Abstract
Chromosome replication in Escherichia coli is initiated from a single origin, oriC. Initiation involves a number of DNA binding proteins, but only DnaA is essential and specific for the initiation process. DnaA is an AAA+ protein that binds both ATP and ADP with similar high affinities. DnaA associated with either ATP or ADP binds to a set of strong DnaA binding sites in oriC, whereas only DnaAATP is capable of binding additional and weaker sites to promote initiation. Additional DNA binding proteins act to ensure that initiation occurs timely by affecting either the cellular mass at which DNA replication is initiated, or the time window in which all origins present in a single cell are initiated, i.e. initiation synchrony, or both. Overall, these DNA binding proteins modulate the initiation frequency from oriC by: (i) binding directly to oriC to affect DnaA binding, (ii) altering the DNA topology in or around oriC, (iii) altering the nucleotide bound status of DnaA by interacting with non-coding chromosomal sequences, distant from oriC, that are important for DnaA activity. Thus, although DnaA is the key protein for initiation of replication, other DNA-binding proteins act not only on oriC for modulation of its activity but also at additional regulatory sites to control the nucleotide bound status of DnaA. Here we review the contribution of key DNA binding proteins to the tight regulation of chromosome replication in E. coli cells.
Keywords: E. coli, chromosome replication, DNA binding proteins, cell mass, initiation synchrony
Timing of initiation of chromosome replication in E. coli
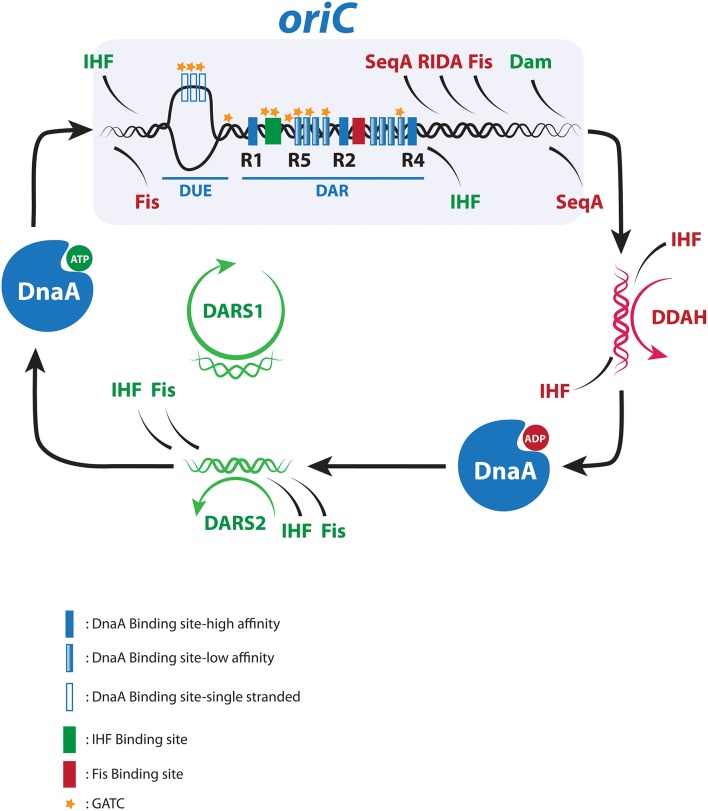
Chromosome replication in Escherichia coli is initiated from a single replication origin, oriC. The oriC-encoded structural and functional instructions for initiation are well-described (Leonard and Mechali, 2013; Skarstad and Katayama, 2013). In brief, the minimal oriC contains two functional regions: the Duplex Unwinding Element (DUE), which comprises three AT-rich repeat sequences of each 13 bp, and the flanking DnaA Assembly Region (DAR) (Figure 1; Mott and Berger, 2007; Ozaki and Katayama, 2012). DnaA is the initiator protein responsible for DUE opening and for the recruitment of replisome components and is the only protein that is both essential and specific for the initiation process (Kaguni, 2011; Leonard and Grimwade, 2011). DnaA belongs to the AAA+ proteins (ATPases Associated with diverse Activities) and can bind both ATP and ADP with similar high affinities (Sekimizu et al., 1987). The DAR region contains high affinity DnaA Boxes (R1, R4, and R2) that bind both DnaAATP and DnaAADP, along with multiple low affinity sites (R3, R5/M, I1, I2, I3, C1, C2, C3, τ1, and τ2) that bind DnaAATP (McGarry et al., 2004; Kawakami et al., 2005; Rozgaja et al., 2011). The DAR region also contains recognition sequences for two additional DNA binding proteins; IHF and Fis (Figure 1; Polaczek, 1990; Gille et al., 1991).
Figure 1.
The chromosome replication cycle. Dynamic binding of activators (green) and inhibitors (red) to oriC and distal regulatory sequences during the replication cycle. For details see text.
Throughout most of the cell cycle oriC is bound by DnaA located at R1, R2, and R4. This origin recognition complex (ORC) serves dual purposes in setting the stage for proper orisome assembly and preventing premature DNA unwinding. The ratio of DnaAATP to DnaAADP varies through the cell cycle and the peak at about 70–80% DnaAATP coincides with replication initiation (Kurokawa et al., 1999). In the current model for orisome formation, two converging DnaAATP filaments are formed (Rozgaja et al., 2011). One filament originates from R4 and grows leftward. This R4-filament displaces Fis from its binding site next to R2, which allows IHF to bind its recognition sequence next to R1. IHF bends the DNA 180° thereby bringing R1 in proximity of R5 and allows for the formation of the rightward filament responsible for duplex opening at the DUE, DnaC assisted helicase loading and assembly of the replisome (Leonard and Grimwade, 2011, 2015; Ozaki et al., 2012). Following initiation, DnaAATP is converted to DnaAADP primarily by a process called regulatory inactivation of DnaA (RIDA), which is dependent on the Hda protein bound to ADP and the DNA-loaded β-clamp of the polymerase III holoenzyme (Kato and Katayama, 2001), and by the less efficient datA-dependent DnaAATP hydrolysis (DDAH). DDAH takes place at datA and is dependent on IHF (Figure 1; Kasho and Katayama, 2013).
Coordination of initiations with cell mass increase
A long standing observation is that initiation of chromosome replication occurs when a certain cellular mass per origin, the initiation mass, is reached (Donachie, 1968; Hill et al., 2012). This coupling of replication initiation to cell growth depends on the DnaA protein. Earlier studies indicate that accumulation of DnaA protein sets the time of initiation in the cell cycle especially around or below wild-type level (Løbner-Olesen et al., 1989). On the other hand, a coordinated increase in DnaAATP and DnaAADP does not significantly increase initiation (Kurokawa et al., 1999; Flatten et al., 2015), suggesting that accumulation of DnaAATP is insufficient to trigger initiation. However, in the absence of RIDA, where DnaA is mainly ATP bound, a modest increase in DnaAATP level leads to excessive initiations from oriC (Riber et al., 2006; Fujimitsu et al., 2008), as does expression of a DnaA mutant protein insensitive to RIDA (Simmons et al., 2004). Together, this indeed suggests that accumulation of DnaAATP triggers initiation, whereas this effect can be offset by a similar increase in DnaAADP (Donachie and Blakely, 2003). The participation of DnaAADP in orisome formation remains unclear (Leonard and Grimwade, 2015), but the above observations suggest that it affects initiation negatively. Overall, accumulation of DnaA protein during steady-state growth, along with the cell cycle specific peak in DnaAATP/DnaAADPratio, determines the onset of initiation with little variation between individual cells.
Coordination of initiations within a single cell
In individual cells, initiation at all origins occurs within approximately 1/10 of the doubling time (Initiation period, IP; Figure 2A). Rapidly growing cells with overlapping replication cycles therefore predominantly contain 2n (n = 1, 2, 3) copies of oriC, referred to as initiation synchrony (Skarstad et al., 1986). Initiation synchrony depends on the immediate inactivation of newly replicated origins by sequestration. oriC contain 11 copies of the sequence GATC that are methylated by Dam methyltransferase and bound, i.e., sequestered, by SeqA when hemimethylated. Sequestration prevents DnaA binding to its weak sites in oriC (Nievera et al., 2006) for approximately 1/3 generation (Sequestration period, SP; Figure 2A) and serves to keep track of which origins have been initiated (Boye and Løbner-Olesen, 1990; Campbell and Kleckner, 1990; Lu et al., 1994). The ability to initiate all origins in synchrony could result from maintaining a high DnaAATP level throughout IP. Alternatively the first origin initiated may release its DnaAATP to assist in triggering successive initiations at remaining origins in a cascade-like manner to ensure that free DnaAATP increases through IP and enforces synchrony (Løbner-Olesen et al., 1994). These models predict different outcomes for sequestration deficient cells. A high DnaAATP level throughout IP would result in re-initiation(s) within IP, asynchrony and overinitiation. The cascade model predicts a delay between successive initiations due to newly initiated origins competing with old origins for a limited amount of DnaAATP. The initiation frequency would be directly proportional with accumulation of DnaAATP resulting in asynchrony but an unchanged overall initiation frequency, which is in accordance with experimental observations for Dam deficient cells (Boye and Løbner-Olesen, 1990; Løbner-Olesen et al., 1994).
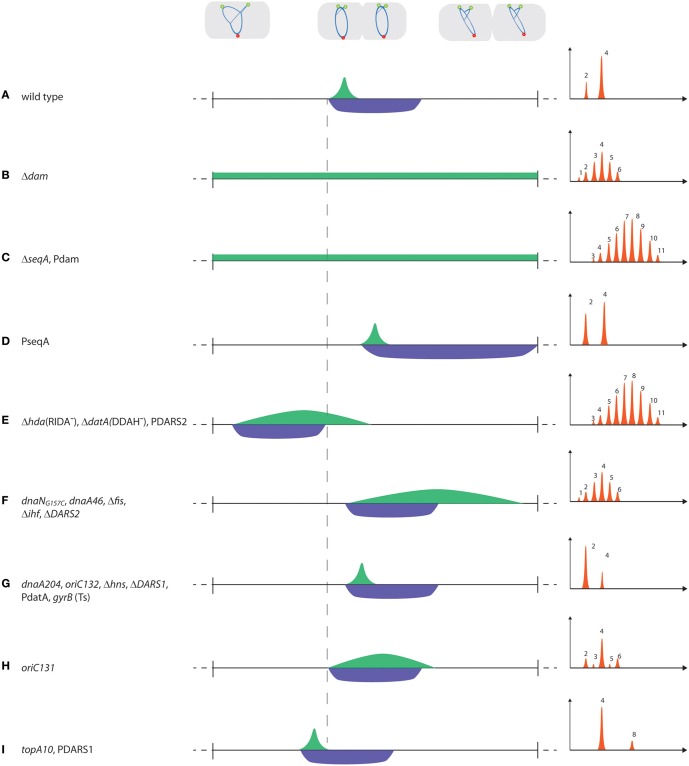
Figure 2.
Timing of replication initiation. Examples of mutants/plasmids with altered initiation (IP; green) and sequestration (SP; blue) periods. The horizontal line represents one doubling time, whereas the vertical (hyphenated) line illustrates the time of initiation of the first origin in wild-type cells. Note that the start of SP always coincides with the first origin initiated, i.e., start of IP. In the graphical representation of initiation synchrony, the number of origins per cell are on the X-axis, whereas the cell number is on the Y-axis of each histogram. When more than one mutation/plasmid is listed for a specific example (e.g., in C,E–G,I), the histograms are representative of the initiation phenotype of each individual mutation/plasmid.
Synchrony is only observed when IP < SP (Figure 2A). In cells with aberrant timing of initiation, the IP and SP periods change, i.e., either start earlier in the cell cycle at a decreased initiation mass, i.e., overinitiation, or are delayed with an increased initiation mass, i.e., underinitiation. Alternatively, the duration of IP and SP may change relative to each other, and when IP > SP, newly initiated origins, released from sequestration, compete with origins not yet initiated. Consequently, some origins are re-initiated while others are not initiated at all, leading to loss of synchrony (Olsson et al., 2003; Skarstad and Løbner-Olesen, 2003). This is exemplified by dam mutants without a sequestration period that initiate throughout the cell cycle (Figure 2B; Boye and Løbner-Olesen, 1990; Lu et al., 1994). seqA mutants are also asynchronous but have a higher origin concentration, possibly because DnaA is increased, relative to dam mutants (Figure 2C; Campbell and Kleckner, 1990; von Freiesleben et al., 1994). Increased levels of Dam will, due to faster re-methylation rates, reduce SP and when this becomes shorter than IP, asynchrony follows (Figure 2C; Boye and Løbner-Olesen, 1990; von Freiesleben et al., 2000a). Excess SeqA protein delays initiation, prolongs the sequestration period but does not affect synchrony (Figure 2D; Bach et al., 2003; Charbon et al., 2011). During sequestration the activity of DnaA is lowered by RIDA and DDAH. RIDA is presumably accelerated by generation of new replication forks at initiation and hence more DNA loaded β-clamps (Moolman et al., 2014). Similarly, DDAH is increased shortly after initiation when the datA locus is duplicated and together they ensure a post-initiation decrease in the DnaAATP/DnaAADP ratio (Figure 1). RIDA (Δhda) and to a lesser degree DDAH (ΔdatA) deficient cells fail to lower the ratio of DnaAATP/DnaAADP to prevent re-initiation following sequestration. This results in asynchrony and early initiation at a reduced cell mass (Figure 2E; Kitagawa et al., 1998; Fujimitsu et al., 2008; Kasho and Katayama, 2013). On the other hand, the dnaNG157C mutant, which is more active in RIDA (dnaN encodes the β-clamp), or extra copies of datA, results in delayed initiation and, for dnaNG157C cells, also produces asynchrony (Figures 2F,G; Morigen et al., 2001; Gon et al., 2006; Charbon et al., 2011; Johnsen et al., 2011). During sequestration, the overall level of free DnaA is reduced by titration (Hansen et al., 1991; Kitagawa et al., 1996, 1998; Ogawa et al., 2002) and by arrest of de novo DnaA synthesis (Campbell and Kleckner, 1990).
Modulation of timing of replication initiation by DNA binding proteins
Several DNA binding proteins affect either the cell mass at initiation, the initiation synchrony, or both. These proteins either bind specifically to oriC to affect DnaA binding, non-specifically to DNA to alter oriC topology, or they bind sequences important for the nucleotide bound status of DnaA.
Proteins that specifically interact with oriC prior to initiation
The most important protein to interact with oriC prior to initiation is DnaA. Mutations in DnaA that affect nucleotide binding, such as dnaA46, are presumably somewhat deficient in formation of DnaA multimers on oriC, which results in delayed initiation and a prolonged initiation period (Skarstad et al., 1988; Boye et al., 1996). As sequestration remains unchanged (IP > SP), dnaA46 cells are asynchronous (Figure 2F; Skarstad and Løbner-Olesen, 2003). Mutations in DnaA that affect DNA binding, but not nucleotide binding (e.g., dnaA204), lead to late but synchronous initiation (Figure 2G; Skarstad et al., 1988; Torheim et al., 2000). The ability to form DnaAATP filaments on oriC therefore seems of greater importance for initiation synchrony than a tight anchoring to DnaA binding sites.
Conflicting data exist on the role of Fis for timing of initiation. Binding Fis to oriC in vitro is reported to either inhibit initiation of replication by inducing conformational changes at oriC that prevent orisome formation (Wold et al., 1996; Ryan et al., 2002, 2004), or have no effect on initiation (Margulies and Kaguni, 1998). Cells with a mutated primary Fis binding in oriC (oriC131) have an origin concentration similar to wild-type (Figure 2H; Weigel et al., 2001; Riber et al., 2009; Flatten and Skarstad, 2013). Fis-deficient cells, on the other hand, have a lowered origin concentration (Flatten and Skarstad, 2013; Kasho et al., 2014), suggesting that initiation is delayed (Figure 2F). However, because Fis affects multiple cellular processes due to its involvement in DNA organization one should be careful in assessing its role in initiation solely based on the behavior of Fis-deficient cells. Both Fis deficiency or loss of its primary oriC binding site result in initiation asynchrony (Figures 2F,H; Riber et al., 2009; Flatten and Skarstad, 2013), indicating that these cells are deficient for proper orisome assembly and/or for preventing premature DNA unwinding. The role of IHF in replication timing is less controversial. An oriC mutant with a disrupted IHF binding site (oriC132) is somewhat deficient in orisome formation and has delayed but synchronous initiation (Figure 2G; Weigel et al., 2001; Skarstad and Løbner-Olesen, 2003; Riber et al., 2009). ihf mutant cells also initiate replication at an increased mass per origin consistent with a stimulatory role of IHF on initiation. Cells deficient in IHF are on the other hand asynchronous (Figure 2F; von Freiesleben et al., 2000b). This is in agreement with an additional role of IHF for DnaAATP generation at DARS2 (see below).
A number of proteins negatively regulate initiation of replication in vitro. These include ArcA that binds to 13 mer AT rich repeats, to DnaA box R1 and to the IHF binding site in oriC, and IciA that binds to 13-mer AT-rich repeats in oriC (Hwang and Kornberg, 1990; Lee et al., 2001). The impact of ArcA and IciA on replication initiation in vivo is modest (Nystrom et al., 1996) or not known, respectively. The stationary-phase induced CspD protein binds ssDNA to inhibit replication initiation and elongation in vitro, whereas no in vivo data are available (Yamanaka et al., 2001). Upon association with Cnu and/or Hha, H-NS (see below) binds to a specific sequence in oriC that overlaps DnaA box R5 (Kim et al., 2005; Yun et al., 2012). Cells deficient in Cnu and/or Hha are, however, similar to wild-type (Kim et al., 2005). Finally, the protein Rob binds to a single site in oriC in vitro, but does not affect initiation in vivo (Skarstad et al., 1993).
DNA binding proteins that affect topology of oriC
In E. coli the genomic DNA is mostly negatively supercoiled (Wang et al., 2013). Unconstrained supercoiling of oriC contributes to the ease of duplex opening and is determined by transcription (not covered here; for review see Magnan and Bates, 2015) along with the actions of topoisomerase I and DNA gyrase enzymes (Wu et al., 1988). Mutations in topoisomerase I, which removes negative supercoils, result in initiation at a slightly reduced mass while synchrony is maintained (Figure 2I; von Freiesleben and Rasmussen, 1992; Olsson et al., 2003). Conversely, temperature sensitive gyrB mutant cells, with moderately reduced negative superhelicity of the chromosome, enhance the temperature sensitivity of a dnaA46 mutant (Filutowicz, 1980) and show delayed synchronous initiations (Figure 2G; von Freiesleben and Rasmussen, 1991; Usongo et al., 2013). This suggests that initiation is facilitated by an increase in negative superhelicity of the chromosome. However, topA-gyr mutations influence chromosome segregation, R-loop formation and possibly induce stable DNA replication independent of oriC (Usongo et al., 2013, 2016) making it difficult to assess the effect of large changes in overall supercoiling on replication initiation. In vivo, nucleoid-associated proteins (NAPs; Dillon and Dorman, 2010), such as IHF, Fis, H-NS, HU, and MukFEB constrain negative supercoils to condense the chromosome and could therefore affect initiation of chromosome replication (Badrinarayanan et al., 2015; Lal et al., 2016). H-NS deficient cells have an increased negative superhelicity of the genome (Mojica and Higgins, 1997; Hardy and Cozzarelli, 2005). Yet, genetic evidence suggests that loss of H-NS hampers initiation (Katayama et al., 1996), and H-NS deficient cells initiate replication in synchrony at an increased cell mass (Figure 2G; Kaidow et al., 1995; Atlung and Hansen, 2002). The HU protein can substitute for IHF in DnaA-mediated unwinding of oriC in vitro (Hwang and Kornberg, 1992) although their mechanisms of action differ (Ryan et al., 2002). In vivo, genetic evidence suggests that loss of HU stimulates initiation despite decreased negative supercoiling (Louarn et al., 1984). Loss of MukB, involved in condensation of the bacterial chromosome (Hiraga et al., 1989; Cui et al., 2008), results in reduced negative supercoiling (Weitao et al., 2000), but initiations remain synchronous (Weitao et al., 1999). It is not known whether MukB affects the initiation mass. Finally, the starvation-induced NAP, Dps, binds non-specifically to oriC, and interacts with the N-terminus of DnaA, inhibiting DNA unwinding in vitro. Loss of Dps does not result in loss of synchrony, but increases the cellular origin content somewhat (Chodavarapu et al., 2008). In summary, it seems that NAPs modulate replication initiation but that the effect is not solely mediated through an effect on DNA supercoiling.
Getting ready for the next round of replication
At later cell cycle stages DnaAATP is regenerated for the next initiation to take place (Figure 1). E. coli can rejuvenate DnaAADP to DnaAATP in a process assisted by acidic phospholipids (Saxena et al., 2013) or at two non-coding chromosomal sites called DARS1 and DARS2 (Fujimitsu et al., 2009). DARS1 and DARS2 are located in each replichore halfway between oriC and terC, and are duplicated after the end of sequestration. Multiple DnaAADP molecules form complexes with DARS to facilitate release of ADP resulting in apo-DnaA, which will primarily rebind ATP as this is more abundant than ADP within the cell (Petersen and Møller, 2000).
DARS1 is not known to be regulated by any proteins, whereas rejuvenation at the more efficient DARS2 locus is dependent on binding of both IHF and Fis (Kasho et al., 2014). While Fis binds DARS2 throughout the cell cycle, IHF provides cell cycle specificity to DARS2 activity by only binding and activating DARS2 immediately prior to initiation to ensure an increase in DnaAATP level (Fujimitsu et al., 2009; Kasho et al., 2014). Extra copies of DARS1 or DARS2 will increase the overall DnaAATP level, which results in early initiation (Figures 2E,I) and for DARS2 also extends IP, thereby resembling RIDA deficiency (Figure 2E; Fujimitsu et al., 2009; Charbon et al., 2011). Deletion of DARS1, DARS2, or both reduces the ability to reactivate DnaA for new initiations in the following cell cycle and results in delayed initiation (Figures 2F,G; Fujimitsu et al., 2009; Kasho et al., 2014; Frimodt-Moller et al., 2015). Loss of DARS2 also increases the relative duration of the initiation period, leading to initiation asynchrony (Figure 2F; Fujimitsu et al., 2009; Frimodt-Moller et al., 2015). This suggests that both DARS1 and DARS2 are important for coupling initiation to cell mass increase, whereas only the cell-cycle regulated DARS2 is crucial for maintaining initiation synchrony.
Concluding remarks
Overall, timing of chromosome replication in E. coli takes place at least at two levels. First, initiation of replication is tightly coupled to cell mass increase through accumulation of DnaAATP. Second, synchrony of initiations within the single cell is not necessarily connected to initiation mass but results from each origin being simultaneously initiated only once per generation, with asynchrony originating from failure to obey this once-and-only-once rule. DnaA remains the only replication protein solely required for initiation at oriC, but additional proteins act on oriC and elsewhere to assist in coupling of replication to cell growth and synchrony. In particular IHF and Fis display complex functions, targeting several regulatory sites. IHF has a dual role on replication initiation, acting both positively (i.e., binding to DARS2 and oriC) and negatively (i.e., binding to datA). Also, IHF binds oriC at the pre-initiation stage and interacts with datA and DARS2 following initiation. Binding of IHF to these regions is suggested to be temporally regulated so that IHF binds to oriC, to datA and to DARS2 in a successive manner during cell cycle progression (Kasho and Katayama, 2013; Kasho et al., 2014). In vivo, ihf mutants display an initiation-compromised phenotype, indicating that the overall role of IHF on initiation of replication appears positive.
For a long time, the contribution of Fis in initiation regulation has been questioned. Recent studies do, however, suggest an overall positive role of Fis in replication initiation (Flatten and Skarstad, 2013; Kasho et al., 2014), which likely results from ensuring ordered orisome formation by preventing premature IHF binding and DNA unwinding (Leonard and Grimwade, 2015) and from stimulating DnaAATP rejuvenation at DARS2. As the cellular Fis level depends on both growth- rate and phase, it could adjust chromosome replication to the bacterial growth rate through its activity on DARS2 (Kasho et al., 2014).
Author contributions
All authors listed, have made substantial, direct and intellectual contribution to the work, and approved it for publication.
Funding
This research was part of the Center for Bacterial Stress Response and Persistence (BASP) funded by a grant from the Danish National Research Foundation (DNRF120) and by a grant from the Novo Nordisk Foundation.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The reviewer MM declared a past co-authorship with the author ALO to the handling Editor, who ensured that the process met the standards of a fair and objective review.
References
- Atlung T., Hansen F. G. (2002). Effect of different concentrations of H-NS protein on chromosome replication and the cell cycle in Escherichia coli. J. Bacteriol. 184, 1843–1850. 10.1128/JB.184.7.1843-1850.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach T., Krekling M. A., Skarstad K. (2003). Excess SeqA prolongs sequestration of oriC and delays nucleoid segregation and cell division. EMBO J. 22, 315–323. 10.1093/emboj/cdg020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badrinarayanan A., Le T. B., Laub M. T. (2015). Bacterial chromosome organization and segregation. Annu. Rev. Cell Dev. Biol. 31, 171–199. 10.1146/annurev-cellbio-100814-125211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boye E., Løbner-Olesen A. (1990). The role of Dam methyltransferase in the control of DNA replication in E. coli. Cell 62, 981–989. 10.1016/0092-8674(90)90272-G [DOI] [PubMed] [Google Scholar]
- Boye E., Stokke T., Kleckner N., Skarstad K. (1996). Coordinating DNA replication initiation with cell growth: differential roles for DnaA and SeqA proteins. Proc. Natl. Acad. Sci. U.S.A. 93, 12206–12211. 10.1073/pnas.93.22.12206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell J. L., Kleckner N. (1990). E. coli oriC and the dnaA gene promoter are sequestered from dam methyltransferase following the passage of the chromosomal replication fork. Cell 62, 967–979. 10.1016/0092-8674(90)90271-F [DOI] [PubMed] [Google Scholar]
- Charbon G., Riber L., Cohen M., Skovgaard O., Fujimitsu K., Katayama T., et al. (2011). Suppressors of DnaA(ATP) imposed overinitiation in Escherichia coli. Mol. Microbiol. 79, 914–928. 10.1111/j.1365-2958.2010.07493.x [DOI] [PubMed] [Google Scholar]
- Chodavarapu S., Gomez R., Vicente M., Kaguni J. M. (2008). Escherichia coli Dps interacts with DnaA protein to impede initiation: a model of adaptive mutation. Mol. Microbiol. 67, 1331–1346. 10.1111/j.1365-2958.2008.06127.x [DOI] [PubMed] [Google Scholar]
- Cui Y., Petrushenko Z. M., Rybenkov V. V. (2008). MukB acts as a macromolecular clamp in DNA condensation. Nat. Struct. Mol. Biol. 15, 411–418. 10.1038/nsmb.1410 [DOI] [PubMed] [Google Scholar]
- Dillon S. C., Dorman C. J. (2010). Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat. Rev. Microbiol. 8, 185–195. 10.1038/nrmicro2261 [DOI] [PubMed] [Google Scholar]
- Donachie W. D. (1968). Relationship between cell size and time of initiation of DNA replication. Nature 219, 1077–1079. 10.1038/2191077a0 [DOI] [PubMed] [Google Scholar]
- Donachie W. D., Blakely G. W. (2003). Coupling the initiation of chromosome replication to cell size in Escherichia coli. Curr. Opin. Microbiol. 6, 146–150. 10.1016/S1369-5274(03)00026-2 [DOI] [PubMed] [Google Scholar]
- Filutowicz M. (1980). Requirement of DNA gyrase for the initiation of chromosome replication in Escherichia coli K-12. Mol. Gen. Genet. 177, 301–309. 10.1007/BF00267443 [DOI] [PubMed] [Google Scholar]
- Flatten I., Fossum-Raunehaug S., Taipale R., Martinsen S., Skarstad K. (2015). The DnaA protein is not the limiting factor for initiation of replication in Escherichia coli. PLoS Genet. 11:e1005276. 10.1371/journal.pgen.1005276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flatten I., Skarstad K. (2013). The Fis protein has a stimulating role in initiation of replication in Escherichia coli in vivo. PLoS ONE 8:e83562. 10.1371/journal.pone.0083562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frimodt-Moller J., Charbon G., Krogfelt K. A., Løbner-Olesen A. (2015). Control regions for chromosome replication are conserved with respect to sequence and location among Escherichia coli strains. Front. Microbiol. 6:1011. 10.3389/fmicb.2015.01011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimitsu K., Senriuchi T., Katayama T. (2009). Specific genomic sequences of E. coli promote replicational initiation by directly reactivating ADP-DnaA. Genes Dev. 23, 1221–1233. 10.1101/gad.1775809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimitsu K., Su'etsugu M., Yamaguchi Y., Mazda K., Fu N., Kawakami H., et al. (2008). Modes of overinitiation, dnaA gene expression, and inhibition of cell division in a novel cold-sensitive hda mutant of Escherichia coli. J. Bacteriol. 190, 5368–5381. 10.1128/JB.00044-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gille H., Egan J. B., Roth A., Messer W. (1991). The FIS protein binds and bends the origin chromosomal DNA replication, oriC, of Escherichia coli. Nucleic Acids Res. 19, 4167–4172. 10.1093/nar/19.15.4167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gon S., Camara J. E., Klungsøyr H. K., Crooke E., Skarstad K., Beckwith J. (2006). A novel regulatory mechanism couples deoxyribonucleotide synthesis and DNA replication in Escherichia coli. EMBO J. 25, 1137–1147. 10.1038/sj.emboj.7600990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen F. G., Christensen B. B., Atlung T. (1991). The Initiator titration model: computer simulation of chromosome and minichromosome control. Res. Microbiol. 142, 161–167. 10.1016/0923-2508(91)90025-6 [DOI] [PubMed] [Google Scholar]
- Hardy C. D., Cozzarelli N. R. (2005). A genetic selection for supercoiling mutants of Escherichia coli reveals proteins implicated in chromosome structure. Mol. Microbiol. 57, 1636–1652. 10.1111/j.1365-2958.2005.04799.x [DOI] [PubMed] [Google Scholar]
- Hill N. S., Kadoya R., Chattoraj D. K., Levin P. A. (2012). Cell size and the initiation of DNA replication in bacteria. PLoS Genet 8:e1002549. 10.1371/journal.pgen.1002549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraga S., Niki H., Ogura T., Ichinose C., Mori H., Ezaki B., et al. (1989). Chromosome partitoning in Escherichia coli: novel mutants producing Anucleate Cells. J. Bacteriol. 171, 1496–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang D. S., Kornberg A. (1990). A novel protein binds a key origin sequence to block replication of an E. coli minichromosome. Cell 63, 325–331. 10.1016/0092-8674(90)90165-B [DOI] [PubMed] [Google Scholar]
- Hwang D. S., Kornberg A. (1992). Opening of the replication origin of Escherichia coli by DnaA protein with protein HU or IHF. J. Biol. Chem. 267, 23083–23086. [PubMed] [Google Scholar]
- Johnsen L., Flatten I., Morigen Dalhus, B., Bjørås M., Waldminghaus T., Skarstad K. (2011). The G157C mutation in the Escherichia coli sliding clamp specifically affects initiation of replication. Mol. Microbiol. 79, 433–446. 10.1111/j.1365-2958.2010.07453.x [DOI] [PubMed] [Google Scholar]
- Kaguni J. M. (2011). Replication initiation at the Escherichia coli chromosomal origin. Curr. Opin. Chem. Biol. 15, 606–613. 10.1016/j.cbpa.2011.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaidow A., Wachi M., Nakamura J., Magae J., Nagai K. (1995). Anucleate cell production by Escherichia coli delta hns mutant lacking a histone-like protein, H-NS. J. Bacteriol. 177, 3589–3592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasho K., Fujimitsu K., Matoba T., Oshima T., Katayama T. (2014). Timely binding of IHF and Fis to DARS2 regulates ATP-DnaA production and replication initiation. Nucleic Acids Res. 42, 13134–13149. 10.1093/nar/gku1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasho K., Katayama T. (2013). DnaA binding locus datA promotes DnaA-ATP hydrolysis to enable cell cycle-coordinated replication initiation. Proc. Natl. Acad. Sci. U.S.A. 110, 936–941. 10.1073/pnas.1212070110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama T., Takata M., Sekimizu K. (1996). The nucleoid protein H-NS facilitates chromosome DNA replication in Escherichia coli dnaA mutants. J. Bacteriol. 178, 5790–5792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato J., Katayama T. (2001). Hda, a novel DnaA-related protein, regulates the replication cycle in Escherichia coli. EMBO J. 20, 4253–4262. 10.1093/emboj/20.15.4253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami H., Keyamura K., Katayama T. (2005). Formation of an ATP-DnaA-specific initiation complex requires DnaA Arginine 285, a conserved motif in the AAA+ protein family. J. Biol. Chem. 280, 27420–27430. 10.1074/jbc.M502764200 [DOI] [PubMed] [Google Scholar]
- Kim M. S., Bae S. H., Yun S. H., Lee H. J., Ji S. C., Lee J. H., et al. (2005). Cnu, a novel oriC-binding protein of Escherichia coli. J. Bacteriol. 187, 6998–7008. 10.1128/JB.187.20.6998-7008.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa R., Mitsuki H., Okazaki T., Ogawa T. (1996). A novel DnaA protein-binding site at 94.7 min on the Escherichia coli chromosome. Mol. Microbiol. 19, 1137–1147. 10.1046/j.1365-2958.1996.453983.x [DOI] [PubMed] [Google Scholar]
- Kitagawa R., Ozaki T., Moriya S., Ogawa T. (1998). Negative control of replication initiation by a novel chromosomal locus exhibiting exceptional affinity for Escherichia coli DnaA protein. Genes Develop. 12, 3032–3043. 10.1101/gad.12.19.3032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurokawa K., Nishida S., Emoto A., Sekimizu K., Katayama T. (1999). Replication cycle-coordinated change of the adenine nucleotide-bound forms of DnaA protein in Escherichia coli. EMBO J. 18, 6642–6652. 10.1093/emboj/18.23.6642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal A., Dhar A., Trostel A., Kouzine F., Seshasayee A. S., Adhya S. (2016). Genome scale patterns of supercoiling in a bacterial chromosome. Nat. Commun. 7:11055. 10.1038/ncomms11055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. S., Han J. S., Jeon Y., Hwang D. S. (2001). The arc two-component signal transduction system inhibits in vitro Escherichia coli chromosomal initiation. J. Biol. Chem. 276, 9917–9923. 10.1074/jbc.M008629200 [DOI] [PubMed] [Google Scholar]
- Leonard A. C., Grimwade J. E. (2011). Regulation of DnaA assembly and activity: taking directions from the genome. Annu. Rev. Microbiol. 65, 19–35. 10.1146/annurev-micro-090110-102934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard A. C., Grimwade J. E. (2015). The orisome: structure and function. Front. Microbiol. 6:545. 10.3389/fmicb.2015.00545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard A. C., Mechali M. (2013). DNA replication origins. Cold Spring Harb. Perspect. Biol. 5:a010116. 10.1101/cshperspect.a010116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Løbner-Olesen A., Hansen F. G., Rasmussen K. V., Martin B., Kuempel P. L. (1994). The initiation cascade for chromosome replication in wild-type and Dam methyltransferase deficient Escherichia coli cells. EMBO J. 13, 1856–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Løbner-Olesen A., Skarstad K., Hansen F. G., von Meyenburg K., Boye E. (1989). The DnaA protein determines the initiation mass of Escherichia coli K- 12. Cell 57, 881–889. 10.1016/0092-8674(89)90802-7 [DOI] [PubMed] [Google Scholar]
- Louarn J., Bouche J. P., Louarn J. M. (1984). Genetic inactivation of topoisomerase I suppresses a defect in initiation of chromosome replication in E. coli. Mol. Gen. Genet. 195, 170–174. 10.1007/BF00332741 [DOI] [PubMed] [Google Scholar]
- Lu M., Campbell J. L., Boye E., Kleckner N. (1994). SeqA: a negative modulator of replication initiation in E. coli. Cell 77, 413–426. 10.1016/0092-8674(94)90156-2 [DOI] [PubMed] [Google Scholar]
- Magnan D., Bates D. (2015). Regulation of DNA replication initiation by Chromosome structure. J. Bacteriol. 197, 3370–3377. 10.1128/JB.00446-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies C., Kaguni J. M. (1998). The FIS protein fails to block the binding of DnaA protein to oriC, the Escherichia coli chromosomal origin. Nucleic Acids Res. 26, 5170–5175. 10.1093/nar/26.22.5170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGarry K. C., Ryan V. T., Grimwade J. E., Leonard A. C. (2004). Two discriminatory binding sites in the Escherichia coli replication origin are required for DNA strand opening by initiator DnaA-ATP. Proc. Natl. Acad. Sci. U.S.A. 101, 2811–2816. 10.1073/pnas.0400340101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojica F. J., Higgins C. F. (1997). In vivo supercoiling of plasmid and chromosomal DNA in an Escherichia coli hns mutant. J. Bacteriol. 179, 3528–3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moolman M. C., Krishnan S. T., Kerssemakers J. W., van den Berg A., Tulinski P., Depken M., et al. (2014). Slow unloading leads to DNA-bound beta2-sliding clamp accumulation in live Escherichia coli cells. Nat. Commun. 5, 5820. 10.1038/ncomms6820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morigen Boye, E., Skarstad K., L8bner-Olesen A. (2001). Regulation of chromosomal replication by DnaA protein availability in Escherichia coli: effects of the datA region. Biochim. Biophys. Acta. 1521, 73–80. 10.1016/S0167-4781(01)00292-5 [DOI] [PubMed] [Google Scholar]
- Mott M. L., Berger J. M. (2007). DNA replication initiation: mechanisms and regulation in bacteria. Nat. Rev. Microbiol. 5, 343–354. 10.1038/nrmicro1640 [DOI] [PubMed] [Google Scholar]
- Nievera C., Torgue J. J., Grimwade J. E., Leonard A. C. (2006). SeqA blocking of DnaA-oriC interactions ensures staged assembly of the E. coli pre-RC. Mol. Cell 24, 581–592. 10.1016/j.molcel.2006.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nystrom T., Larsson C., Gustafsson L. (1996). Bacterial defense against aging: role of the Escherichia coli ArcA regulator in gene expression, readjusted energy flux and survival during stasis. EMBO J. 15, 3219–3228. [PMC free article] [PubMed] [Google Scholar]
- Ogawa T., Yamada Y., Kuroda T., Kishi T., Moriya S. (2002). The datA locus predominantly contributes to the initiator titration mechanism in the control of replication initiation in Escherichia coli. Mol. Microbiol. 44, 1367–1375. 10.1046/j.1365-2958.2002.02969.x [DOI] [PubMed] [Google Scholar]
- Olsson J. A., Nordstrom K., Hjort K., Dasgupta S. (2003). Eclipse-synchrony relationship in Escherichia coli strains with mutations affecting sequestration, Initiation of replication and superhelicity of the bacterial chromosome. J. Mol. Biol. 334, 919–931. 10.1016/j.jmb.2003.10.029 [DOI] [PubMed] [Google Scholar]
- Ozaki S., Katayama T. (2012). Highly organized DnaA-oriC complexes recruit the single-stranded DNA for replication initiation. Nucleic Acids Res. 40, 1648–1665. 10.1093/nar/gkr832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozaki S., Noguchi Y., Hayashi Y., Miyazaki E., Katayama T. (2012). Differentiation of the DnaA-oriC subcomplex for DNA unwinding in a replication initiation complex. J. Biol. Chem. 287, 37458–37471. 10.1074/jbc.M112.372052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen C., Møller L. B. (2000). Invariance of the nucleoside triphosphate pools of Escherichia coli with growth rate. J. Biol. Chem. 275, 3931–3935. 10.1074/jbc.275.6.3931 [DOI] [PubMed] [Google Scholar]
- Polaczek P. (1990). Bending of the origin of replication of E. coli by binding of IHF at a specific site. New Biol. 2, 265–271. [PubMed] [Google Scholar]
- Riber L., Fujimitsu K., Katayama T., Løbner-Olesen A. (2009). Loss of Hda activity stimulates replication initiation from I-box, but not R4 mutant origins in Escherichia coli. Mol. Microbiol. 71, 107–122. 10.1111/j.1365-2958.2008.06516.x [DOI] [PubMed] [Google Scholar]
- Riber L., Olsson J. A., Jensen R. B., Skovgaard O., Dasgupta S., Marinus M. G., et al. (2006). Hda-mediated inactivation of the DnaA protein and dnaA gene autoregulation act in concert to ensure homeostatic maintenance of the Escherichia coli chromosome. Genes Dev. 20, 2121–2134. 10.1101/gad.379506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozgaja T. A., Grimwade J. E., Iqbal M., Czerwonka C., Vora M., Leonard A. C. (2011). Two oppositely oriented arrays of low-affinity recognition sites in oriC guide progressive binding of DnaA during Escherichia coli pre-RC assembly. Mol. Microbiol. 82, 475–488. 10.1111/j.1365-2958.2011.07827.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan V. T., Grimwade J. E., Camara J. E., Crooke E., Leonard A. C. (2004). Escherichia coli prereplication complex assembly is regulated by dynamic interplay among Fis, IHF and DnaA. Mol. Microbiol. 51, 1347–1359. 10.1046/j.1365-2958.2003.03906.x [DOI] [PubMed] [Google Scholar]
- Ryan V. T., Grimwade J. E., Nievera C. J., Leonard A. C. (2002). IHF and HU stimulate assembly of pre-replication complexes at Escherichia coli oriC by two different mechanisms. Mol. Microbiol. 46, 113–124. 10.1046/j.1365-2958.2002.03129.x [DOI] [PubMed] [Google Scholar]
- Saxena R., Fingland N., Patil D., Sharma A. K., Crooke E. (2013). Crosstalk between DnaA protein, the initiator of Escherichia coli chromosomal replication, and acidic phospholipids present in bacterial membranes. Int. J. Mol. Sci. 14, 8517–8537. 10.3390/ijms14048517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekimizu K., Bramhill D., Kornberg A. (1987). ATP activates dnaA protein in initiating replication of plasmids bearing the origin of the E. coli chromosome. Cell 50, 259–265. 10.1016/0092-8674(87)90221-2 [DOI] [PubMed] [Google Scholar]
- Simmons L. A., Breier A. M., Cozzarelli N. R., Kaguni J. M. (2004). Hyperinitiation of DNA replication in Escherichia coli leads to replication fork collapse and inviability. Mol. Microbiol. 51, 349–358. 10.1046/j.1365-2958.2003.03842.x [DOI] [PubMed] [Google Scholar]
- Skarstad K., Boye E., Steen H. B. (1986). Timing of initiation of chromosome replication in individual E. coli cells. EMBO J. 5, 1711–1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skarstad K., Katayama T. (2013). Regulating DNA replication in bacteria. Cold Spring Harb. Perspect. Biol. 5:a012922. 10.1101/cshperspect.a012922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skarstad K., Løbner-Olesen A. (2003). Stable co-existence of separate replicons in Escherichia coli is dependent on once-per-cell-cycle initiation. EMBO J. 22, 140–150. 10.1093/emboj/cdg003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skarstad K., Thöny B., Hwang D. S., Kornberg A. (1993). A novel binding protein of the origin of the Escherichia coli chromosome. J. Biol. Chem. 268, 5365–5370. [PubMed] [Google Scholar]
- Skarstad K., von Meyenburg K., Hansen F. G., Boye E. (1988). Coordination of chromosome replication initiation in Escherichia coli: effects of different dnaA alleles. J. Bacteriol. 170, 852–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torheim N. K., Boye E., Løbner-Olesen A., Stokke T., Skarstad K. (2000). The Escherichia coli SeqA protein destabilizes mutant DnaA204 protein. Mol. Microbiol. 37, 629–638. 10.1046/j.1365-2958.2000.02031.x [DOI] [PubMed] [Google Scholar]
- Usongo V., Martel M., Balleydier A., Drolet M. (2016). Mutations reducing replication from R-loops suppress the defects of growth, chromosome segregation and DNA supercoiling in cells lacking topoisomerase I and RNase HI activity. DNA Repair 40, 1–17. 10.1016/j.dnarep.2016.02.001 [DOI] [PubMed] [Google Scholar]
- Usongo V., Tanguay C., Nolent F., Bessong J. E., Drolet M. (2013). Interplay between type 1A topoisomerases and gyrase in chromosome segregation in Escherichia coli. J. Bacteriol. 195, 1758–1768. 10.1128/JB.02001-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Freiesleben U., Krekling M. A., Hansen F. G., Løbner-Olesen A. (2000a). The eclipse period of Escherichia coli. EMBO J. 19, 6240–6248. 10.1093/emboj/19.22.6240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Freiesleben U., Rasmussen K. V. (1991). DNA replication in Escherichia coli gyrB(Ts) mutants analysed by flow cytometry. Res. Microbiol. 142, 223–227. 10.1016/0923-2508(91)90034-8 [DOI] [PubMed] [Google Scholar]
- von Freiesleben U., Rasmussen K. V. (1992). The level of supercoiling affects the regulation of DNA replication in Escherichia coli. Res. Microbiol. 143, 655–663. 10.1016/0923-2508(92)90060-2 [DOI] [PubMed] [Google Scholar]
- von Freiesleben U., Rasmussen K. V., Atlung T., Hansen F. G. (2000b). Rifampicin resistant initiation of chromosome replication from oriC in ihf mutants. Mol. Microbiol. 37, 1–8. 10.1046/j.1365-2958.2000.02060.x [DOI] [PubMed] [Google Scholar]
- von Freiesleben U., Rasmussen K. V., Schaechter M. (1994). SeqA limits DnaA activity in replication from oriC in Escherichia coli. Mol. Microbiol. 14, 763–772. 10.1111/j.1365-2958.1994.tb01313.x [DOI] [PubMed] [Google Scholar]
- Wang X., Montero L. P., Rudner D. Z. (2013). Organization and segregation of bacterial chromosomes. Nat. Rev. Genet. 14, 191–203. 10.1038/nrg3375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel C., Messer W., Preiss S., Welzeck M., Boye M., Boye E. (2001). The sequence requirements for a functional Escherichia coli replication origin are different for the chromosome and a minichromosome. Mol. Microbiol. 40, 498–507. 10.1046/j.1365-2958.2001.02409.x [DOI] [PubMed] [Google Scholar]
- Weitao T., Nordstrom K., Dasgupta S. (1999). Mutual suppression of mukB and seqA phenotypes might arise from their opposing influences on the Escherichia coli nucleoid structure. Mol. Microbiol. 34, 157–168. 10.1046/j.1365-2958.1999.01589.x [DOI] [PubMed] [Google Scholar]
- Weitao T., Nordström K., Dasgupta S. (2000). Escherichia coli cell cycle control genes affect chromosome superhelicity. EMBO Rep. 1, 494–499. 10.1093/embo-reports/kvd106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wold S., Crooke E., Skarstad K. (1996). The Escherichia coli Fis protein prevents initiation of DNA replication from oriC in vitro. Nucleic Acids Res. 24, 3527–3532. 10.1093/nar/24.18.3527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H. Y., Shyy S. H., Wang J. C., Liu L. F. (1988). Transcription generates positively and negatively supercoiled domains in the template. Cell 53, 433–440. 10.1016/0092-8674(88)90163-8 [DOI] [PubMed] [Google Scholar]
- Yamanaka K., Zheng W., Crooke E., Wang Y. H., Inouye M. (2001). CspD, a novel DNA replication inhibitor induced during the stationary phase in Escherichia coli. Mol. Microbiol. 39, 1572–1584. 10.1046/j.1365-2958.2001.02345.x [DOI] [PubMed] [Google Scholar]
- Yun S. H., Ji S. C., Jeon H. J., Wang X., Kim S. W., Bak G., et al. (2012). The CnuK9E H-NS complex antagonizes DNA binding of DicA and leads to temperature-dependent filamentous growth in E. coli. PLoS ONE 7:e45236. 10.1371/journal.pone.0045236 [DOI] [PMC free article] [PubMed] [Google Scholar]