Abstract
Cholangiocarcinoma is the second most common primary liver malignancy with extremely poor prognosis due to early invasion and widespread metastasis. The invasion and metastasis are regulated by multiple factors including CXCR4 chemokine receptor and multiple microRNAs. The goal of this study was to test the hypothesis that inhibition of CXCR4 combined with the action of miR-200c mimic will cooperatively enhance the inhibition of the invasion of human cholangiocarcinoma cells. The results show that CXCR4-inhibition polycation PCX can effectively deliver miR-200c mimic and that the combination treatment consisting of PCX and miR-200c results in cooperative antimigration activity, most likely by coupling the CXCR4 axis blockade with epithelial-to-mesenchymal transition inhibition in the cholangiocarcinoma cells. The ability of the combined PCX/miR-200c treatment to obstruct two migratory pathways represents a promising antimetastatic strategy in cholangiocarcinoma.
Keywords: microRNA, polyplexes, CXCR4, cholangiocarcinoma, poly(amido amine)
Graphical abstract
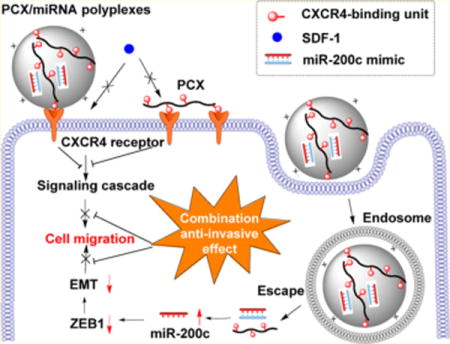
1. INTRODUCTION
MicroRNAs are small (~22 nucleotide) noncoding RNAs that regulate gene expression at the post-transcriptional level.1,2 MicroRNA targeting involves imperfect base-pairing between the microRNA and its mRNA target, which results in decreased protein production.3 MicroRNAs regulate a broad range of cellular pathways and control the expression of nearly 30% of all human proteins.4 Dysregulation of microRNA often leads to pathological states such as cancer.5 In fact, aberrant microRNA expression is found in nearly all human cancers. MicroRNAs function as tumor suppressors or oncogenes and play an important role in tumorigenesis, tumor growth, angiogenesis, and metastasis.6–8 By targeting different genes simultaneously, a single microRNA is capable of regulating multiple biological pathways that are essential for a cancer cell phenotype. Hence, inhibition of overexpressed oncogenic microRNAs or restitution of downregulated tumor-suppressor microRNAs provides a highly promising approach to treat cancer.
Cholangiocarcinoma is a malignant neoplasm of the biliary duct system. Intrahepatic cholangiocarcinoma arises from the epithelial cells of the intrahepatic bile ducts.9,10 Cholangiocarcinoma is the second most common primary liver malignancy after hepatocellular carcinoma and accounts for 10–25% of all primary hepatic malignancies. The incidence rate of cholangiocarcinoma has increased worldwide over the past decade. Despite advances in surgical techniques, chemotherapy, and radiotherapy, 5-year survival of patients after diagnosis remains at about 10%.11 Cholangiocarcinoma is fatal mainly due to early invasion, widespread metastasis, and the lack of effective therapeutic options.12,13 Among others, therapeutic strategies that focus on addressing the invasive character of cholangiocarcinoma promise to improve the treatment outcomes. A growing number of studies confirm the important role of microRNAs in formation and progression of cholangiocarcinoma.14–17 Recent evidence suggests that migration, invasion, and metastasis of cholangiocarcinoma are regulated by multiple microRNAs including miR-21,18 miR-200c,19 and miR-214.20 For example, increasing levels of miR-200c decreased the extent of the epithelial-to-mesenchymal transition (EMT) and inhibited cell migration and invasion in cholangiocarcinoma cells.19 Despite the potential of microRNA for cancer therapy, the clinical translation of therapeutic microRNA is hindered by a lack of efficient delivery systems.21,22 The negative charge and low molecular weight of microRNAs make them suitable for formulation in nanoscale delivery systems, thus enabling their use in clinical cancer therapy.23 Among others, polymers are widely used as nucleic acid carriers suitable for cancer therapy.24,25 Simultaneous delivery of miR-21 and miR-10b using PLGA-b-PEG polymer nanoparticles resulted in significant reduction in the growth of triple-negative breast cancer.26 Polyurethane derivatives of cationic poly(ethylene imine) (PEI) were used to successfully deliver miR-145 to xenograft tumors to reduce tumor growth and metastasis.27
The chemokine receptor CXCR4 is a potential target in antimetastatic therapies because of its crucial role in metastatic spread of multiple types of cancer.28 Current evidence already supports the potential of improving chemotherapy and radiotherapy through its combination with CXCR4 antagonists. In cholangiocarcinoma, CXCR4 was overexpressed in approximately 65% of 23 hilar cholangiocarcinoma samples but was not detected in normal biliary epithelium.29 Increased CXCR4 mRNA was observed in samples from patients with lymph node metastasis as well as perineural invasion.29 Signaling through CXCR4 increases migration of human cholangiocarcinoma cells and inhibition of CXCR4 signaling decreased motility and invasion.30,31 The CXCR4 ligand SDF-1 (CXCL12) is preferentially expressed in the liver, lymph nodes, lung, and bone marrow,32 a pattern that overlaps with the metastatic profile of cholangiocarcinoma. Binding of SDF-1 to CXCR4 activates multiple intracellular signaling transduction pathways that regulate migration and invasion of cancer cells.33–35 CXCR4 then facilitates metastatic spread of the primary tumor cells to sites where SDF-1 is highly expressed. The antimetastatic effect of CXCR4 inhibition can be enhanced by simultaneous use of nucleic acids that target additional pathways involved in cancer cell migration and invasion. This was demonstrated in a recent study, which combined inhibition of the CXCR4 axis with siRNA knockdown of Lipocalin-2 (Lcn2) as a way of synergistically reducing migration in metastatic human breast cancer cells.36
We have recently developed a series of polymeric CXCR4 antagonists (PCX) capable of delivering various types of nucleic acids including DNA and siRNA.37–40 The developed PCX polymers effectively block cancer cell invasion by inhibiting the CXCR4/SDF-1 axis, while at the same time, they deliver nucleic acids into the cancer cells for improved anticancer effect. The PCX polymers were synthesized from either FDA-approved CXCR4 antagonist AMD3100 (Scheme 1A) or novel CXCR4-inhibiting monocyclam inhibitors (Scheme 1B). In the present study, we hypothesized that combining the CXCR4 axis blockade with the action of miR-200c mimic would enhance the inhibition of the migration of metastatic cholangiocarcinoma cells more efficiently than either one of these treatments alone. We expected that in addition to CXCR4 antagonism, the PCX could deliver miR-200c into cholangiocarcinoma cells and inhibit EMT by inhibiting zinc finger E-box-binding homeobox 1 (ZEB1) expression, thus enhancing the inhibitory effect on cancer cell migration and invasion (Scheme 1C). The ability of the combined PCX/miR-200c treatment to obstruct two migratory pathways represents a promising antimetastatic strategy in cholangiocarcinoma.
Scheme 1.
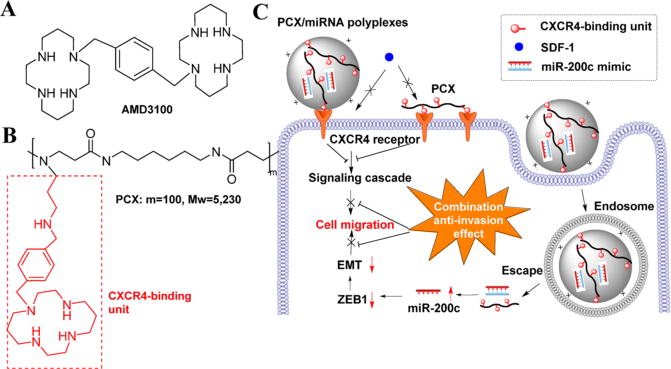
Chemical Structure of (A) AMD3100 (Plerixafor) and (B) PCX. (C) Mechanism of Action of PCX/miR-200c Polyplexes
2. MATERIALS AND METHODS
2.1. Materials
Dulbecco’s modified Eagle medium (DMEM), Dulbecco’s phosphate buffered saline (PBS), and fetal bovine serum (FBS) were from ThermoScientific (Waltham, MA). Cell culture inserts (for 24-well plates, 8.0 μm pores, Translucent PET Membrane, cat# 353097) were purchased from BD Biosciences (Billerica, MA). Human SDF-1α was from Shenandoah Biotechnology, Inc. (Warwick, PA). Oligofectamine was from Invitrogen (Carlsbad, USA) and used as suggested by the supplier. BLOCK-iT Fluorescent Oligo (FITC-Oligo) was supplied by ThermoFisher Scientific. MicroRNA-200c mimic (mature microRNA sequence: 5′-UAAUACUGCCGGGUAAUGAUGGA-3′) and negative control miR-NC mimic (mature microRNA sequence: 5′-UCACAACCUCCUAGAAAGAGUAGA–3′) were purchased from Dharmacon (Lafayette, CO). Polymeric CXCR4 antagonist (PCX, Mw = 5230, Mw/Mn = 1.27) was synthesized and characterized as previously described.38 All other reagents were from Fisher Scientific and used as received unless otherwise noted.
2.2. Cell Culture
Human malignant intrahepatic cholangiocarcinoma HuCCT1 cell line was kindly provided by Dr. Gregory Gores, Mayo Clinic, Rochester MN. The cell line was derived previously from the malignant ascites fluid from a 56-year-old male patient with intrahepatic cholangiocarcinoma.41 HuCCT1 cells were grown in high-glucose DMEM supplemented with 10% FBS, penicillin (100 U/mL), streptomycin (100 μg/mL), G418 (50 μg/mL), and insulin (0.5 μg/mL) at 37 °C with 5% CO2 in a humidified chamber.41,42
2.3. Surface Expression of CXCR4
HuCCT1 cells were detached with enzyme-free Cell Dissociation Buffer (Thermo Scientific) and suspended in a staining buffer. Cells were stained live with allophycocyanin (APC)-conjugated anti-CXCR4 antibody (Abcam, USA) for 1 h at 4 °C. Isotype-matched negative control was used in the panel of mAb to assess background fluorescence intensity. Samples were analyzed on a BD FACSCalibur flow cytometer (BD Bioscience, Bedford, MA). The results were processed using FlowJo software (Tree Star Inc., Ashland, OR).
2.4. Preparation and Physicochemical Characterization of PCX/microRNA Polyplexes
The ability of PCX to condense microRNA was determined by electrophoresis in a 2% agarose gel containing 0.5 μg/mL of ethidium bromide (EtBr). PCX/microRNA polyplexes were formed by adding predetermined volume of PCX to a microRNA solution (20 μM in 10 mM HEPES pH 7.4) to achieve the desired w/w ratio and vigorously vortexed for 10 s. Polyplexes were then incubated at room temperature for 30 min before further use. Polyplexes prepared at different PCX-to-microRNA weight ratios were loaded (20 μL of the sample containing 1.0 μg of microRNA) and run for 30 min at 100 V in 0.5 × Tris/Borate/EDTA buffer. The gels were visualized under UV illumination on a KODAK Gel Logic 100 imaging system.
MicroRNA release from polyplexes was analyzed by heparin displacement assay. The polyplexes were prepared at a w/w ratio of 12 and incubated with increasing concentrations of heparin for 30 min at room temperature. The samples (20 μL of the sample containing 0.5 μg of microRNA) were then analyzed by agarose gel electrophoresis.
Hydrodynamic diameter and zeta potential of the polyplexes were determined by dynamic light scattering (DLS) using a ZEN3600 Zetasizer Nano-ZS (Malvern Instruments Ltd., Massachusetts, United States).
2.5. Cellular Uptake and Intracellular Trafficking of Polyplexes
Flow cytometry analysis was used to study the cellular uptake of polyplexes. HuCCT1 cells (5 × 104) were seeded in 24-well plates and cultured to reach about 50% confluence. The cells were incubated at 37 °C with PCX/FITC-Oligo polyplexes at a FITC-Oligo concentration of 200 nM for 4 h. The cells were then trypsinized, washed with cold PBS, filtered through 35 μm nylon mesh, and subjected to analysis using a BD FACSCalibur flow cytometer (BD Bioscience, Bedford, MA). The results were processed using FlowJo software.
Intracellular localization was observed by confocal laser scanning microscope. Cells were cultured on 20 mm glassbottom cell culture dish (Nest) at 1 × 105 cells/dish. After 24 h, the medium was exchanged with fresh medium, and PCX/FITC-Oligo polyplexes were added (200 nM FITC-Oligo). After incubation for 4 h, the cells were washed twice with PBS and stained with LysoTracker Red DND-99 (Life Technology, USA) for 30 min, fixed with 4% paraformaldehyde for 10 min, and stained with Hoechst 33258 for 10 min. The cells were rinsed three times with PBS and visualized by LSM 710 Laser Scanning Microscope (Zeiss, Jena, Germany).
2.6. Quantitative Real-Time PCR (qRT-PCR)
The expression levels of miR-200c were evaluated by TaqMan qRT-PCR. mirVana miRNA Isolation Kit (Ambion, USA) was used for total RNA extraction from cultured cells. Ten nanograms of total RNA was converted into cDNA using specific primers for miR-200c (or the internal control Z30 (Applied Biosystems, Foster City, CA)) and the TaqMan microRNA reverse transcription kit (Applied Biosystems). qRT-PCR was performed using TaqMan Universal Master Mix II, No AmpErase UNG (2×) and specific primers for miR-200c or Z30 (Applied Biosystems, Foster City, CA) on a Rotor-Gene Q instrument (QIAGEN) according to the manufacturer’s instructions. MicroRNA expression levels were expressed relative to the internal control according to the comparative threshold cycle (Ct) method.
2.7. Western Blot
Cultured cells were lysed with RIPA Lysis buffer by incubation on ice for 10 min. After centrifugation at 12 000g for 10 min, the supernatants were collected, and the concentrations of proteins were quantified by the BCA protein assay kit (Promega, USA). The protein samples were denatured by boiling for 5 min, loaded onto 10% SDS-PAGE gel for electrophoresis (at 120 V for 2 h), and then transferred (at 300 mA for 1 h) to a nitrocellulose membrane. After blocking with 5% nonfat dried milk at room temperature for 1 h, the membrane was incubated overnight at 4 °C with ZEB1 rabbit monoclonal antibody (Cell Signaling Technology, USA) and then washed and incubated with the secondary antirabbit IgG HRP-linked antibody (Cell Signaling Technology, USA) for 1 h. Finally, membranes were again washed and visualized using Pierce ECL Western Blotting Substrate (Thermo Scientific, USA). Quantification of Western blot bands was performed using ImageJ software (National Institutes of Health, Bethesda, MD), and the data were expressed as relative ZEB1 level compared with untreated cells.
2.8. Wound Healing Assay
HuCCT1 cells (2 × 105) were seeded into six-well plates and cultured in complete DMEM to reach about 50% confluence. Cells were then treated with PCX/miR-200c polyplexes (w/w = 12) containing 200 nM miR-200c for 4 h. The polyplex solution was then removed and replaced with fresh medium. Oligofectamine/microRNA lipoplexes were transfected into cells according to the manufacturer’s protocol. When the cells reached confluence at 48 h post-transfection, an artificial wound was created in the monolayer with a sterile plastic 1 mL micropipette tip. Next, the cell monolayers were rinsed gently with PBS and further incubated. Pictures of the wounds were taken using a phase-contrast microscope at different time points.
2.9. Transwell Migration Assay
HuCCT1 cells (2 × 105) were seeded into six-well plates and cultured in complete DMEM to reach 50% confluence. The cultured cells were subsequently treated with PBS, oligofectamine/miR-NC, oligofectamine/miR-200c, PCX/miR-200c, and PCX/miR-200c at microRNA concentration of 200 nM. After 48 h of incubation, the cells were trypsinized and suspended in medium without serum. Subsequently, 5 × 104 cells were seeded in the top chambers in 300 μL of serum-free medium and 500 μL of complete medium containing 10% FBS was added to the lower transwell chambers. After 24 h, the nonmigrated cells in the top chamber were removed with a cotton swab. The migrated cells were fixed in 100% methanol and stained with 0.2% Crystal Violet solution for 10 min at room temperature. The images were taken by EVOS xl microscope. Three 20× visual fields were randomly selected for each insert, and each group was conducted in triplicate.
2.10. Cytotoxicity
Toxicity of the polyplexes was evaluated by Cell Titer Blue assay in HuCCT1 cells. The cells were plated in 96-well microplates at a density of 5000 cells/well. After 24 h, the cultured cells were treated with PBS, oligofectamine/miR-NC, oligofectamine/miR-200c, PCX/miR-200c, and PCX/miR-200c at microRNA concentration of 200 nM. After further 48 h of incubation, the medium was removed and replaced with a mixture of 100 μL of serum-free media and 20 μL of CellTiter-Blue reagent (CellTiter-Blue Cell Viability Assay, Promega). After 2 h of incubation, the fluorescence (560/590 nm) was measured on a Synergy 2 Microplate Reader (BioTek, VT). The relative cell viability (%) was calculated as [fluorescence]sample/[fluorescence]untreated × 100.
2.11. Statistical Analysis
Data are presented as the mean ± SD. The statistical significance was determined using ANOVA followed by Bonferroni post hoc correction with p < 0.05 as the minimal level of significance.
3. RESULTS AND DISCUSSION
3.1. CXCR4 Expression and CXCR4-Mediated Migration in HuCCT1 Cells
Surface expression of CXCR4 in HuCCT1 cells was confirmed by flow cytometry (Figure 1A). Over 36% of the HuCCT1 cells were CXCR4-positive with enhanced fluorescence intensity per cell. We then assessed the involvement of CXCR4 in the migration of the cells. A migration assay was performed to test whether SDF-1 induced migration of HuCCT1 cells and whether this migration could be inhibited by CXCR4 antagonists. As shown in Figure 1, panel B, substantially increased migration across the transwell insert membrane was observed in HuCCT1 cells stimulated with the chemoattractant SDF-1. In agreement with previous reports in other cholangiocarcinoma cells, the migration could be significantly inhibited by CXCR4 antagonist AMD3100.30
Figure 1.
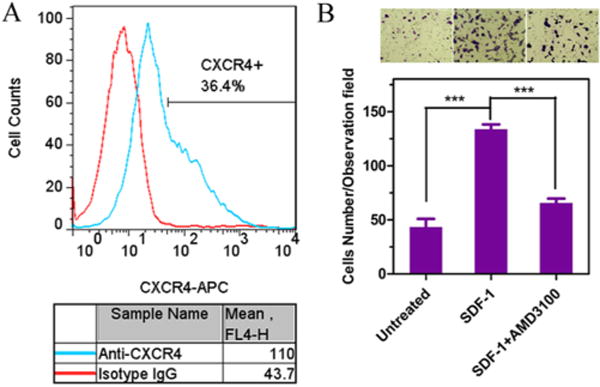
Characterization of the CXCR4 status of HuCCT1 cells. (A) Flow cytometric histograms show CXCR4 expression on HuCCT1 cell surface. The percent of CXCR4-positive cells and mean fluorescence intensity were analyzed using FlowJo software. (B) Inhibition of CXCR4-mediated cell migration. HuCCT1 cells were treated with AMD3100 (300 nM) and allowed to migrate through transwell membranes upon stimulation with SDF-1 for 24 h. Three 20× imaging areas were randomly selected for each insert, and each group was conducted in triplicate. Data are shown as mean ± SD (n = 3). ***, p < 0.001.
3.2. Preparation and Physicochemical Characterization of PCX/microRNA Polyplexes
The ability of PCX to form polyplexes with microRNA was first evaluated by agarose gel electrophoresis. As shown in Figure 2, panel A, PCX was able to fully condense microRNA above a PCX/microRNA (w/w) ratio of 2. PCX condensation of the microRNA was observed already at low w/w ratios (0.5–1) as indicated by the smear of the ethidium bromide-stained microRNA and by the strong fluorescence in the starting well of the gel. At higher PCX/microRNA w/w ratios (above 2), condensed microRNA was completely protected from ethidium bromide binding, and no fluorescence signal was observed. The ability of the PCX/microRNA polyplexes to release microRNA was then assessed by heparin displacement assay (Figure 2B). For PCX/microRNA polyplexes prepared at w/w 12, heparin was able to dissociate the polyplexes and completely release microRNA above 200 μg/mL of heparin.
Figure 2.
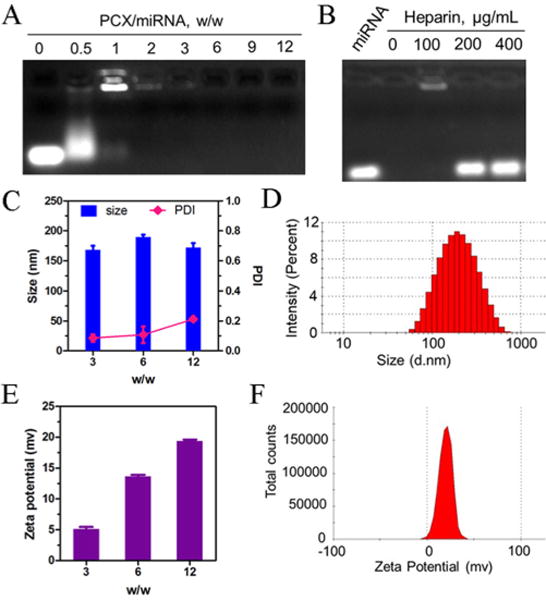
Physicochemical characterization of PCX/microRNA polyplexes. (A) MicroRNA condensation by PCX in 10 mM HEPES buffer (pH 7.4) using agarose gel electrophoresis. (B) Heparin induced microRNA release from the polyplexes. Polyplexes were prepared at w/w 12 and incubated with increasing concentrations of heparin. (C) Hydrodynamic size of PCX/microRNA polyplexes. (D) Size distribution of PCX/microRNA (w/w = 12). (E) Zeta-potential of PCX/microRNA polyplexes. (F) Zeta-potential of PCX/microRNA (w/w = 12) as determined by dynamic light scattering. Data shown as mean ± SD (n = 3).
Hydrodynamic size and zeta-potential of PCX/microRNA polyplexes prepared at various w/w ratios were measured by dynamic light scattering. Polyplexes with all the tested w/w ratios exhibited size in a narrow range from 160–180 nm with polydispersity indexes less than 0.2 (Figure 2C). The size distribution of polyplexes showed a log-transformed normal distribution (Figure 2D). As expected, increasing the w/w ratio used in the preparation of the polyplexes resulted in an increase of the zeta potential (Figure 3E).
Figure 3.
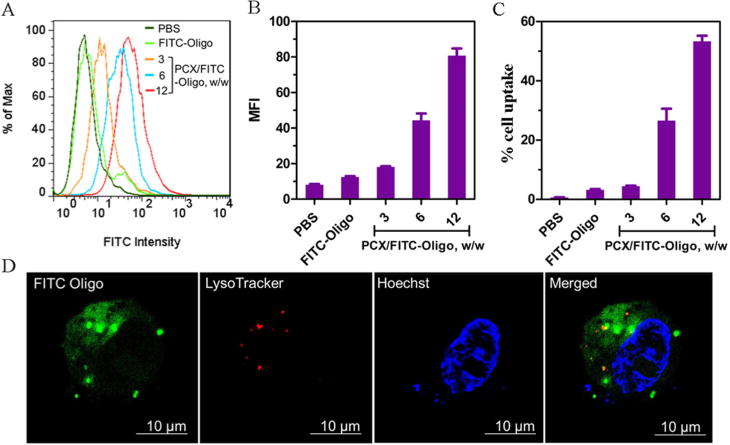
Cellular uptake and intracellular trafficking of PCX polyplexes. (A) Overlayed histogram of flow cytometry analysis of cells treated with PCX/FITC-Oligo polyplexes at various w/w ratios (200 nM FITC-Oligo). Quantification of cellular uptake is shown by (B) mean fluorescence intensity (MFI) and (C) % cell uptake. Data are shown as mean ± SD (n = 3). (D) Intracellular trafficking of PCX/FITC-Oligo in HuCCT1 cells by CLSM after 4 h of incubation.
3.3. Cellular Uptake and Intracellular Trafficking
To study the cellular uptake and intracellular trafficking of the polyplexes, we used a fluorescently labeled FITC-Oligo (200 nM) instead of microRNA in the preparation of the polyplexes. HuCCT1 cells were treated with PCX/FITC-Oligo for 4 h before flow cytometry analysis. As shown in Figure 3, panel A, PCX polyplexes exhibited significant cellular uptake in HuCCT1 cells as indicated by the enhanced fluorescence intensity when compared with untreated cells or cells treated with free FITC-Oligo. Increasing the w/w ratios in preparation of the polyplexes resulted in enhanced cell uptake both in terms of the mean fluorescence intensity per cell (Figure 3B) and the percentage of cells that have taken up the polyplexes (Figure 3C). PCX polyplexes prepared at the highest tested w/w = 12 showed the highest cell uptake and were thus selected for subsequent studies.
We further evaluated the intracellular trafficking of the PCX polyplexes using confocal microscopy. PCX/FITC-Oligo polyplexes (green) prepared at w/w 12 were incubated with the cells for 4 h. Lysosomes were stained with Lysotracker Red (red), and cell nuclei were stained with Hoechst 33258 (blue). As shown in Figure 3, panel D, the fluorescence of the FITC-Oligo was distributed mainly in the cytoplasm, and no FITC-Oligo signal was found in the cell nucleus. Limited extent of the colocalization of the FITC-Oligo signal with the LysoTracker signal in lysosomes (red) indicated efficient endosomal escape of the PCX polyplexes.
3.4. MicroRNA Transfection
The miR-200c and its negative control miR-NC were used to evaluate the microRNA transfection efficiency of the PCX polyplexes. The levels of miR-200c in HuCCT1 cells were measured using TaqMan qRT-PCR (Figure 4). PCX polyplexes exhibited high micro-RNA transfection efficiency, as indicated by a nearly 9500-fold increase in intracellular miR-200c levels when polyplexes prepared at w/w = 12 were used. Similar to the results of the cell uptake experiment (Figure 3), increasing the w/w ratio in preparing the polyplexes resulted in significantly enhanced transfection efficiency.
Figure 4.
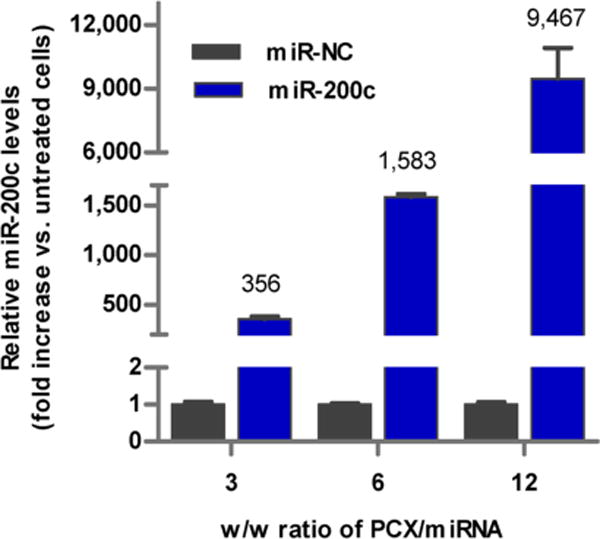
Transfection activity of PCX/microRNA polyplexes. miR-200c level was detected by TaqMan qRT-PCR in HuCCT1 cells. Data are shown as mean ± SD (n = 3).
Having confirmed the ability of PCX to effectively deliver miR-200c to the HuCCT1 cells, we then evaluated the effect of the delivered miR-200c on the target intracellular pathway. We used Western blot to analyze the cellular levels of one of the main downstream targets of miR-200c, namely the zinc finger E-box-binding homeobox 1 (ZEB1). ZEB1 is an inducer of the EMT in cancer cells, and its overexpression is associated with cancer cell migration and invasion.19,43,44 As shown in Figure 5, delivery of miR-200c using PCX polyplexes resulted in a significant decrease (46%) in cellular ZEB1 protein levels in the HuCCT1 cells when compared with the control PCX/miR-NC polyplexes. This finding confirms that the miR-200c was delivered by the PCX polyplexes into the cytoplasm and efficiently released in its active state to successfully downregulate the target ZEB1 protein.
Figure 5.
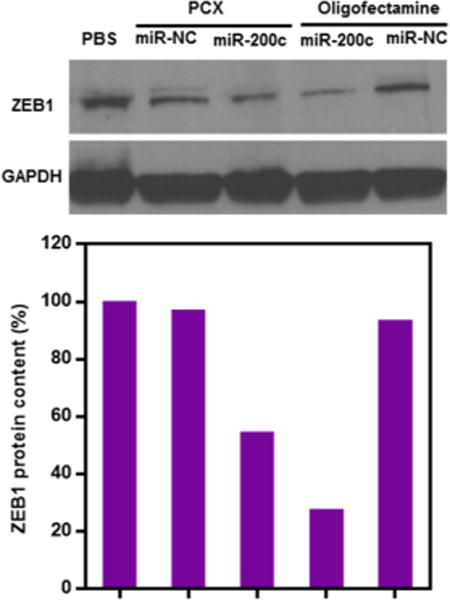
Effect of miR-200c delivery on the expression of ZEB1 protein. Quantification of Western blot bands was performed using ImageJ software, and the data are expressed as relative ZEB1 levels relative to untreated cells (the order of samples corresponds to the gel above)
3.5. Cell Migration
After the ability of the PCX polyplexes to deliver functional microRNA to the human cholangiocarcinoma cells was confirmed, we evaluated the cooperative effect of the inhibition of ZEB1 by miR-200c and CXCR4 inhibition by PCX on the migration of the cells. Before proceeding, we first confirmed that the selected polyplex formulations have no significant adverse effect on cell viability that could negatively affect their migratory properties. As shown in Figure 6, the cells treated with all the PCX polyplexes as well as the control oligofectamine lipoplexes exhibited nearly 100% viability after 48 h of incubation, indicating no adverse effects on cell proliferation. The migration of the cancer cells was then assessed using a wound healing assay and a transwell cell migration assay.
Figure 6.
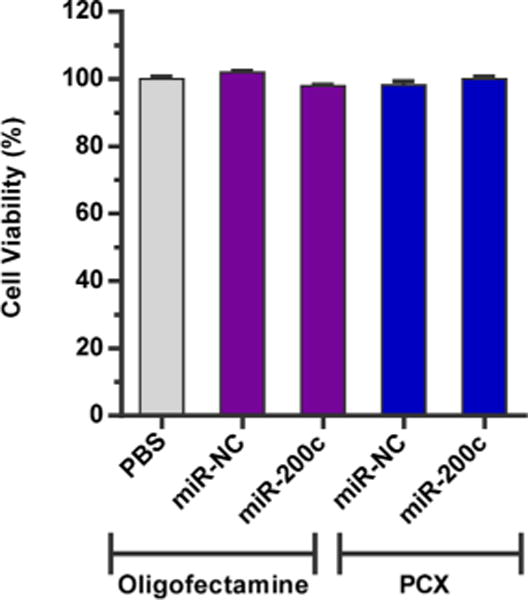
Cell viability was measured using Cell Titer Blue assay in HuCCT1 cells. HuCCT1 cells were treated with PCX polyplexes or control oligofectamine complexes for 48 h. Data are shown as mean ± SD (n = 3).
Wound healing assay was conducted to study the combined inhibitory activity of PCX/miR-200c polyplexes on migration of HuCCT1 cells. The cells were treated with PCX/miR-200c polyplexes, and an artificial wound was created 48 h post-transfection. The healing status of the wound, which represents the extent of cell migration, was measured after 24 and 48 h. As shown in Figure 7, the untreated (PBS) wound reached nearly complete closure after 48 h. The cells treated with control PCX/miR-NC exhibited significant inhibition of wound healing (56% closure) after 48 h, which is consistent with the CXCR4 antagonistic activity of PCX and its effect on cell migration. The combination treatment with PCX/miR-200c polyplexes further enhanced the extent of inhibition (40% closure) due to the cooperative activity of the CXCR4 antagonism of PCX and the effect of miR-200c on ZEB1. ZEB1 has previously been implicated in the migration-inhibitory effect of miR-200c, suggesting that ZEB1 may be a functional mediator of this effect in HuCCT1 cells. As expected, no inhibition of wound healing was observed when the control miR-NC was delivered using oligofectamine. When used to deliver miR-200c, oligofectamine lipoplexes exhibited partial inhibition of wound closure (61% closure).
Figure 7.
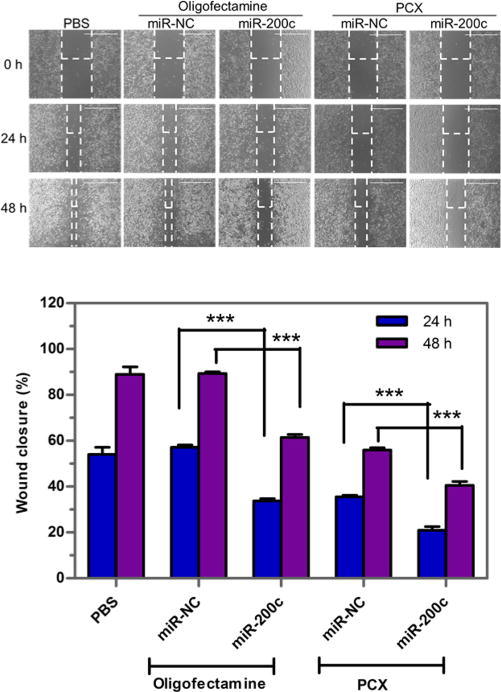
Inhibition of wound healing. Cells were treated with formulations for 48 h. Then an artificial wound was created in the monolayer with a 1 mL pipet tip. The 4× imaging areas of the wounds were taken using a microscope at different time points. Wound closure was expressed as % initial wound size (mean ± SD; n = 3) (Scale bar = 1000 μm). Data are shown as mean ± SD (n = 3). ***, p < 0.001.
To further confirm the cooperative activity of PCX and miR-200c on the inhibition of cancer cell migration, transwell assay was also performed. HuCCT1 cells were transfected with PCX/miR-200c polyplexes as before, and 10% FBS was applied to the lower chamber as the chemoattractant to induce the transwell cell migration. As shown in Figure 8, the migration of HuCCT1 cells was significantly inhibited following treatment with control oligofectamine/miR-200c. Treatment with another control, PCX/miR-NC, resulted in marked migration inhibition due to the CXCR4 antagonism of PCX. Combined treatment with PCX/miR-200c achieved the highest inhibition level of cell migration (~81%) among all the tested formulations, which confirmed the cooperative effect of PCX and miR-200c.
Figure 8.
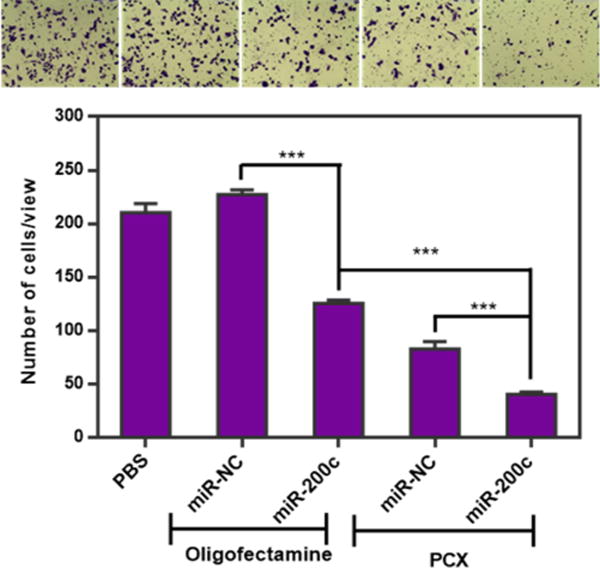
Inhibition of cancer cell migration. HuCCT1 cells were transfected with oligofectamine lipoplexes or PCX polyplexes for 48 h and then allowed to migrate through transwell membrane inserts upon stimulation with 10% FBS for 24 h. The number of migrated cells per 20× imaging area was counted, and the results are shown as mean ± SD of triplicate samples (***, p < 0.001). The photographs show representative images of the stained migrated cells arranged in the same order as the samples in the graph below.
4. CONCLUSIONS
In this study, CXCR4-inhibiting polycation PCX was evaluated as a delivery vector of miR-200c mimic with the goal of improving the inhibition of cholangiocarcinoma cell migration. The results show that PCX can inhibit cancer cell migration due to its CXCR4 antagonism. The ability of PCX to form polyplexes with nucleic acids was used for simultaneous delivery of miR-200c mimic into cells. The delivery of miR-200c resulted in reduced expression of the EMT inducer ZEB1. The combination treatment consisting of PCX and miR-200c resulted in cooperative antimigration activity, most likely by coupling the CXCR4 axis blockade with EMT inhibition in the cholangiocarcinoma cells. Our results suggest a promising antimetastatic strategy for a combination therapy involving multiple migration pathways in cholangiocarcinoma.
Acknowledgments
This work was supported in part by the University of Nebraska Medical Center and in part by grants from the National Institutes of Health (R01 EB015216, R21 EB020308). Support from the Nebraska Center For Nanomedicine (P20 GM103480) for J.M. is also acknowledged.
Footnotes
Notes
The authors declare no competing financial interest.
References
- 1.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Li Z, Rana TM. Therapeutic targeting of microRNAs: current status and future challenges. NatRev Drug Discovery. 2014;13:622–638. doi: 10.1038/nrd4359. [DOI] [PubMed] [Google Scholar]
- 3.Grimson A, Farh KK-H, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 5.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 6.Esquela-Kerscher A, Slack FJ. Oncomirs—microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 7.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kasinski AL, Slack FJ. MicroRNAs en route to the clinic: progress in validating and targeting microRNAs for cancer therapy. Nat Rev Cancer. 2011;11:849–864. doi: 10.1038/nrc3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patel T. Cholangiocarcinoma—controversies and challenges. Nat Rev Gastroenterol Hepatol. 2011;8:189–200. doi: 10.1038/nrgastro.2011.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khan SA, Thomas HC, Davidson BR, Taylor-Robinson SD. Cholangiocarcinoma. Lancet. 2005;366:1303–1314. doi: 10.1016/S0140-6736(05)67530-7. [DOI] [PubMed] [Google Scholar]
- 11.Rizvi S, Gores GJ. Pathogenesis, diagnosis, and management of cholangiocarcinoma. Gastroenterology. 2013;145:1215–1229. doi: 10.1053/j.gastro.2013.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Okuda K, Nakanuma Y, Myazaki M. Cholangiocarcinoma: recent progress. Part 2: molecular pathology and treatment. J Gastroenterol Hepatol. 2002;17:1056–1063. doi: 10.1046/j.1440-1746.2002.02780.x. [DOI] [PubMed] [Google Scholar]
- 13.Sia D, Tovar V, Moeini A, Llovet J. Intrahepatic cholangiocarcinoma: pathogenesis and rationale for molecular therapies. Oncogene. 2013;32:4861–4870. doi: 10.1038/onc.2012.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mott JL. MicroRNAs involved in tumor suppressor and oncogene pathways: implications for hepatobiliary neoplasia. Hepatology. 2009;50:630–637. doi: 10.1002/hep.23010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Razumilava N, Bronk SF, Smoot RL, Fingas CD, Werneburg NW, Roberts LR, Mott JL. miR-25 targets TNF-related apoptosis inducing ligand (TRAIL) death receptor-4 and promotes apoptosis resistance in cholangiocarcinoma. Hepatology. 2012;55:465–475. doi: 10.1002/hep.24698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meng F, Henson R, Lang M, Wehbe H, Maheshwari S, Mendell JT, Jiang J, Schmittgen TD, Patel T. Involvement of human micro-RNA in growth and response to chemotherapy in human cholangiocarcinoma cell lines. Gastroenterology. 2006;130:2113–2129. doi: 10.1053/j.gastro.2006.02.057. [DOI] [PubMed] [Google Scholar]
- 17.Selaru FM, Olaru AV, Kan T, David S, Cheng Y, Mori Y, Yang J, Paun B, Jin Z, Agarwal R, et al. MicroRNA-21 is overexpressed in human cholangiocarcinoma and regulates programmed cell death 4 and tissue inhibitor of metalloproteinase 3. Hepatology. 2009;49:1595–1601. doi: 10.1002/hep.22838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chusorn P, Namwat N, Loilome W, Techasen A, Pairojkul C, Khuntikeo N, Dechakhamphu A, Talabnin C, Chan-On W, Ong C, et al. Overexpression of microRNA-21 regulating PDCD4 during tumorigenesis of liver fluke-associated cholangiocarcinoma contributes to tumor growth and metastasis. Tumor Biol. 2013;34:1579–1588. doi: 10.1007/s13277-013-0688-0. [DOI] [PubMed] [Google Scholar]
- 19.Oishi N, Kumar MR, Roessler S, Ji J, Forgues M, Budhu A, Zhao X, Andersen JB, Ye QH, Jia HL, et al. Transcriptomic profiling reveals hepatic stem-like gene signatures and interplay of miR-200c and epithelial-mesenchymal transition in intrahepatic cholangiocarcinoma. Hepatology. 2012;56:1792–1803. doi: 10.1002/hep.25890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li B, Han Q, Zhu Y, Yu Y, Wang J, Jiang X. Down-regulation of miR-214 contributes to intrahepatic cholangiocarcinoma metastasis by targeting Twist. FEBS J. 2012;279:2393–2398. doi: 10.1111/j.1742-4658.2012.08618.x. [DOI] [PubMed] [Google Scholar]
- 21.Chen Y, Gao D-Y, Huang L. In vivo delivery of miRNAs for cancer therapy: challenges and strategies. Adv Drug Delivery Rev. 2015;81:128–141. doi: 10.1016/j.addr.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheng CJ, Saltzman WM. Polymer nanoparticle-mediated delivery of microRNA inhibition and alternative splicing. Mol Pharmaceutics. 2012;9:1481–1488. doi: 10.1021/mp300081s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y, Wang Z, Gemeinhart RA. Progress in microRNA delivery. J Controlled Release. 2013;172:962–974. doi: 10.1016/j.jconrel.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li J, Wang Y, Zhu Y, Oupický D. Recent advances in delivery of drug–nucleic acid combinations for cancer treatment. J Controlled Release. 2013;172:589–600. doi: 10.1016/j.jconrel.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumar V, Mondal G, Slavik P, Rachagani S, Batra SK, Mahato RI. Codelivery of small molecule hedgehog inhibitor and miRNA for treating pancreatic cancer. Mol Pharmaceutics. 2015;12:1289–98. doi: 10.1021/mp500847s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Devulapally R, Sekar NM, Sekar TV, Foygel K, Massoud TF, Willmann JrK, Paulmurugan R. Polymer nanoparticles mediated codelivery of antimiR-10b and antimiR-21 for achieving triple negative breast cancer therapy. ACS Nano. 2015;9:2290–2302. doi: 10.1021/nn507465d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chiou G-Y, Cherng J-Y, Hsu H-S, Wang M-L, Tsai C-M, Lu K-H, Chien Y, Hung S-C, Chen Y-W, Wong C-I, Tseng L-M, Huang P-I, Yu C-C, Hsu W-H, Chiou S-H. Cationic polyurethanes-short branch PEI-mediated delivery of Mir145 inhibited epithelial-mesenchymal transdifferentiation and cancer stem-like properties and in lung adenocarcinoma. J Controlled Release. 2012;159:240–250. doi: 10.1016/j.jconrel.2012.01.014. [DOI] [PubMed] [Google Scholar]
- 28.Zhao H, Guo L, Zhao H, Zhao J, Weng H, Zhao B. CXCR4 over-expression and survival in cancer: A system review and meta-analysis. Oncotarget. 2015;6:5022–40. doi: 10.18632/oncotarget.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tan X-Y, Chang S, Liu W, Tang H-H. Silencing of CXCR4 Inhibits Tumor Cell Proliferation and Neural Invasion in Human Hilar Cholangiocarcinoma. Gut Liver. 2014;8:196–204. doi: 10.5009/gnl.2014.8.2.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohira S, Sasaki M, Harada K, Sato Y, Zen Y, Isse K, Kozaka K, Ishikawa A, Oda K, Nimura Y, et al. Possible regulation of migration of intrahepatic cholangiocarcinoma cells by interaction of CXCR4 expressed in carcinoma cells with tumor necrosis factor-α and stromal-derived factor-1 released in stroma. Am J Pathol. 2006;168:1155–1168. doi: 10.2353/ajpath.2006.050204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leelawat K, Keeratichamroen S, Leelawat S, Tohtong R. CD24 induces the invasion of cholangiocarcinoma cells by upregulating CXCR4 and increasing the phosphorylation of ERK1/2. Oncol Lett. 2013;6:1439–1446. doi: 10.3892/ol.2013.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muller A, Homey B, Soto H, Ge N, Catron D, Buchanan ME, McClanahan T, Murphy E, Yuan W, Wagner SN, et al. Involvement of chemokine receptors in breast cancer metastasis. Nature. 2001;410:50–56. doi: 10.1038/35065016. [DOI] [PubMed] [Google Scholar]
- 33.Kucia M, Reca R, Miekus K, Wanzeck J, Wojakowski W, Janowska-Wieczorek A, Ratajczak J, Ratajczak MZ. Trafficking of Normal Stem Cells and Metastasis of Cancer Stem Cells Involve Similar Mechanisms: Pivotal Role of the SDF-1/CXCR4 Axis Stem. Cells. 2005;23:879–894. doi: 10.1634/stemcells.2004-0342. [DOI] [PubMed] [Google Scholar]
- 34.Smith MC, Luker KE, Garbow JR, Prior JL, Jackson E, Piwnica-Worms D, Luker GD. CXCR4 regulates growth of both primary and metastatic breast cancer. Cancer Res. 2004;64:8604–8612. doi: 10.1158/0008-5472.CAN-04-1844. [DOI] [PubMed] [Google Scholar]
- 35.Balkwill F. Cancer and the chemokine network. Nat Rev Cancer. 2004;4:540–550. doi: 10.1038/nrc1388. [DOI] [PubMed] [Google Scholar]
- 36.Guo P, You J-O, Yang J, Jia D, Moses MA, Auguste DT. Inhibiting metastatic breast cancer cell migration via the synergy of targeted, pH-triggered siRNA delivery and chemokine axis blockade. Mol Pharmaceutics. 2014;11:755–765. doi: 10.1021/mp4004699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li J, Zhu Y, Hazeldine ST, Li C, Oupicky D. Dual-function CXCR4 antagonist polyplexes to deliver gene therapy and inhibit cancer cell invasion. Angew Chem, Int Ed. 2012;51:8740–3. doi: 10.1002/anie.201203463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Y, Hazeldine ST, Li J, Oupicky D. Development of Functional Poly(amido amine) CXCR4 Antagonists with the Ability to Mobilize Leukocytes and Deliver Nucleic Acids. Adv Healthcare Mater. 2015;4:729–38. doi: 10.1002/adhm.201400608. [DOI] [PubMed] [Google Scholar]
- 39.Wang Y, Li J, Chen Y, Oupicky D. Balancing polymer hydrophobicity for ligand presentation and siRNA delivery in dual function CXCR4 inhibiting polyplexes. Biomater Sci. 2015;3:1114–23. doi: 10.1039/C5BM00003C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li J, Oupicky D. Effect of biodegradability on CXCR4 antagonism, transfection efficacy and antimetastatic activity of polymeric Plerixafor. Biomaterials. 2014;35:5572–5579. doi: 10.1016/j.biomaterials.2014.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miyagiwa M, Ichida T, Tokiwa T, Sato J, Sasaki H. A new human cholangiocellular carcinoma cell line (HuCC-T1) producing carbohydrate antigen 19/9 in serum-free medium. In Vitro Cell Dev Biol. 1989;25:503–510. doi: 10.1007/BF02623562. [DOI] [PubMed] [Google Scholar]
- 42.Wehrkamp CJ, Gutwein AR, Natarajan SK, Phillippi MA, Mott JL. XIAP Antagonist Embelin Inhibited Proliferation of Cholangiocarcinoma Cells. PLoS One. 2014;9:e90238. doi: 10.1371/journal.pone.0090238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 44.Korpal M, Lee ES, Hu G, Kang Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J Biol Chem. 2008;283:14910–14914. doi: 10.1074/jbc.C800074200. [DOI] [PMC free article] [PubMed] [Google Scholar]


