Abstract
Using fluorescence resonance energy transfer technology and Lightcycler analysis, we developed a real-time PCR assay with primers and probes designed by using IS900 which allowed rapid detection of Mycobacterium avium subsp. paratuberculosis DNA in artificially contaminated milk. Initially, the PCR parameters (including primer and probe levels, assay volume, Mg2+ concentration, and annealing temperature) were optimized. Subsequently, the quantitative ability of the assay was tested and was found to be accurate over a broad linear range (3 × 106 to 3 × 101 copies). The assay sensitivity when purified DNA was used was determined to be as low as five copies, with excellent reproducibility. A range of DNA isolation strategies was developed for isolating M. avium subsp. paratuberculosis DNA from spiked milk, the most effective of which involved the use of 50 mM Tris HCl, 10 mM EDTA, 2% Triton X-100, 4 M guanidinium isothiocyante, and 0.3 M sodium acetate combined with boiling, physical grinding, and nucleic acid spin columns. When this technique was used in conjunction with the real-time PCR assay, it was possible to consistently detect <100 organisms per ml of milk (equivalent to 2,000 organisms per 25 ml). Furthermore, the entire procedure (extraction and PCR) was performed in less than 3 h and was successfully adapted to quantify M. avium subsp. paratuberculosis in spiked milk from heavily and mildly contaminated samples.
For over 100 years, the slowly growing acid-fast bacillus Mycobacterium avium subsp. paratuberculosis has been implicated as the causative agent of a chronic granulomatous enteritis in cattle called Johne's disease (15). Although a role in human disease remains to be elucidated, the potential association of M. avium subsp. paratuberculosis with Crohn's disease has been the subject of many high-profile reviews (2, 14, 27). Despite the uncertainty regarding its potential hazard to human health, this organism has been shown to be pathogenic for many species; therefore, it may be prudent to adopt a precautionary approach towards reducing human exposure to M. avium subsp. paratuberculosis until further, unambiguous evidence is available.
As milk has been shown to be involved in the transmission of Johne's disease to calves (30) and previous studies have shown that M. avium subsp. paratuberculosis is present in retail milk (21), regular screening of milk for human consumption should be encouraged. This would have the effect of tracking contaminated herds and farms and would ultimately contribute to reducing exposure. Although conventional culturing remains the “gold standard,” it is an impractical method for detection of this slowly growing organism as there is a requirement for a harsh decontamination step that significantly reduces the viable titer and countable colonies seldom appear on agar slopes before 8 to 12 weeks. Other tests for M. avium subsp. paratuberculosis can also be employed, such as enzyme-linked immunosorbent assays, agar gel imunodiffusion, complement fixation, and fecal culturing; however, in general, these assays all have sensitivity and/or specificity issues.
PCR detection has proven to be a popular and sensitive method for screening milk and clinical samples for M. avium subsp. paratuberculosis DNA (4, 5, 9, 10, 11, 21, 23, 26, 32). In particular, the IS900 transposon has been accepted by researchers as the most suitable and specific target, although its specificity has been called into doubt recently (6, 16) due to the discovery of an IS900-like element in non-M. avium subsp. paratuberculosis strains. Despite this however, the IS900 target is particularly suited to sensitive detection as each organism contains between 14 and 18 copies of this insertion element and reports of non-M. avium subsp. paratuberculosis strains containing the transposon are exceedingly rare.
In general, PCR-based assays provide a rapid means of qualitatively assessing the presence of M. avium subsp. paratuberculosis in milk. The development of real-time PCR assays has offered researchers the potential to determine initial levels of nucleic acid, which can be extrapolated to estimate the microbial load. Such real-time assays are based on determining the crossing point or cycle threshold (cycle number at which the first significant increase in fluorescence above a threshold level is detected) for a known set of DNA standards and have recently been described for many of the common food-borne bacterial pathogens (3, 18, 28, 29), as well as several mycobacterial species (19, 22, 31). Quantitative assays for slowly growing bacteria, such as members of the genus Mycobacterium, are particularly useful as they offer a convenient alternative means to rapidly quantify organisms from a wide variety of sources. In the recent past three such assays for M. avium subsp. paratuberculosis have been described (7, 16, 25), although quantitative analysis from milk has not been reported previously.
The objective of this study was to develop a rapid and convenient extraction strategy for M. avium subsp. paratuberculosis in whole milk based on quantitative real-time PCR by using fluorescence resonance energy transfer-based probes. The final procedure (including extraction and PCR) facilitates quantitative analysis of artificially contaminated milk samples and can be performed in less than 3 h.
MATERIALS AND METHODS
Growth of bacterial strains.
All strains of M. avium subsp. paratuberculosis were subcultured on Middlebrook agar with the OADC supplement (Becton Dickinson) and 2 mg of mycobactine (Synbiotics) per ml. Liquid cultures were grown in continuously stirred Middlebrook broth with the OADC supplement and mycobactine (2% inoculum, 100 ml), and the organisms were enumerated on Middlebrook agar in 25-cm3 tissue culture flasks (Sarstedt) to prevent dehydration as described previously (25). The identities of the slowly growing cultures were routinely tested by using a Ziehl-Neelsen acid-fast stain, checking for mycobactine dependence, and confirming the presence of the IS900 element by PCR.
Isolation of DNA and generation of quantification standard.
DNA was isolated from growing cultures and was purified by using freeze-thawing, enzymatic degradation, lysis, phenol-chloroform treatment, and isopropanol precipitation as described previously (25). A conventional PCR was carried out with isolated M. avium subsp. paratuberculosis DNA by using primers F1 and R1 (designed with the IS900 transposon). Thirty PCR cycles consisting of 95°C for 1 min, 58°C for 1 min, and 72°C for 1 min were carried out, and the product was gel excised (QIAGEN) and quantified with appropriate standards. The standards were diluted, divided into aliquots, and frozen before they were used. The sequences of the primers used are as follows: primer F1, CGGGTATGGCTTTCATGTGGT; and primer R1, GTCGATCGCCCACGTGAC. The product size was 354 bp.
Real-time PCR and sensitivity.
All real-time experiments were carried out with a Lightcycler-Faststart DNA master hybridization probe kit (Roche). The assay volume, Mg2+ concentration, probe and primer concentrations, and annealing temperature were optimized initially. Subsequently, the sensitivity of the assay and the linear range were determined by using known amounts of purified template DNA (generated as described above). A typical optimized reaction mixture (total volume, 15 μl) contained 1.5 μl of buffer, 5 mM Mg2+, each probe at a concentration of 0.1 μM, each primer at a concentration of 0.25 μM, water, and 1 to 5 μl of DNA. The contents were placed in a glass capillary, briefly centrifuged, capped, and placed in the Lightcycler. Each run consisted of an 8-min hot start which activated the conjugated polymerase, followed by 40 cycles of 95°C for 2 s, 58°C for 5 s, and 72°C for 10 s. In order to ascertain the sensitivity of the method, standards generated as described above were diluted and assayed by using optimal real-time PCR conditions (5 × 105 to 5 × 100 copies). Fluorescence acquisition occurred within the F2/F1 channel after the annealing stage of each cycle, and all subsequent analysis was carried out by using the second derivative maximum option of the Lightcycler software (version 3.01). The sequences of the probes used are as follows: FL-labeled probe, CCACCTCCGTAACCGTCATTGTCCAGA; and LC Red 640-labeled probe, CAACCCAGCAGACGACCACGC. All primers and probes were designed and synthesized by TIB-Molbiol (Berlin, Germany).
Quantitation.
The ability to quantitate unknown targets was evaluated by using five separate DNA samples containing various amounts of template (range, 6 × 105 to 6 × 101 copies) that were analyzed by using real-time PCR in conjunction with known standards in this range in duplicate. The ability to accurately determine the starting template was evaluated by using the Lightcycler software, and the results were compared to the actual known values.
Reproducibility.
To evaluate the variability between experiments, four different known concentrations of DNA were amplified by performing the real-time assay described above in triplicate on three separate days. For each experiment the crossing point, average crossing point, standard deviation, and coefficient of variation for each assay were calculated.
Extraction of mycobacterial DNA from cultures in spiked milk.
Due to the complex, difficult matrix involved, several strategies were evaluated to isolate mycobacterial DNA from cultures in 25-ml samples of spiked milk (this volume was chosen for logistical reasons). Prior to addition of M. avium subsp. paratuberculosis to milk, the bacterial cultures were grown with continuous stirring and the organisms were enumerated on Middlebrook agar in 25-cm3 tissue culture flasks as described above and previously (25). This procedure had the effect of drastically reducing clumps and was useful for spiking studies. Every effort was made to reduce clumping in order to obtain accurate values for the number of cells as determined by subsequent analysis of the number of CFU per milliliter, although inevitably some clumps could have remained which could have slightly reduced the accuracy of the method. However, as M. avium subsp. paratuberculosis cells invariably aggregate when they are in close contact, absolute quantification of the bacteria is impossible by any method.
(i) Method 1: proteinase K treatment and lysis.
For proteinase K treatment and lysis, 24-ml samples of retail milk (previously shown to be M. avium subsp. paratuberculosis free) were spiked with 104 cells of M. avium subsp. paratuberculosis/ml (final concentration) and left at room temperature for 30 min. Subsequently, 250 μg of proteinase K was added to each preparation prior to incubation for 1 h at 37°C. Each sample was then centrifuged at 6,000 rpm for 5 min in an Eppendorf microcentrifuge. The liquid fraction was carefully removed, and the cream and pellet were resuspended in 750 μl of lysis buffer (2% Triton X-100, 1% sodium dodecyl sulfate, 100 mM NaCl, 10 mM Tris [pH 8.0], 1 mM EDTA) and subsequently transferred to a screw-cap Eppendorf tube containing 100 mg of acid-washed glass beads (diameter, 425 to 600 μm; Sigma, St. Louis, Mo.). The tubes were then processed in a bead beater for 3 min and later boiled for 5 min. After a brief centrifugation step (14,000 rpm for 3 min), 650 μl of the clear supernatant was removed from each tube, precipitated with an equal volume of isopropanol, washed with 70% ethanol, dried, and resuspended in 50 μl of Tris-EDTA (TE) buffer.
(ii) Method 2: treatment with lysozyme and proteinase K.
For treatment with lysozyme and proteinase K, samples were treated as described above for method 1 except that each spiked milk sample was initially incubated with 250 μg of proteinase K and 50 μg of lysozyme for 1 h.
(iii) Method 3: chloroform-methanol treatment.
For chloroform-methanol treatment, the spiked milk was centrifuged (as described for method 1) and resuspended (pellet and cream) in 1 ml of chloroform-methanol (50:50). After 1 min, the sample was centrifuged again (14,000 rpm for 5 min) and resuspended in lysis buffer as described above. The remainder of the procedure was the same as the method 1 procedure.
(iv) Method 4: basic lysis.
The basic lysis procedure was exactly the same as the method 1 procedure except that the initial proteinase K step was omitted.
(v) Method 5: alternative buffer.
For alternative buffer treatment, the spiked milk was centrifuged as described above for method 1. Subsequently, the pellet and cream were resuspended in an alternative lysis buffer as described by Abolmatty et al. (1) prior to bead beating, boiling, precipitation, washing, and resuspension as described above for method 1.
(vi) Method 6: GITC lysis.
For guanidinium isothiocyanate (GITC) lysis, after the spiked milk was centrifuged, the liquid fraction was carefully removed, and the cream and pellet were resuspended in 750 μl of lysis buffer (50 mM Tris HCl, 10 mM EDTA, 2% Triton X-100, 4 M GITC, 0.3 M sodium acetate). This mixture was adapted from the mixture described by Odumeru et al.(24) and helped to effectively liquefy the cream. The lysate was subsequently transferred to screw-cap Eppendorf tubes containing 100 mg of acid-washed glass beads (diameter, 425 to 600 μm) for bead beating. After boiling (5 min) and a brief centrifugation step (14,000 rpm for 3 min), 650 μl of the clear supernatant was removed from each tube and precipitated with an equal volume of isopropanol.
(vii) Method 7: clarifying solution.
For the clarifying solution treatment, the spiked milk was centrifuged, the whey was removed, and the pellet and cream fractions were resuspended in 1 ml of clarifying reagent for dairy products (Fluka GmbH, Buchs, Switzerland). The preparation was subsequently centrifuged (14,000 rpm for 5 min), and then the pellet was resuspended in 750 μl of lysis solution, processed in the bead beater, boiled, centrifuged, and precipitated as described above for method 1.
In all seven procedures described above the DNA was resuspended in 50 μl of TE buffer to facilitate comparative analysis of the individual methods. A real-time PCR was performed with 5 μl of DNA prepared by each method as described above. The performance of each method was evaluated by monitoring the strength of the individual fluorescent signals and by analyzing the intensities of the corresponding bands on an agarose gel.
Subsequently, it was found that a further improvement could be added to the extraction protocol with nucleic acid purifying spin columns. After addition of isopropanol, the precipitate was passed through a spin column (Roche). After removal of inhibitors and two washing steps (as recommended by the manufacturer), the DNA was recovered from the column in 50 μl of preheated elution buffer and processed immediately by real-time PCR as described above.
Sensitivity of extraction from milk.
In order to establish the sensitivity of the best method for extraction from milk, a dilution series of M. avium subsp. paratuberculosis was prepared in 1-ml portions of Ringer's diluent. Each 1-ml portion was then added to 24 ml of whole pasteurized milk (M. avium subsp. paratuberculosis free) to obtain between 4 × 106 and 4 × 101 CFU/ml. DNA was extracted by using method 6 (see above), purified with a spin column, and eluted in 50 μl. For each dilution, 5 μl of DNA was removed and amplified by using the real-time PCR protocol described above, and the overall sensitivity was determined.
Quantitation from milk.
Artificially spiked samples of milk containing between 1 × 106 and 5 × 101 CFU/ml were prepared as standards. DNA was extracted from these samples by using the optimized procedure (method 6) and was purified with spin columns as outlined above. By using the same batch of milk, a different dilution series was prepared which contained spiked M. avium subsp. paratuberculosis at concentrations ranging from 2 × 105 to 6.4 × 101 CFU/ml. DNA were also isolated and purified from these dilutions as outlined above and treated as unknown samples. A real-time PCR was carried out concurrently with 5 μl of DNA from each dilution by using the DNA from the first series as standards and DNA generated from the second series as unknown samples. This experiment was repeated on three separate days, and the ability of the Lightcycler to estimate the average microbial loads in the unknown samples after the real-time assay based on the crossing point values was determined.
RESULTS
Sensitivity of real-time PCR.
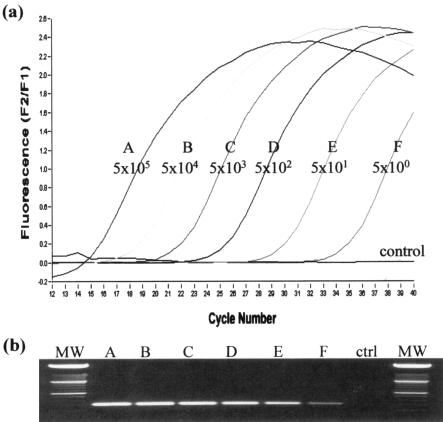
By using known amounts of template, several parameters of the assay were optimized initially. Addition of extra Mg2+ to the reaction buffer was found to be essential and had a significant impact on assay performance. We found that a final concentration of 5 mM was optimal. Similarly, the most effective annealing temperature (58°C), most effective primer and probe concentrations (0.25 μM for each primer and 0.1 μM for each probe), and most effective economic assay volume (15 μl) were determined. Subsequently, the sensitivity of the assay was evaluated by using standard concentrations of template. The reproducible experimental limit was found to be five copies as detected by 40 cycles of PCR in less than 30 min (Fig. 1a). The specificity of the reaction was confirmed by observing a product of the predicted size (354 bp) upon gel electrophoresis (Fig. 1b).
FIG. 1.
(a) Graph showing the sensitivity of the real-time PCR assay with purified PCR product from the IS900 target as the template (5 × 105 to 5 × 100 copies). (b) Corresponding gel showing the single specific 354-bp PCR product generated with the F1-R1 primer set. Lanes MW contained molecular weight markers.
Quantitation and reproducibility.
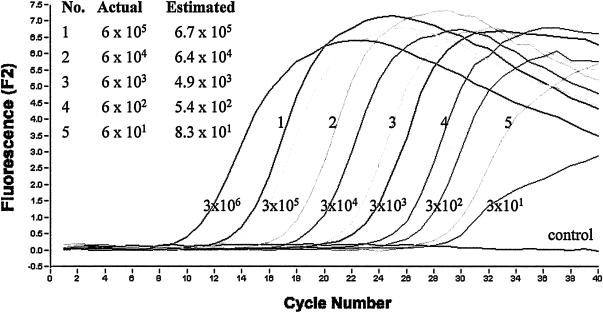
One of the main advantages of real-time PCR is the ability to quantitate unknown samples. With this assay it was possible to carry out a rapid quantitative analysis of DNA over a wide linear range with an unknown template. By using standards containing from 3 × 106 to 3 × 101 copies, accurate results for a series of samples were obtained (Fig. 2) based on data generated from experiments carried out in duplicate. The R2 value for the associated standard curve was 0.99 (data not shown), indicating that the crossing threshold values for the standards fell within accurate and acceptable experimental limits.
FIG. 2.
Quantitative analysis of purified PCR product from the IS900 target by real-time PCR. Known standards containing from 3 × 106 to 3 × 101 copies were used to estimate the numbers of targets in five unknown samples. Plots of the actual values versus measured values for the unknown standards (lines 1 to 5) are also shown. The estimated values represent the averages for two separate real-time PCR experiments carried out as described in Materials and Methods. In the negative control, DNA was replaced with water.
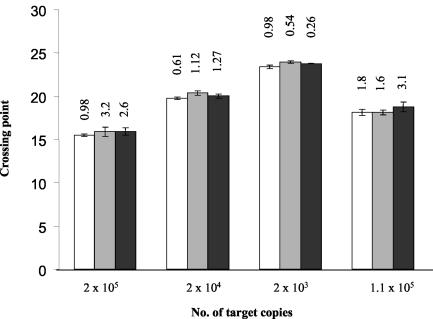
To determine the reproducibility of the assay system, amplification of DNA from four different standards was carried out nine times (triplicate experiments on three separate days), and the crossing threshold values were recorded. Analysis of these values proved that the assay was reproducible as the coefficient of variation was statistically low (average, 1.5%; standard deviation, 0.99%) (Fig. 3).
FIG. 3.
Reproducibility of the real-time PCR assay. The crossing points for four different concentrations of purified DNA were calculated in triplicate on three separate days. Each column shows the results for an assay carried out in triplicate. Each block of three columns shows the results for the same concentration on three separate days. The interassay coefficient of variation is indicated above the column for each assay.
Extraction of mycobacterial DNA from milk samples.
Considerable effort was devoted to evaluating and optimizing the most efficient protocol for extraction of mycobacterial DNA from milk. Previously, in our laboratory it was determined that inclusion of bead beating and boiling was critical for extraction of DNA from M. avium subsp. paratuberculosis (data not shown), and these procedures were thus included in each method for isolation of DNA from milk. Several other parameters were varied and evaluated, and real-time PCR analysis was carried out with the resultant DNA from each extraction. We found that the performance of each method varied greatly, and a summary of the results is presented in Fig. 4. As determined by direct comparison, it is evident that while a basic lysis solution (method 4) performed adequately and produced a relatively strong PCR signal, addition of extra steps, such as the use of proteinase K, lysozyme, or chloroform-methanol, significantly reduced the efficiency of extraction. Inclusion of sodium azide in the lysis buffer also slightly reduced the efficiency of amplification (method 5), as did inclusion of a milk-clarifying solution (method 7). The most effective strategy involved the use of a mixture containing 50 mM Tris HCl, 10 mM EDTA, 2% Triton X-100, 4 M GITC, and 0.3 M sodium acetate (method 6). This extraction solution produced the strongest fluorescent signal and most intense band on a gel and was used exclusively for all subsequent analyses of M. avium subsp. paratuberculosis in milk. It was later found that the use of nucleic acid spin columns greatly improved the quality of the DNA and, consequently, the sensitivity of the assay.
FIG. 4.
Evaluation of a variety of procedures for extraction of DNA from milk (see Materials and Methods). 1, proteinase K and lysis; 2, lysozyme, proteinase K, and lysis; 3, chloroform-methanol and lysis; 4, basic lysis; 5, alternative lysis; 6, GITC lysis; 7, clarifying solution and lysis (see text for details). Each spiked milk sample contained 104 CFU of M. avium subsp. paratuberculosis per ml, and the resulting DNA was resuspended in 50 μl of TE buffer. After PCR, the final fluorescent reading for each method was indicative of the yield or quality of DNA recovered. There is a direct correlation between peak height and corresponding band intensity on an agarose gel. Lane MW contained molecular weight markers.
Sensitivity and quantitation from milk.
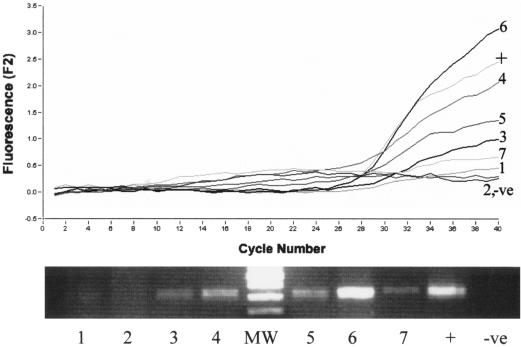
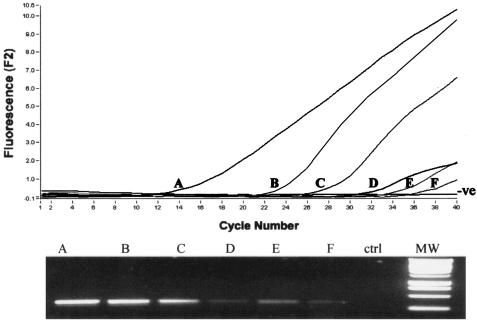
In order to critically test the potential application of the extraction protocol, the sensitivity of the assay was determined by using DNA extracted from M. avium subsp. paratuberculosis in spiked milk. Pasteurized whole milk was spiked with known dilutions of M. avium subsp. paratuberculosis, creating a series of artificially contaminated samples with concentrations of M. avium subsp. paratuberculosis ranging from 4 × 106 to 4 × 101 CFU/ml. After DNA extraction (method 6) and purification through spin columns, the real-time PCR assay was able to consistently detect less than 100 CFU/ml and had a sensitivity limit of 40 CFU/ml (Fig. 5). Moreover, when less than 10 samples were analyzed together, the entire procedure, including extraction and PCR, could be carried out in less than 3 h. The assay also exhibited excellent linearity for the range of samples analyzed, indicating that it is an appropriate procedure for both heavily and mildly contaminated samples.
FIG. 5.
Determination of the detection limit for M. avium subsp. paratuberculosis in milk by real-time PCR. The 24-ml samples were spiked with M. avium subsp. paratuberculosis to obtain between 4 × 106 and 4 × 101 CFU/ml of milk. A, 4 × 106 CFU/ml; B, 4 × 105 CFU/ml; C, 4 × 104 CFU/ml; D, 4 × 103 CFU/ml; E, 4 × 102 CFU/ml; F, 4 × 101 CFU/ml. The DNA was subsequently extracted and analyzed by real-time PCR and also by agarose gel electrophoresis. Lane MW contained molecular weight markers. ctrl, control.
The ability to rapidly quantitate M. avium subsp. paratuberculosis in milk should offer significant advantages to workers interested in screening for this organism in potentially contaminated samples. By using a wide range of standards to simulate various levels of contamination (1 × 106 to 5 × 101 CFU/ml), it was possible to ascertain M. avium subsp. paratuberculosis concentrations within acceptable limits by using the extraction and assay conditions outlined above. By using data obtained on three separate days, initial crossing point values were successfully determined with the Lightcycler software and were used to calculate unknown M. avium subsp. paratuberculosis concentrations in milk based on concurrently analyzed standards. The results of these experiments, along with the average of these values and associated standard deviations, are shown in Table 1. Furthermore, when 10 or fewer samples were processed, quantitative results for milk were obtained in less than 3 h.
TABLE 1.
Quantitation of M. avium subsp. paratuberculosis from milk by real-time PCR
| Titer (CFU/ml)a | Concn (CFU/ml)b
|
SD (%) | |||
|---|---|---|---|---|---|
| Day 1 | Day 2 | Day 3 | Avg | ||
| 200,000 | 167,400 | 111,400 | 158,700 | 145,833 | 16.8 |
| 40,000 | 14,300 | 13,980 | 15,800 | 14,693 | 5.4 |
| 8,000 | 11,270 | 12,430 | 12,970 | 12,223 | 5.8 |
| 1,600 | 5,300 | 5,373 | 5,333 | 5,335 | 0.5 |
| 320 | 63 | 1,007 | 1,007 | 882 | 20 |
| 64 | 82 | 102 | 119 | 101 | 15 |
Known titers used to generate standards for the real-time PCR assay.
Values determined with the Lightcycler software based on crossing point values for standards and unknowns.
DISCUSSION
For many years the dairy industry has been faced with a significant problem regarding M. avium subsp. paratuberculosis. This organism causes chronic disease in cattle, is difficult to grow, has been shown to survive pasteurization when it is present at high concentrations (8, 12), and has been implicated as a potential factor in the development of Crohn's disease. Consequently, the demand for rapid and sensitive milk screening assays should be a priority in the dairy farming and milk-producing industries. Ideally, any analytical method should be able to establish presence or absence, microbial load, and viability. Unfortunately, within the shelf life of liquid milk these factors are difficult to establish for M. avium subsp. paratuberculosis due to its slow growth. In order to address the quantitative difficulties, it was our intention to take technology previously developed in our laboratory (25) and design a real-time PCR assay for M. avium subsp. paratuberculosis following extraction from milk which would be rapid, sensitive, and ultimately quantitative. Recently, two such assays have been reported for M. avium subsp. paratuberculosis from bovine fecal samples, which illustrated the potential and application of this technology for slow-growing pathogens (7, 16).
In this study, the target chosen was a region of the IS900 transposon as this region is the most widely used target for M. avium subsp. paratuberculosis from milk (5, 9, 11, 26) and also multiple copies (14 to 18 copies) are present in each cell, thereby increasing the sensitivity of the assay. We decided to use fluorescent probes in place of SYBR Green due to the specificity of the probes and the greater sensitivity afforded by them. After the various PCR parameters were optimized, the assay proved to be sensitive, quantitative, and reproducible, providing a suitable alternative to previously developed methods. As expected, the real-time PCR worked exceptionally well with purified template (Fig. 1) (sensitivity, five copies, which is equivalent to less than one organism), was reproducible (Fig. 3), and was completed in less than 30 min, which is significantly faster than conventional PCR assays. Furthermore, the analysis of each sample was carried out in real time and in closed capillaries, offering the advantages of convenience and assurance of sample integrity. However, despite the apparent benefits of this procedure with purified DNA, the diagnostic applications are limited in the absence of a robust and effective procedure for extraction of M. avium subsp. paratuberculosis DNA from milk.
To this end, several extraction strategies were evaluated, and despite significant variation in yield, a strong PCR signal was observed for most methods with a moderate inoculum of M. avium subsp. paratuberculosis (104 CFU/ml) (Fig. 4). This allowed us to refine the strategy and improve the sensitivity even further. Surprisingly, pretreatment of the milk with enzymes, solvents, and clarifying agents reduced the efficiency, and simple lysis solutions proved to be the most effective treatment. In particular, the use of GITC in the lysis mixture (adopted from the method of Odumeru et al. [24]) and inclusion of nucleic acid spin columns were extremely useful in purifying the DNA.
Therefore, the combination of centrifugation, harsh lysis, physical grinding, boiling, nucleic acid purification, and real-time PCR pushed the detection limit to 40 CFU/ml of milk, which is comparable to or better than the detection limits in previously reported studies (5, 11, 21, 24, 26). Furthermore, results could be obtained in less than 3 h, which is significantly faster than results previously obtained in studies with milk. The most notable advantage of this strategy over previously reported methods is the ability to quantitate the initial titer of M. avium subsp. paratuberculosis in milk by using predetermined standards. Quantitation of DNA from microbes by using real-time PCR has been reported previously (13, 17, 20, 29) and is based on determining the crossing point for each sample (the cycle number at which the fluorescence is notably increased above a baseline level). This crossing point or threshold is unique for a particular concentration of DNA and can be used to construct a standard curve that is used to determine the corresponding concentrations of unknown samples. By using software available with the Lightcycler system, the crossing points of standards were plotted and used to accurately measure the amounts of DNA targets (Fig. 2) or the numbers of CFU per milliliter (Table 1). In both cases, quantitation was linear over a broad range and the method could be used for heavily or mildly contaminated products.
Prior to this study it was difficult to accurately determine the microbial load of M. avium subsp. paratuberculosis in milk as conventional PCR is not quantitative and decontamination procedures necessary to eliminate other organisms during culturing are lethal for a variable proportion of the M. avium subsp. paratuberculosis population. Determining the presence and number of M. avium subsp. paratuberculosis organisms in a milk sample provides a useful means for identifying contaminated product. The presence of M. avium subsp. paratuberculosis in milk samples as determined by PCR has been reported by many workers, including workers in the United Kingdom (21), the United States (26), Canada (8), and Switzerland (5), providing evidence that M. avium subsp. paratuberculosis DNA or whole cells are entering the human food chain. Using the optimized assay described above should facilitate rapid, sensitive, and quantitative screening for M. avium subsp. paratuberculosis in milk, as shown in Table 1. Although this assay was evaluated solely with spiked milk, the initial experimental results indicate that our extraction and detection methodology is equally effective in detecting M. avium subsp. paratuberculosis in naturally contaminated milk (data not shown). Further laboratory work and surveillance are being performed in order to prove this observation.
In summary, we developed and optimized a real-time PCR assay for M. avium subsp. paratuberculosis with fluorescence resonance energy transfer probes which, when combined with a strategic extraction method, generate qualitative or quantitative data for the presence of M. avium subsp. paratuberculosis in milk in less than 3 h. Implementation of this assay could lead to early identification of contaminated product and allow control measures to be put in place to protect the public from unnecessary exposure to M. avium subsp. paratuberculosis until further unambiguous evidence is made available regarding the dangers associated with this organism for humans.
Acknowledgments
This project was funded by the Irish Government under National Development Plan 2000-2006.
REFERENCES
- 1.Abolmatty, A., C. Vu, J. Oliver, and R. E. Levin. 2000. Development of a new lysis solution for releasing genomic DNA from bacterial cells for DNA amplification by polymerase chain reaction. Microbios 101:181-189. [PubMed] [Google Scholar]
- 2.Anonymous. 2000. Possible links between Crohn's disease and paratuberculosis. European Commission draft report SANCO/B3/R16/2000.1-76. Directorate, General Health and Consumer Protection, Brussels, Belgium.
- 3.Bhagwatt, A. A. 2003. Simultaneous detection of Escherichia coli O157:H7, Listeria monocytogenes and Salmonella strains by real-time PCR. Int. J. Food Microbiol. 84:217-224. [DOI] [PubMed] [Google Scholar]
- 4.Bull, T. J., E. J. McMinn, K. Sidi-Boumedine, A. Skull, D. Durkin, P. Neild, G. Rhodes, R. Pickup, and J. Hermon-Taylor. 2003. Detection and verification of Mycobacterium avium subsp. paratuberculosis in fresh ileocolonic mucosal biopsy specimens from individuals with and without Crohn's disease. J. Clin. Microbiol. 41:2915-2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Corti, S., and R. Stephan. 2002. Detection of Mycobacterium avium subspecies paratuberculosis specific IS900 insertion sequences in bulk-tank milk samples obtained from different regions throughout Switzerland. BMC Microbiol. 2:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Englund. S., G. Bolske, and K. E. Johansson. 2002. An IS900-like sequence found in a Mycobacterium sp. other than Mycobacterium avium subsp. paratuberculosis. FEMS Microbiol Lett. 209:267-271. [DOI] [PubMed] [Google Scholar]
- 7.Fang, Y., W. H. Wu, J. L. Pepper, J. L. Larsen, S. A. Marras, E. A. Nelson, W. B. Epperson, and J. Christopher-Hennings. 2002. Comparison of real-time, quantitative PCR with molecular beacons to nested PCR and culture methods for detection of Mycobacterium avium subsp. paratuberculosis in bovine fecal samples. J. Clin. Microbiol. 40:287-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao, A., L. Mutharia, S. Chen, K. Rahn, and J. Odumeru. 2002. Effect of pasteurization on survival of Mycobacterium paratuberculosis in milk. J. Dairy Sci. 85:3198-3205. [DOI] [PubMed] [Google Scholar]
- 9.Giese, S. B., and P. Ahrens. 2000. Detection of Mycobacterium avium subsp. paratuberculosis in milk from clinically affected cows by PCR and culture. Vet. Microbiol. 77:291-297. [DOI] [PubMed] [Google Scholar]
- 10.Grant, I. R., H. J. Ball, and M. T. Rowe. 1998. Isolation of Mycobacterium paratuberculosis from milk by immunomagnetic separation. Appl. Environ. Microbiol. 64:3153-3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grant, I. R., C. Pope, L. O'Riordan, H. Ball, and M. Rowe. 2000. Improved detection of Mycobacterium avium subsp. paratuberculosis in milk by immunomagnetic PCR. Vet. Microbiol. 77:369-378. [DOI] [PubMed] [Google Scholar]
- 12.Grant, I. R., E. I. Hitchings, A. McCartney, F. Ferguson, and M. T. Rowe. 2002. Effect of commercial-scale high-temperature, short-time pasteurization on the viability of Mycobacterium paratuberculosis in naturally infected cows' milk. Appl. Environ. Microbiol. 68:602-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hein, I., A. Lehner, P. Rieck, K. Klein, E. Bandl, and M. Wagner. 2001. Comparison of different approaches to quantify Staphylococcus aureus cells by real-time quantitative PCR and application of this technique for examination of cheese. Appl. Environ. Microbiol. 67:3122-3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hermon-Taylor, J., T. Bull, J. Sheridan, J. Cheng, M. Stellakis, and N. Sumar. 2000. The causation of Crohn's disease by Mycobacterium avium subspecies paratuberculosis. Can. J. Gastroenterol. 14:521-539. [DOI] [PubMed] [Google Scholar]
- 15.Johne, H. A., and L. Frothingham. 1895. Ein eigenthumlicher Fall von Tuberculose beim. Rind. Dtsch. Zeitschr. Tiern. Pathol. 21:438-454. [Google Scholar]
- 16.Kim, S. G., S. J. Shin, R. H. Jacobson, L. J. Miller, P. R. Harpending, S. M. Stehman, C. A. Rossiter, and D. A. Lein. 2002. Development and application of quantitative polymerase chain reaction assay based on the ABI 7700 system (TaqMan) for detection and quantification of Mycobacterium avium subsp. paratuberculosis. J. Vet. Diagn. Investig. 14:126-131. [DOI] [PubMed] [Google Scholar]
- 17.Kimura, B., S. Kawasaki, H. Nakano, and T. Fujii. 2001. Rapid, quantitative PCR monitoring of growth of Clostridium botulinum type E in modified-atmosphere-packaged fish. Appl. Environ. Microbiol. 67:206-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koo, K., and L. A. Jaykus. 2003. Detection of Listeria monocytogenes from a model food by fluorescence resonance energy transfer-based PCR with an asymmetric fluorogenic probe set. Appl. Environ. Microbiol. 69:1082-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kraus, G., T. Cleary, N. Miller, R. Seivright, A. K. Young, G. Spruill, and H. J. Hnatyszyn. 2001. Rapid and specific detection of the Mycobacterium tuberculosis complex using fluorogenic probes and real-time PCR. Mol. Cell Probes 15:375-383. [DOI] [PubMed] [Google Scholar]
- 20.Loeffler, J., N. Henke, H. Hebart, D. Schmidt, L. Hagmeyer, U. Schumacher, and H. Einsele. 2000. Quantification of fungal DNA by using fluorescence resonance energy transfer and the light cycler system. J. Clin. Microbiol. 38:586-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Millar, D. J., J. Ford, J. Senderson, S. Withey, M. Tizard, T. Doran, and J. Hermon-Taylor. 1996. IS900 PCR to detect Mycobacterium paratuberculosis in retail supplies of whole pasteurized cow's milk in England and Wales. Appl. Environ. Microbiol. 62:3446-3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller, N., T. Cleary, G. Kraus, A. K. Young, G. Spruill, and H. J. Hnatyszyn. 2002. Rapid and specific detection of Mycobacterium tuberculosis from acid-fast bacillus smear-positive respiratory specimens and BacT/ALERT MP culture bottles by using fluorogenic probes and real-time PCR. J. Clin. Microbiol. 40:4143-4147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moss, M. T., J. Sanderson, M. Tizard, J. Hermon-Taylor, F. EL-Zaatari, D. Markeisch, and D. Graham. 1992. PCR detection of Mycobacterium paratuberculosis in long term cultures from Crohn's disease tissues. Gut 33:1209-1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Odumeru, J., A. Gao, S. Chen, M. Raymond, and L. Muthuria. 2001. Use of the bead beater for preparation of Mycobacterium paratuberculosis template DNA in milk. Can. J. Vet. Res. 65:201-205. [PMC free article] [PubMed] [Google Scholar]
- 25.O'Mahony, J., and C. Hill. 2002. A real-time PCR assay for the detection and quantitation of Mycobacterium avium subsp. paratuberculosis using SYBR green and the Lightcycler. J. Microbiol. Methods 51:282-293. [DOI] [PubMed] [Google Scholar]
- 26.Pillai, S. R., and B. M. Jayarao. 2002. Application of IS900 PCR for detection of Mycobacterium avium subsp. paratuberculosis directly from raw milk. J. Dairy Sci. 85:1052-1057. [DOI] [PubMed] [Google Scholar]
- 27.Rubery, E. 2001. A review of the evidence for a link between exposure to Mycobacterium paratuberculosis and Crohn's disease in humans. Food Standard Agency Report. [Online.] Food Standards Agency, London, United Kingdom. http://www.foodstandards.gov.uk/multimedia/pdfs/mapcrohnreport.pdf.
- 28.Sails, A. D., A. J. Fox, F. J. Bolton, D. R. Wareing, and D. L. Greenway. 2003. A real-time PCR assay for the detection of Campylobacter jejuni in foods after enrichment culture. Appl. Environ. Microbiol. 69:1383-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sharma, V. K. 2002. Detection and quantitation of enterohemorrhagic Escherichia coli O157, O111, and O26 in beef and bovine feces by real-time polymerase chain reaction. J. Food Prot. 65:1371-1380. [DOI] [PubMed] [Google Scholar]
- 30.Sweeney, R. W. 1996. Transmission of paratuberculosis. Vet. Clin. N. Am. Food Anim. Pract. 12:305-312. [DOI] [PubMed] [Google Scholar]
- 31.Taylor, M. J., M. Hughes, R. Skuce, and S. Neill. 2001. Detection of Mycobacterium bovis in bovine clinical specimens using real-time fluorescence and fluorescence resonance energy transfer probe rapid-cycle PCR. J. Clin. Microbiol. 39:1272-1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Whittington, R. J., L. Reddacliff, I. Marsh, and V. Saunders. 1999. Detection of Mycobacterium avium subsp. paratuberculosis in formalin-fixed paraffin-embedded intestinal tissue by IS900 polymerase chain reaction. Aust. Vet. J. 77:392-397. [DOI] [PubMed] [Google Scholar]