (1 ml per liter) (27). To obtain 10−1 most-probable-number (MPN) dilutions, soil (0.2 g [dry weight basis]) was weighed directly into ignition tubes. After incubation (9 weeks, 20°C), the remaining radioactivity was quantified, and tubes containing ≤60% of that for the noninoculated controls were scored positive. MPN2,4-D estimates were derived by reference to MPN tables (1). A dilution series constructed from 25-day harvest soil that had previously been exposed to 2,4-D (50 μg g−1) for 36 days was also used to inoculate MPN tubes.
[U-14C]2,4-D and [U-14C]2,4-dichlorophenol fate.
[14C]2,4-D mineralization was determined for soil harvested after 0, 25, 60, or 116 days of plant growth. [14C]2,4-dichlorophenol mineralization was determined for the 25-day harvest sample only. In addition, [14C]2,4-D mineralization kinetics were also recorded for nonplanted soil exposed previously to 2,4-D (50 μg g−1; 36 days). Soil subsamples (4 g [dry weight basis]) in EPA vials (40 ml; gas-tight tetrafluoroethylene-silicone septa; Sigma-Aldrich Co., Ltd.) were amended with [U-14C]2,4-D or [U-14C]2,4-dichlorophenol (50 μg g−1; 250 Bq g−1) in a sufficient volume of sterile distilled water to bring the soil moisture content to 70% of the maximum moisture-holding capacity. A test tube (internal diameter, 75 by 9 mm) containing NaOH (1 ml; 1 M) was placed on the surface of the soil to trap the 14CO2 released. Vials were incubated at 25°C. On sampling days, the NaOH solution was taken for quantification of radioactivity and replaced with fresh NaOH solution. At the end of the mineralization experiment (36 days), soil subsamples (0.5 g [wet weight]) were extracted with water (1 ml) by vortexing (30 s), the soil was removed by centrifugation (16,100 × g, 5 min), 800 μl of the supernatant was decanted, and 200 μl was used for 14C determination. The water-extracted soil pellet was further extracted with methanol (1 ml, 30 s), 1 ml of the supernatant was decanted, and radioactivity in a 400-μl aliquot was determined. The remaining aqueous and methanolic supernatants were stored at −20°C for high-performance liquid chromatography (HPLC) analysis. The recovery efficiency of this sequential extraction method was tested by using soil freshly spiked with 2,4-D and was found to be 99.7% ± 0.5%.
The extracted soil pellet was dried (105°C), and levels of nonextractable residues were determined by CrO3 oxidation as described by Dalal (7) by using NaOH (2 ml; 4 M) to trap the 14CO2 evolved. In all cases, radioactivity was quantified by using a Beckman LS6000TA Liquid Scintillation system programmed to count each sample for 5 min. UltimaGold (Packard Bioscience, Groningen, The Netherlands) was used as the scintillant in a 1:4 (for aqueous extract and 1 M NaOH trap samples) or 1:10 (for methanolic samples) sample-to-cocktail ratio. HionicFluor (Packard) was used as the scintillant for 4 M NaOH traps (sample-to-cocktail ratio, 1:12).
HPLC analysis of extracts was performed as described previously (44).
DNA template isolation.
DNA was extracted from soil exactly according to the method of Griffiths et al. (19) except that bead beating was performed using white quartz sand (0.2 g) mixed with glass beads (0.3 g; 425 to 600 U) (both from Sigma-Aldrich Co. Ltd.) as the matrix and a mini-beadbeater (Biospec Products) set to beat for 30 s at 2,500 rpm.
PCR amplification and cloning.
Two primer sets were used: those designed by Vallaeys et al. (52) by sequence alignment of the tfdA genes from R. eutropha JMP134(pJP4) and Burkholderia sp. strain RASC and those (tfdAα1 [5′-CCGGCGTCGATCTGCGCAAG-3′] and tfdAα2 [5′-GTTGACGACGCGCGCCGACA-3′]) designed to amplify a 359-bp region of the tfdA-like gene (encoding 2,4-D α-ketoglutarate-dependent dioxygenase) recently characterized in α-Proteobacteria (25) by alignment of complete tfdA-like sequences from strains RD5-C2 (GenBank accession number AB074490), HWK12 (AB074491), and HW13 (AB074492). Reaction mixtures contained 1× PCR buffer, 2.5 mM MgCl2, 1× Q solution, 1 U of HotStarTaq DNA polymerase (QIAGEN, Crawley, United Kingdom), deoxynucleoside triphosphate solution (200 μM [each] dATP, dCTP, dGTP, and dTTP), forward and reverse primers (0.4 μM each), 0.5 μl of template DNA, and sterile distilled water up to 25 μl. Reaction mixtures were incubated at 95°C for 15 min to activate the HotStarTaq prior to the start of the temperature program. For the tfdAα primer set, the temperature program was 40 cycles of denaturation (1 min at 94°C), annealing (45 s at 67°C), extension (1 min at 72°C), and final extension (10 min at 72°C). For the Vallaeys et al. (52) primer set, we used a touchdown program: 23 cycles of denaturation (1 min at 94°C), annealing (1 min at 65°C, decreasing by 1°C cycle−1 to 42°C), and extension (1.5 min at 72°C), followed by 17 cycles of denaturation (1 min at 94°C), annealing (1 min at 42°C), extension (1.5 min at 72°C), and final extension (10 min at 72°C). The ability of tfdAα1 and -2 to amplify tfdA-like gene fragments from soil was confirmed by sequencing. Putative partial tfdA-like fragments obtained by PCR amplification of DNA extracted from nonplanted soil before and after 2,4-D treatment (Fig. 5) were ligated into the pGEM-T Easy vector (Promega, Southampton, United Kingdom) and transformed into E. coli JM109 competent cells (Promega), according to the manufacturer's instructions. White colonies were checked for inserts by PCR using primers tfdAα1 and -2. Plasmid DNA extracted (QIAprep Spin Miniprep kit; QIAGEN) from six unique (as determined by single-strand conformational polymorphism [SSCP]) randomly selected clones was used as the basis for sequencing (Comfort Read; MWG Biotech, Ebersberg, Germany) using M13 primers.
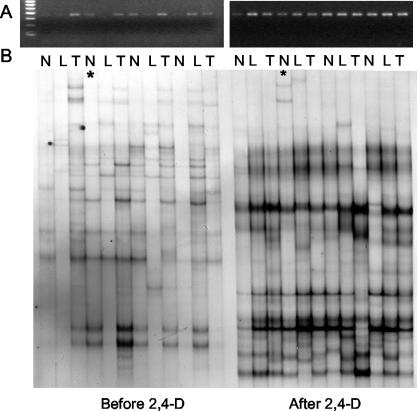
FIG. 5.
PCR amplification using tfdAα primers (A) and subsequent SSCP analysis (B) of tfdA-like genes in Sourhope soil before and after 2,4-D treatment. Each lane contains a PCR product obtained from DNA extracted from an independent, destructively sampled soil sample that either was not planted (N) or was planted with L. perenne (L) or T. pratense (T) and was harvested after 25 days. Asterisked lanes contain putative partial tfdA-like fragments that were cloned subsequently; clones were sequenced to check primer fidelity.
Analysis of cloned tfdA-like sequences.
Cloned tfdA-like sequences were compared with selected reference tfdA-like sequences in the GenBank database. Partial cloned sequences and full reference sequences were aligned by using ClustalX (version 1.81), and reference sequences were trimmed so that only contiguous nucleotides were used for subsequent analysis. A phylogenetic tree was constructed by using the neighbor-joining method and Jukes-Cantor distances (Phylip, version 3.6a3). Bootstrap analysis with 100 replicates was used to place confidence estimates on the tree. E. coli tauD (encoding taurine/α-ketoglutarate dioxygenase [53]) was used as an outgroup.
SSCP analysis.
The SSCP analysis protocol was based on the method described by Schwieger and Tebbe (41). Prior to electrophoretic analysis, aliquots of PCR products were mixed with denaturing loading buffer (95% formamide, 10 mM NaOH, 0.25% [wt/vol] bromophenol blue, 0.25% [wt/vol] xylene cyanol [41]) in a 1:5 sample-to-buffer ratio. Samples were denatured (95°C for 3 min) and then immediately cooled on ice water. Denatured samples (4 μl) were loaded onto a 0.6X SequaGel MD (National Diagnostics, Hull, United Kingdom) gel (40 by 33 cm with 0.4-mm spacers) and run at 600 V and 21 ± 2°C for 20 h by using a Hoefer SQ3 sequencing tank and an EPS601 (Amersham Pharmacia Biotech) power pack. The gel was silver stained according to the Promega Silver Sequence DNA sequencing system (39). The SSCP gels were composed of sample lanes with several conformer bands per lane. Band positions were assigned and scored either 1, for present, or 0, for absent. The resulting binary matrix was used as the basis for cluster analysis using the matching binary similarity coefficient and single linkage method (Intercooled Stata 7 for Windows; Stata Corporation, College Station, Tex.).
Nucleotide sequence accession numbers.
The nucleotide sequences determined in this study have been deposited in the GenBank database under accession numbers AY193866 to AY193870.
RESULTS
Culturable numbers and dehydrogenase activity.
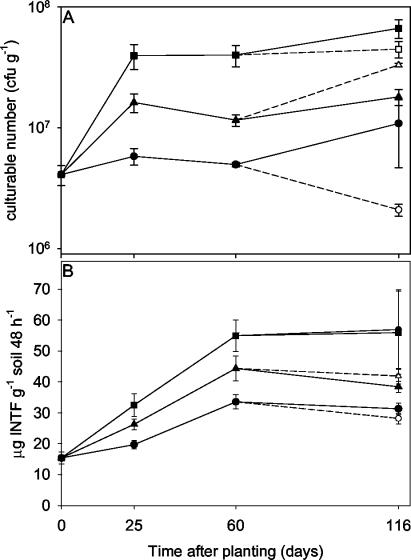
In soil harvested after only 25 days of plant growth, plant treatment had a significant effect (P = 0.006 by analysis of variance [ANOVA]) on culturable numbers (Fig. 1A). The effect was most pronounced in the T. pratense rhizosphere (CFU were 4.0 × 107, 1.6 × 107, and 5.8 × 106 g−1 for T. pratense, L. perenne, and nonplanted soils, respectively). The T. pratense rhizosphere also showed significantly increased dehydrogenase activity on day 25 (P = 0.017 by ANOVA) and day 60 (P = 0.013 by ANOVA) (Fig. 1B). Results for both culturable number and dehydrogenase activity followed the same treatment order (from highest to lowest, T. pratense, L. perenne, and nonplanted soils).
FIG. 1.
Effects of plant species and plant age on the number of culturable bacteria (A) and dehydrogenase activity (B) in soil sampled from the rhizosphere of L. perenne (▴) or T. pratense (▪) or in nonplanted controls (•). Soils were fertilized on days 87 and 106 with NH4NO3 and KH2PO4 to give 35 μg of N g−1, 25 μg of P g−1, and 30 μg of K g−1. Open symbols, fertilized treatments. Error bars, standard errors.
MPN2,4-D.
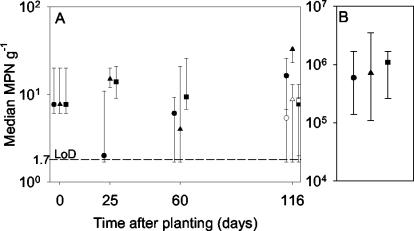
Before the pristine soil was exposed to 2,4-D, MPN2,4-D were low (<100 g−1; below detectable limits in some replicates) for material harvested either from nonplanted soil or from L. perenne- or T. pratense-planted soil (Fig. 2A). In contrast to culturable numbers or dehydrogenase activity (Fig. 1), MPN2,4-D were not related to planting treatment (P = 0.063 by the Kruskal-Wallis test) or harvest day (P = 0.33 by the Kruskal-Wallis test) (Fig. 2A). However, soil sampled from 25-day-old rhizospheres or nonplanted controls and incubated in the presence of 2,4-D (50 μg g−1, 36 days) had significantly (P < 0.001 by the Kruskal-Wallis test) higher MPN2,4-D (Fig. 2B) than non-2,4-D-exposed soil (Fig. 2A). In 2,4-D-exposed soil, median MPN2,4-D ranged from 6 × 105 g−1 (nonplanted) to 1 × 106 g−1 (T. pratense), although plant treatment did not have a significant effect (P = 0.91 by the Kruskal-Wallis test).
FIG. 2.
(A) Effects of plant species and plant age on MPN2,4-D in pristine soil sampled from the rhizosphere of L. perenne (▴) or T. pratense (▪) or from nonplanted controls (•). Soils were fertilized on days 87 and 106 with NH4NO3 and KH2PO4 to give 35 μg of N g−1, 25 μg of P g−1, and 30 μg of K g−1. Open symbols, fertilized treatments. (B) MPN2,4-D in soil sampled from 25-day-old rhizospheres or nonplanted controls and incubated in the presence of 50 μg of 2,4-D g−1 for 36 days. Error bars, maximum and minimum values. LoD, limit of detection.
2,4-D and 2,4-dichlorophenol mineralization and 2,4-D 14C mass balance.
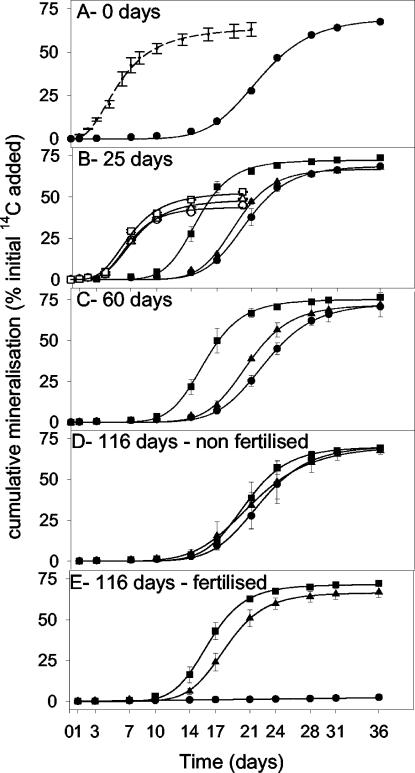
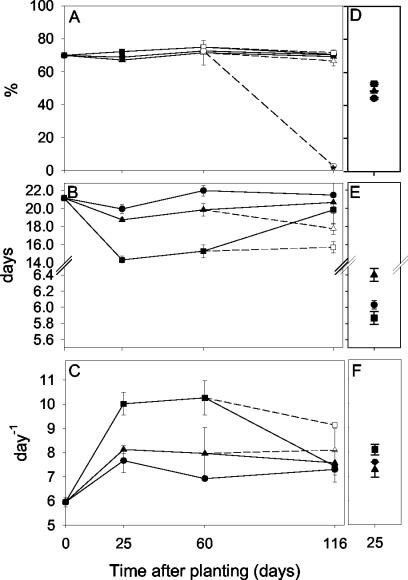
Cumulative 14CO2 evolution from nonplanted, L. perenne-planted, or T. pratense-planted soil amended either with [U-14C]2,4-D over the entire time course (0 to 116 days) (Fig. 3A to E) or with [U-14C]2,4-dichlorophenol at the day-25 harvest (Fig. 3B) was determined. Mineralization of 2,4-D by nonplanted soil previously exposed to 2,4-D (50 μg g−1, 36 days) is shown for comparison (Fig. 3A). To enable a quantitative comparison of mineralization kinetics, curves were fitted to the function Y = a[1 + (t/t0)b]−1, and parameters (asymptote [a], time at which the maximum rate is reached [t1], and maximum rate) were compared for each sampling time and treatment (Fig. 4). The t1 and rate parameters were calculated from the function as described previously (42). Generally, cumulative mineralization curves were sigmoidal, except for that of 2,4-D in fertilized nonplanted soil (day 116 [Fig. 3E]), where mineralization was minimal (<2.5% mineralized in 36 days). For soils not previously exposed to 2,4-D, 2,4-D mineralization curves were characterized by a lag phase prior to the onset of exponential 14CO2 evolution, during which time mineralization occurred at a low and linear rate (∼0.2% day−1). This is in contrast to results for soil preexposed to 2,4-D (containing an enriched 2,4-D-degrading population of 6 × 105 MPN g−1) (Fig. 2B), where exponential 14CO2 evolution was immediate (Fig. 3A). Nevertheless, independently of whether the soil had previously been exposed to 2,4-D, the percent 14CO2 evolution always approached an asymptote of ∼70 (Fig. 3 and 4A). The mineralization asymptote was also independent of planting treatment (Fig. 3 and 4A). However, there was a pronounced reduction in the length of the lag phase for T. pratense-planted versus L. perenne-planted or nonplanted soil, particularly on days 25 and 60 (Fig. 3B and C). The reduction in the lag phase length is reflected by the parameter t1 (expressed in days). For example, t1 is 22.0 ± 0.6 or 15.3 ± 0.7 days for day-60-harvested nonplanted or T. pratense-planted soil, respectively (Fig. 4B). The rhizosphere of T. pratense also had a significant (P < 0.05 by ANOVA) effect on the maximum rate of 2,4-D mineralization on harvest days 25 and 60 (e.g., for day 25, the maximum rate for the T. pratense rhizosphere was 10.0% day−1 while that for nonplanted soil was 7.6% day−1 [Fig. 4C]). By day 116, the T. pratense rhizosphere effect persisted only in fertilized treatments (Fig. 4B and C).
FIG. 3.
Effects of plant species, plant age at harvest (0, 25, 60, or 116 days old), and NPK fertilization on mineralization of 2,4-D (solid symbols) or 2,4-dichlorophenol (open symbols) at 50 μg g−1 in soil sampled from the rhizosphere of L. perenne (▴), the T. pratense rhizosphere (▪), or nonplanted controls (•). Dashed line in panel A shows mineralization of 2,4-D in nonplanted soil previously exposed to 50 μg of 2,4-D g−1 for 36 days. Soils were fertilized on days 87 and 106 with NH4NO3 and KH2PO4 to give 35 μg of N g−1, 25 μg of P g−1, and 30 μg of K g−1. Error bars, standard errors.
FIG. 4.
Effects of plant species and plant age on 2,4-D (A to C) and 2,4-dichlorophenol (D to F) mineralization parameters in soil sampled from the rhizosphere of L. perenne (▴), T. pratense (▪), or nonplanted controls (•). Effects are expressed as asymptotic percentage mineralized (A and D), time point at which the maximum mineralization rate is reached (t1) (B and E), or calculated maximum rate (C and F). Soils were fertilized on days 87 and 106 with NH4NO3 and KH2PO4 to give 35 μg of N g−1, 25 μg of P g−1, and 30 μg of K g−1. Open symbols represent fertilized treatments. Error bars, standard errors.
In contrast to that of 2,4-D, 2,4-dichlorophenol mineralization (Fig. 3B) proceeded after a shorter lag phase (t1, 5.8 to 6.4 days [Fig. 4B]), but there was no significant effect (P > 0.05 by ANOVA) of either L. perenne or T. pratense planting on any of the parameters describing mineralization (Fig. 4D to F).
Quantification of the fate of the 2,4-D 14C (as 14CO2, water-extractable 14C, methanol-extractable 14C, and nonextractable residues released by wet oxidation) at the end of the experiment revealed, except for the fertilized nonplanted soil, no significant difference (P > 0.1) in the partitioning of the 14C among the four fractions tested (data not shown). Less than 3% of the 14C could be water or methanol extracted, and the remainder of the 14C (15 to 27%) could be recovered only by wet oxidation. Thus, it is likely that the 2,4-D was completely degraded and that the remaining nonextractable 14C fraction was associated with the soil organic matter and microbial biomass. For nonplanted fertilized soils (day 116), where mineralization was inhibited, the 14C could be accounted for as follows: water extractable, 36.7% ± 2%; methanol extractable, 20.2% ± 2%; nonextractable but released by wet oxidation, 50.2% ± 2%. Subsequent HPLC analysis revealed that the water- and methanol-extractable 14C was 2,4-D associated. The reason for the inhibition of 2,4-D transformation and mineralization by NPK fertilization in nonplanted soil is not known, although we speculate that addition of relatively high concentrations of inorganic salts caused osmotic stress. In planted soil, NPK did not have the same detrimental effect, presumably because the plants took up a significant proportion of the fertilizer.
PCR-SSCP analysis of diversity of tfdA-like genes.
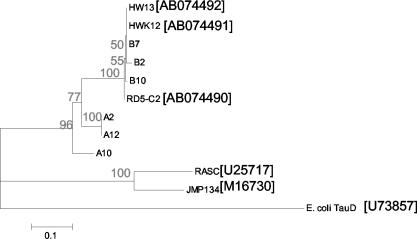
For PCR of DNA extracted from soil, we initially used primers designed by sequence alignment of tfdA genes from the β-proteobacterial degraders R. eutropha JMP134(pJP4) and Burkholderia sp. strain RASC (52). However, we were not able to obtain any PCR products with these primers, even for soil samples that had previously been exposed to 2,4-D (50 μg g−1, 36 days) and, according to the MPN2,4-D analysis, contained approximately 106 degraders g−1 (Fig. 2B). In preliminary experiments using template DNA extracted from soil inoculated with a 10-fold dilution series of Burkholderia sp. strain RASC (50), we determined that the detection limit for the PCR was approximately 104 tfdA copies g−1 of soil (data not shown). Therefore, if the 2,4-D-degrading population present in the soil possessed the β- and γ-proteobacterial tfdA-like gene, we would expect to be able to obtain a PCR product in the 2,4-D-exposed soil. Even when we used DNA extracted from positive MPN2,4-D tubes as a template, we did not achieve amplification. However, PCR of DNA extracted from soil using the primers (tfdAα1 and -2) designed to target tfdA-like genes from α-Proteobacteria yielded faint PCR products of the expected size (359 bp) from soil not previously exposed to 2,4-D and more-distinct products from soil exposed to 2,4-D (50 μg g−1) for 36 days (Fig. 5A). Furthermore, DNA was extracted from the highest-dilution (10−5 and 10−6) positive MPN2,4-D tubes set up with 2,4-D-exposed soil sampled at the end of the mineralization experiments. This DNA was used as a basis for PCR with the tfdAα primers; of the 26 samples tested, 22 yielded a product of the correct size, suggesting that the α-proteobacterial tfdA-like gene was numerically important in soil exposed to 2,4-D. To check primer fidelity, putative soil tfdA-like PCR products were cloned and six clones were sequenced. Phylogenetic comparisons with reference strain tfdA genes (representatives of β- and α-proteobacterial degraders) placed the clones on the α-proteobacterial branch of the tfdA tree (Fig. 6). The partial tfdA-like DNA sequences cloned from soil had 83.2 to 99.4% identity to the common partial tfdA-like sequence of the α-proteobacterial strain RD5-C2 (25) (GenBank accession number AB074490).
FIG. 6.
Phylogenetic positions of tfdA sequences cloned from Sourhope soil among tfdA sequences from R. eutropha JMP134(pJP4), Burkholderia sp. strain RASC, and reference strains HW12, HW13, and RD2-C5. Strains HW12, HW13, and RD2-C5 are members of the BANA cluster of the α-Proteobacteria most closely related to Bradyrhizobium spp. E. coli tauD encodes taurine/α-ketoglutarate dioxygenase. The neighbor-joining dendrogram (Jukes-Cantor distances) was constructed from reference sequences and common partial sequences (357 bp) of tfdA PCR amplified by using primers tfdAα1 and tfdAα2. Soil clone designations begin with “B ” (for “before 2,4-D application”) or “A ” (for “after 2,4-D application”). Bootstrap confidence limits (percentages) are given at each branch. Scale bar represents a Jukes-Cantor distance of 0.1. GenBank accession numbers are given in brackets after strain designations.
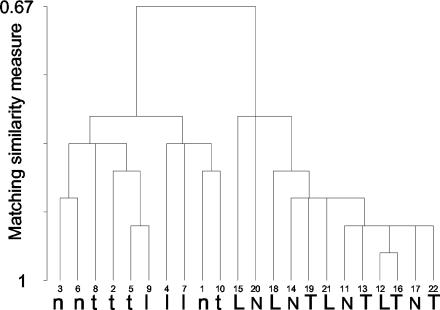
SSCP analysis of the resulting PCR products revealed differences in the position and abundance of tfdA-like conformer bands between non-2,4-D-exposed and 2,4-D-exposed soils which were discernible by eye (Fig. 5B). Indeed, hierarchical cluster analysis of SSCP band patterns (Fig. 7) confirmed that the samples clustered according to 2,4-D exposure history but that plant treatment had no effect on the diversity of tfdA-like genes either in non-2,4-D-exposed or in 2,4-D-exposed soils.
FIG. 7.
Hierarchical cluster analysis (using matching binary similarity distances and the single linkage method) of the tfdA-like SSCP banding patterns shown in Fig. 5B. N, L, and T, replicate sample lanes from nonplanted, L. perenne-planted, and T. pratense-planted soils, respectively. Lowercase letters, non-2,4-D-exposed samples; capital letters, 2,4-D-exposed samples.
DISCUSSION
After only 25 days of plant growth, culturable microbial numbers and activity were increased in soil planted with T. pratense or L. perenne. This is known as the “rhizosphere effect,” documented by many authors who report the 2- to 10-fold increases in microbial number and activity in planted soil over nonplanted soil seen in our present study (4, 43). The rhizosphere effect was more pronounced in the T. pratense than in the L. perenne rhizosphere despite the fact that L. perenne produced a root system that was more extensive and had greater biomass than that of T. pratense (data not shown). The reason for the difference between the microbial properties of L. perenne- and T. pratense-planted soils is unknown. We were not able to detect any effect of the rhizospheres on the organic carbon content of the soil (Table 1). However, it can be supposed that T. pratense produced exudates either in greater quantity (although this was not detected by the wet oxidation organic carbon method) or of superior quality to those of L. perenne, or both.
Aside from the rhizodeposit-mediated impact on gross microbial parameters, such as culturable heterotrophs or activity of a global respiratory enzyme (dehydrogenase), our main aim was to dissect the impact of plant roots on 2,4-D mineralization and numbers and diversity of 2,4-D degraders. Our hypothesis was that plant roots deposit compounds to the rhizosphere that are natural analogs of the 2,4-D pathway and enhance 2,4-D biodegradation by serving as either (i) a substrate for 2,4-D catabolic enzymes, (ii) a cooxidized substrate during cometabolism, or (iii) an effector molecule in the induction the pathway. The results of the mineralization assays show that there was indeed an effect of the rhizosphere on 2,4-D biodegradation but that the effect was dependent on plant species (T. pratense) and plant age (for nonfertilized soil, the effect was confined to younger [<60-day] plants).
If enhanced 2,4-D mineralization was due to the presence of rhizodeposit 2,4-D analogs that could be broken down by Tfd enzymes and used as a source of carbon for growth, then we would expect T. pratense to have an impact on the numbers and diversity of 2,4-D degraders. Although activities and culturable numbers increased in the rhizosphere, MPN2,4-D in the pristine soil were low (<100 g−1) and were not affected by plant treatment. In contrast, soil previously exposed to 2,4-D (50 μg g−1) gave MPN2,4-D estimates of ∼1 × 106 g−1, showing that the MPN2,4-D method used was effective at detecting increases in 2,4-D degrader numbers in response to a selective pressure. Other workers (8) have also shown that MPN assays can sensitively detect the enrichment of 2,4-D degraders in aquifer sediments exposed to phenoxy acid herbicides. If rhizodeposition was selectively enriching the pristine soil, then the rhizosphere-to-nonrhizosphere ratio (R:S) for 2,4-D degraders would exceed that for the total heterotrophs. In an early study, Sandmann and Loos (40) used an MPN method to determine R:S values for 2,4-D degraders in the rhizospheres of sugarcane and African clover. They found that rhizosphere soil sampled from sugarcane, with no previous exposure to phenoxyacetic acid herbicides, had an enriched 2,4-D degrading population (R:S2,4-D = 105; R:Sheterotroph = 6). Similarly, Fang et al. (12) reported an enrichment of phenanthrene degraders in soil planted with various grass species. The results of those studies are in contrast to our findings, since we calculate an R:S2,4-D of 4.27 and a R:Sheterotroph of 7.44 based on day-25 and -60 mean (heterotroph) or median (MPN2,4-D) values for T. pratense-planted and nonplanted soils.
The soil used for our experiments was collected from an upland grassland with no history of 2,4-D application. The field site was remote from areas used for arable agriculture and therefore could not have received 2,4-D inputs from short-range spray drift. Since 2,4-D is nonvolatile and hydrophilic, it is unlikely to undergo long-range atmospheric transport and global distillation such as has been reported for other more volatile and hydrophobic xenobiotics (58). Thus, the fact that that this pristine soil had intrinsic 2,4-D mineralization ability (an observation common to other pristine soils [17]) is consistent with the idea that naturally occurring analogs of 2,4-D do exist, can be used directly as substrates for growth (17), and are responsible for the maintenance of the low numbers of 2,4-D-mineralizing microorganisms recorded here. However, the MPN evidence suggests that the putative analog was not an exclusive property of rhizosphere soil and therefore did not result directly from rhizodeposition.
Evidence from PCR analysis of DNA extracted from soil and positive MPN2,4-D tubes with two different primer sets (that targeting the β- and γ-proteobacterial tfdA and that targeting α-proteobacterial tfdA) suggests that 2,4-D degraders possessing the α-proteobacterial tfdA-like genes were present and numerically important in Sourhope soil. As indicated earlier, tfdA-like genes have been characterized only recently (25) in oligotrophic α-proteobacterial 2,4-D degraders isolated from pristine soils of Hawaii, Saskatchewan, and Japan (25, 27). 2,4-D mineralizers have proved difficult to isolate from pristine soils (17, 27), probably because of their slow-growing habits and sensitivity to high concentrations of organic nutrients (27). Indeed, despite many attempts, we were not able to resolve 2,4-D mineralizers from active enrichment cultures. However, using culture-independent PCR and subsequent SSCP analysis of tfdA-like PCR products, we were able to demonstrate the presence and also the diversity of tfdA-like genes in a pristine Scottish soil, adding further weight to the claim of Kamagata et al. (27) that this new class of 2,4-D degraders is widespread in nature.
We used SSCP analysis to examine the effect of the rhizosphere on the sequence diversity of tfdA-like genes. In our analysis, we also included soil from the three plant treatments that had previously been exposed to 2,4-D. We found that prior 2,4-D exposure had a pronounced effect on the diversity of 2,4-D-degradative genes, thus demonstrating the ability of the method to detect the impact of a selective pressure on the 2,4-D-degrading population. However, no plant treatment effect on diversity was detectable either in nonexposed or in 2,4-D-exposed soils. Taken together, the MPN2,4-D and PCR-SSCP evidence suggests that the rhizosphere does not exert a selective pressure on 2,4-D-degrading microorganisms. Rather the 2,4-D degraders present in Sourhope soil form a near-constant proportion of the soil community (both in numbers and in composition) and, in the absence of 2,4-D, are oblivious to the presence of a plant root. When 2,4-D is added to the soil, MPN2,4-D and PCR-SSCP evidence suggests that a subpopulation of 2,4-D degraders initially present in soil utilizes 2,4-D for growth; however, the composition of this population of 2,4-D degraders also is not affected by plant treatment. In other words, the same subpopulation is degrading 2,4-D in the three treatments, but it is able to mineralize with a shorter lag time and at a higher rate in soil previously planted with T. pratense.
If the plant root (at least in the plant-soil system used in the present study) does not exert a direct selective pressure as a growth substrate on either the number or the diversity of 2,4-D degraders, an alternative explanation for the enhanced mineralization of 2,4-D in soil harvested from the T. pratense rhizosphere is required. Alternative explanations (alluded to earlier) include the involvement of an analog as an inducer or cometabolite, rhizodeposit-mediated promotion of the growth of 2,4-D degraders, or the operation of less-specific mechanisms such as rhizosphere-enhanced horizontal transfer of 2,4-D catabolic genes or the action of phytosurfactants to increase the concentration and bioavailability of 2,4-D in the soil aqueous phase. We think that some of these explanations are unlikely. First, the exponential kinetics of 2,4-D mineralization and the recorded increase in MPN2,4-D point toward a growth-linked, not a cometabolic process. Second, mating experiments with α-proteobacterial 2,4-D-degrading strains (HW13, HWK12, HWK13, and BTH) have shown that the tfdA-like gene is not transmissible (27). Third, there was no evidence of a bioavailability effect, as we could detect no difference in the parameters describing the partitioning of 2,4-dichlorophenol between the soil solid and aqueous phases (Table 1).
Therefore, one explanation not discounted relates to rhizodeposit-mediated promotion of the growth of 2,4-D degraders. For example, since it has been shown that TfdA has affinity for nonchlorinated substrates (11, 25), it may be that, once induced by the presence of 2,4-D, TfdA (or other 2,4-D pathway enzymes) was able to accept Trifolium rhizodeposits as substrates, thereby enhancing the growth rate of 2,4-D degraders. However, the explanation we favor is that a component of the rhizodeposits is able to act as an inducer of the tfd pathway. The regulation of the pathway in the newly discovered tfdA-like-gene-possessing α-proteobacterial degraders has not yet been characterized. Until we know more, we can only discuss the possibility of a rhizodeposit inducer in the context of our knowledge of the canonical tfd pathway in β-Proteobacteria. We know this pathway to be regulated by a LysR-type transcriptional regulatory protein encoded by tfdR (and, in JMP134, the identical tfdS gene) (31, 33, 54). The inducing effector molecule of the pathway is thought to be the intermediate 2,4-dichloromuconate (14). Presumably, on initial exposure to 2,4-D, low-level constitutive expression of the tfd pathway results in intracellular accumulation of the effector molecule and full induction of the pathway. In the present study, the low linear rate of 14CO2 evolution during the lag phase and prior to the onset of exponential mineralization (Fig. 3) would be consistent with this process. It is reasonable to hypothesize that muconate derivatives will be present in the rhizosphere as a result of microbial metabolism of aromatic rhizodeposits. Leigh et al. (30) have shown that a variety of phenolic compounds are produced in the rhizosphere, particularly during the decay of dead roots. cis,cis-Muconate is the product of catechol metabolism via the central ortho-cleavage pathway (37), and many substituted phenols and aromatic acids (produced by rhizodeposition) could be degraded through this route, thus producing nonchlorinated cis,cis-muconates (21). Ogawa et al. (36) have reported that the LysR-type regulator of chlorocatechol degradation in R. eutropha can be activated by both chlorinated and nonchlorinated muconate (36). Thus, we consider it likely that an analog of 2,4-dichloromuconate produced in the T. pratense rhizosphere may activate transcription of the tfd genes by interaction with the regulatory TfdR-like protein.
2,4-D mineralization kinetics were both plant species and plant age specific. Boyle and Shann (3) also reported a specific plant effect on 2,4-D biodegradation: mineralization was enhanced to a greater extent in soil collected from a monocotyledon (e.g., L. perenne) rhizosphere than in soil from a dicotyledon (e.g., T. pratense) rhizosphere. This is the inverse of the findings of our present study. Because Boyle and Shann (3) did not determine the numbers or diversity of 2,4-D degraders in their study, it is difficult to explain the discrepancy in the results. However, in agreement with our work, Boyle and Shann found that the rhizosphere effect was specific for the first (tfdA-encoded) step of 2,4-D degradation, as there was no effect of either the monocot or dicot rhizosphere on the mineralization of the first metabolite of 2,4-D breakdown, 2,4-dichlorophenol. The quality and quantity of rhizodeposition are known to differ between plant species and also to be dependent on the growth stage of the plant (18). Thus, the rhizosphere effect of T. pratense may be due not solely to root death, as suggested by Leigh et al. (30), but also to more actively extruded components of the rhizodeposits. In order to narrow down the identity of the active compound(s) in the T. pratense rhizodeposits, we are currently conducting experiments to identify in which fraction of the rhizodeposits it predominates (exudates versus root structural compounds) and to examine what edaphic factors (e.g., pH, nitrogen content) control its production.
Acknowledgments
This research was funded by the EU FP 5 grant “HERBICBIOREM” (proposal QLRT-1999-00041) and a Natural Environmental Research Council grant (NER/B/S/2002/00518).
We thank Jem Stach for help with SSCP and phylogenetic analysis.
REFERENCES
- 1.Alexander, M. 1982. Most probable number method for microbial populations, p. 815-820. In A. L. Page, R. H. Miller, and D. R. Keeney (ed.), Methods of soil analysis, part 2. Chemical and microbiological properties, 2nd ed. American Society for Agronomy, Madison, Wis.
- 2.Binet, P., J. M. Portal, and C. Leyval. 2000. Dissipation of 3-6-ring polycyclic aromatic hydrocarbons in the rhizosphere of ryegrass. Soil Biol. Biochem. 32:2011-2017. [Google Scholar]
- 3.Boyle, J., and J. R. Shann. 1995. Biodegradation of phenol, 2,4-DCP, 2,4-D and 2,4,5-T in field-collected rhizosphere and non-rhizosphere soils. J. Environ. Qual. 24:782-785. [Google Scholar]
- 4.Buyer, J. S., D. P. Roberts, and E. Russek-Cohen. 2002. Soil and plant effects on microbial community structure. Can. J. Microbiol. 48:955-964. [DOI] [PubMed] [Google Scholar]
- 5.Chaineau, C. H., J. L. Morel, and J. Oudot. 2000. Biodegradation of fuel oil hydrocarbons in the rhizosphere of maize. J. Environ. Qual. 29:569-578. [Google Scholar]
- 6.Crowley, D. E., S. Alvey, and E. S. Gilbert. 1997. Rhizosphere ecology of xenobiotic-degrading microorganisms, p. 20-36. In E. L. Kruger, T. A. Anderson, and J. R. Coats (ed.), Phytoremediation of soil and water contaminants. American Chemical Society, Washington, D.C.
- 7.Dalal, R. C. 1979. Simple procedure for the determination of total carbon and its radioactivity in soils and plant materials. Analyst 104:151-154. [Google Scholar]
- 8.de Lipthay, J. R., N. Tuxen, K. Johnsen, L. H. Hansen, H.-J. Albrechtsen, P. L. Bjerg, and J. Aamand. 2003. In situ exposure to low herbicide concentrations affects microbial population composition and catabolic gene frequency in an aerobic shallow aquifer. Appl. Environ. Microbiol. 69:461-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Don, R. H., and J. M. Pemberton. 1981. Properties of six pesticide degradation plasmids isolated from Alcaligenes paradoxus and Alcaligenes eutrophus. J. Bacteriol. 145:681-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dorn, E., M. Hellwig, W. Reineke, and H. J. Knackmuss. 1974. Isolation and characterisation of a 3-chlorobenzoate degrading pseudomonad. Arch. Microbiol. 99:61-70. [DOI] [PubMed] [Google Scholar]
- 11.Dunning Hotopp, J. C., and R. P. Hausinger. 2001. Alternative substrates of 2,4-dichlorophenoxyacetate/α-ketoglutarate dioxygenase. J. Mol. Catal. B 15:155-162. [Google Scholar]
- 12.Fang, C., M. Radosevich, and J. J. Fuhrmann. 2001. Atrazine and phenanthrene degradation in grass rhizosphere soil. Soil Biol. Biochem. 33:671-678. [Google Scholar]
- 13.Ferro, A. M., R. C. Sims, and B. Bugbee. 1994. Hycrest crested wheatgrass accelerates the degradation of pentachlorophenol in soil. J. Environ. Qual. 23:272-279. [DOI] [PubMed] [Google Scholar]
- 14.Filer, K., and A. R. Harker. 1997. Identification of the inducing agent of the 2,4-dichlorophenoxyacetic acid pathway encoded by plasmid pJP4. Appl. Environ. Microbiol. 63:317-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fukumori, F., and R. P. Hausinger. 1993. Alicaligenes eutrophus JMP134 “2,4-dichlorophenoxyacetate monooxygenase” is an α-ketoglutarate-dependent dioxygenase. J. Bacteriol. 175:2083-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fukumori, F., and R. P. Hausinger. 1993. Purification and characterization of 2,4-dichlorophenoxyacetate/α-ketoglutarate dioxygenase. J. Biol. Chem. 268:24311-24317. [PubMed] [Google Scholar]
- 17.Fulthorpe, R. R., A. N. Rhodes, and J. M. Tiedje. 1996. Pristine soils mineralize 3-chlorobenzoate and 2,4-dichlorophenoxyacetate via different microbial populations. Appl. Environ. Microbiol. 62:1159-1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gransee, A., and L. Wittenmayer. 2000. Qualitative and quantitative analysis of water-soluble root exudates in relation to plant species and development. J. Plant Nutr. Soil Sci. 163:381-385. [Google Scholar]
- 19.Griffiths, R. I., A. S. Whiteley, A. O'Donnell, and M. J. Bailey. 2000. Rapid method for coextraction of DNA and RNA from natural environments for analysis of ribosomal DNA and rRNA-based microbial community composition. Appl. Environ. Microbiol. 66:5488-5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haby, P. A., and D. E. Crowley. 1996. Biodegradation of 3-chlorobenzoate as affected by rhizodeposition and selected carbon substrates. J. Environ. Qual. 25:304-310. [Google Scholar]
- 21.Harwood, C. S., and R. E. Parales. 1996. The β-ketoadipate pathway and the biology of self-identity. Annu. Rev. Microbiol. 50:553-590. [DOI] [PubMed] [Google Scholar]
- 22.Hogan, D. A., D. H. Buckley, C. H. Nakatsu, T. M. Schmidt, and R. P. Hausinger. 1997. Distribution of the tfdA gene in soil bacteria that do not degrade 2,4-dichlorophenoxyacetic acid (2,4-D). Microb. Ecol. 34:90-96. [DOI] [PubMed] [Google Scholar]
- 23.Hsu, T.-S., and R. Bartha. 1979. Accelerated mineralization of two organophosphate insecticides in the rhizosphere. Appl. Environ. Microbiol. 37:36-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Itoh, K., R. Kanda, Y. Momoda, Y. Sumita, Y. Kamagata, K. Suyama, and H. Yamamoto. 2000. Presence of 2,4-D-catabolizing bacteria in a Japanese arable soil that belong to BANA (Bradyrhizobium-Agromonas-Nitrobacter-Afipia) cluster in α-Proteobacteria. Microb. Environ. 15:113-117. [Google Scholar]
- 25.Itoh, K., R. Kanda, Y. Sumita, H. Kim, Y. Kamagata, K. Suyama, H. Yamamoto, R. P. Hausinger, and J. M. Tiedje. 2002. tfdA-like genes in 2,4-dichlorophenoxyacetic acid-degrading bacteria belonging to the Bradyrhizobium-Agromonas-Nitrobacter-Afipia cluster in α-Proteobacteria. Appl. Environ. Microbiol. 68:3449-3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ka, J. O., W. E. Holben, and J. M. Tiedje. 1994. Genetic and phenotypic diversity of 2,4-dichlorophenoxyacetic acid (2,4-D)-degrading bacteria isolated from 2,4-D-treated fields. Appl. Environ. Microbiol. 60:1106-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kamagata, Y., R. R. Fulthorpe, K. Tamura, H. Takami, L. J. Forney, and J. M. Tiedje. 1997. Pristine environments harbor a new group of oligotrophic 2,4-dichlorophenoxyacetic acid-degrading bacteria. Appl. Environ. Microbiol. 63:2266-2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kitagawa, W., S. Takami, K. Miyauchi, E. Masai, Y. Kamagata, J. M. Tiedje, and M. Fukuda. 2002. Novel 2,4-dichlorophenoxyacetic acid degradation genes from oligotrophic Bradyrhizobium sp. strain HW13 isolated from a pristine environment. J. Bacteriol. 184:509-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Knaebel, D. B., and J. R. Vestal. 1992. Effects of intact rhizosphere microbial communities on the mineralization of surfactants in surface soils. Can. J. Microbiol. 38:643-653. [Google Scholar]
- 30.Leigh, M. B., J. S. Fletcher, X. Fu, and F. J. Schmitz. 2002. Root turnover: an important source of microbial substrates in rhizosphere remediation of recalcitrant contaminants. Environ. Sci. Technol. 36:1579-1583. [DOI] [PubMed] [Google Scholar]
- 31.Leveau, J. H. J., and J. R. van der Meer. 1996. The tfdR gene product can successfully take over the role of the insertion element-inactivated tfdT protein as a transcriptional activator of the tfdCDEF gene cluster, which encodes chlorocatechol degradation in Ralstonia eutropha JMP134(pJP4). J. Bacteriol. 178:6824-6832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marchand, A.-L., S. Piutti, B. Lagacherie, and G. Soulas. 2002. Atrazine mineralization in bulk soil and maize rhizosphere. Biol. Fertil. Soil 35:288-292. [Google Scholar]
- 33.McFall, S. M., S. A. Chugani, and A. M. Chakrabarty. 1998. Transcriptional activation of the catechol and chlorocatechol operons: variations on a theme. Gene 223:257-267. [DOI] [PubMed] [Google Scholar]
- 34.McGowan, C., R. Fulthorpe, A. Wright, and J. M. Tiedje. 1998. Evidence for interspecies gene transfer in the evolution of 2,4-dichlorophenoxyacetic acid degraders. Appl. Environ. Microbiol. 64:4089-4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miya, R. K., and M. K. Firestone. 2000. Phenanthrene-degrader community dynamics in rhizosphere soil from a common annual grass. J. Environ. Qual. 29:584-592. [Google Scholar]
- 36.Ogawa, N., S. M. McFall, T. J. Klem, K. Miyashita, and A. M. Chakrabarty. 1999. Transcriptional activation of the chlorocatechol degradative genes of Ralstonia eutropha NH9. J. Bacteriol. 181:6697-6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parsek, M. R., D. L. Shinabarger, R. K. Rothmel, and A. M. Chakrabarty. 1992. Roles of CatR and cis,cis-muconate in activation of the catBC operon, which is involved in benzoate degradation in Pseudomonas putida. J. Bacteriol. 174:7798-7806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pichtel, J., and P. Liskanen. 2001. Degradation of diesel fuel in rhizosphere soil. Environ. Eng. Sci. 18:145-157. [Google Scholar]
- 39.Promega. 2000. Silver Sequence DNA sequencing system. Technical manual 023. [Online.] http://www.shpromega.com.cn/tm023.pdf.
- 40.Sandmann, E. R., and M. A. Loos. 1984. Enumeration of 2,4-D-degrading microorganisms in soils and crop plant rhizospheres using indicator media: high populations associated with sugarcane (Saccharum officinarum). Chemosphere 13:1073-1084. [Google Scholar]
- 41.Schwieger, F., and C. C. Tebbe. 1998. A new approach to utilize PCR-single-strand-conformation polymorphism for 16S rRNA gene-based microbial community analysis. Appl. Environ. Microbiol. 64:4870-4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shaw, L. J., Y. Beaton, S. Sousa, L. A. Glover, K. Killham, and A. A. Meharg. 2002. Mineralisation of 2,4-dichlorophenol and glucose placed into the same or different hydrological domains as a bacterial inoculant. Soil Biol. Biochem. 34:531-539. [Google Scholar]
- 43.Shaw, L. J., and R. G. Burns. 2003. Biodegradation of organic pollutants in the rhizosphere. Adv. Appl. Microbiol. 53:1-60. [DOI] [PubMed] [Google Scholar]
- 44.Shaw, L. J., and R. G. Burns. 1998. Biodegradation-transport interactions of pulse applied 2,4-D in repacked soil column microcosms. J. Environ. Qual. 27:1472-1478. [Google Scholar]
- 45.Siciliano, S. D., N. Fortin, A. Mihoc, G. Wisse, S. Labelle, D. Beaumier, D. Ouellette, R. Roy, L. G. Whyte, M. K. Banks, P. Schwab, K. Lee, and C. W. Greer. 2001. Selection of specific endophytic bacterial genotypes by plants in response to soil contamination. Appl. Environ. Microbiol. 67:2469-2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Siciliano, S. D., and J. J. Germida. 1997. Bacterial inoculants of forage grasses that enhance degradation of 2-chlorobenzoic acid in soil. Environ. Toxicol. Chem. 16:1098-1104. [Google Scholar]
- 47.Siciliano, S. D., and J. J. Germida. 1998. Mechanisms of phytoremediation: biochemical and ecological interactions between plants and bacteria. Environ. Rev. 6:65-79. [Google Scholar]
- 48.Siciliano, S. D., J. J. Germida, K. Banks, and C. W. Greer. 2003. Changes in microbial community composition and function during a polyaromatic hydrocarbon phytoremediation field trial. Appl. Environ. Microbiol. 69:483-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Streber, W. R., K. N. Timmis, and M. H. Zenk. 1987. Analysis, cloning, and high-level expression of 2,4-dichlorophenoxyacetate monooxygenase gene tfdA of Alcaligenes eutrophus JMP134. J. Bacteriol. 169:2950-2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Suwa, Y., A. D. Wright, F. Fukumori, K. A. Nummy, R. P. Hausinger, W. E. Holben, and L. J. Forney. 1996. Characterization of a chromosomally encoded 2,4-dichlorophenoxyacetic acid/α-ketoglutarate dioxygenase from Burkholderia sp. strain RASC. Appl. Environ. Microbiol. 62:2464-2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Trevors, J. T. 1984. Effect of substrate concentration, inorganic nitrogen, O2 concentration, temperature and pH on dehydrogenase activity in soil. Plant Soil 77:285-293. [Google Scholar]
- 52.Vallaeys, T., R. R. Fulthorpe, A. M. Wright, and G. Soulas. 1996. The metabolic pathway of 2,4-dichlorophenoxyacetic acid degradation involves different families of tfdA and tfdB genes according to PCR-RFLP analysis. FEMS Microbiol. Ecol. 20:163-172. [Google Scholar]
- 53.van der Ploeg, J. R., M. A. Weiss, E. Saller, H. Nashimoto, N. Saito, M. A. Kertesz, and T. Leisinger. 1996. Identification of sulfate starvation-regulated genes in Escherichia coli: a gene cluster involved in the utilization of taurine as a sulfur source. J. Bacteriol. 178:5438-5446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vedler, E., V. Koiv, and A. Heinaru. 2000. TfdR, the LysR-type transcriptional activator, is responsible for the activation of the tfdCB operon of Pseudomonas putida 2,4-dichlorophenoxyacetic acid degradative plasmid pEST4011. Gene 245:161-168. [DOI] [PubMed] [Google Scholar]
- 55.vonMersi, W., and F. Schinner. 1991. An improved and accurate method for determining the dehydrogenase activity of soils with iodonitrotetrazolium chloride. Biol. Fertil. Soil 11:216-220. [Google Scholar]
- 56.Walker, T., H. Bias, E. Grotewold, and J. Vivanco. 2003. Root exudation and rhizosphere biology. Plant Physiol. 132:44-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Walton, B. T., A. M. Hoylman, M. M. Perez, T. A. Anderson, T. R. Johnson, E. A. Guthrie, and R. F. Christman. 1994. Rhizosphere microbial communities as a plant defense against toxic substances in soils, p. 82-92. In T. A. Anderson and J. R. Coats (ed.), Bioremediation through rhizosphere technology. American Chemical Society, Washington, D.C.
- 58.Wania, F., and D. MacKay. 1996. Tracking the distribution of persistent organic pollutants. Environ. Sci. Technol. 30:390A-396A. [DOI] [PubMed] [Google Scholar]