Abstract
We describe a genetic instability found in natural wine yeasts but not in the common laboratory strains of Saccharomyces cerevisiae. Spontaneous cyh2R/cyh2R mutants resistant to high levels of cycloheximide can be directly isolated from cyh2S/cyh2S wine yeasts. Heterozygous cyh2R/cyh2S hybrid clones vary in genetic instability as measured by loss of heterozygosity at cyh2. There were two main classes of hybrids. The lawn hybrids have high genetic instability and generally become cyh2R/cyh2R homozygotes and lose the killer phenotype under nonselective conditions. The papilla hybrids have a much lower rate of loss of heterozygosity and maintain the killer phenotype. The genetic instability in lawn hybrids is 3 to 5 orders of magnitude greater than the highest loss-of-heterozygosity rates previously reported. Molecular mechanisms such as DNA repair by break-induced replication might account for the asymmetrical loss of heterozygosity. This loss-of-heterozygosity phenomenon could be economically important if it causes sudden phenotype changes in industrial or pathogenic yeasts and of more basic importance to the degree that it influences the evolution of naturally occurring yeast populations.
Saccharomyces cerevisiae is a good model for the study of genetic instability. Genetic stability is important for industrial (fermentation) and health (genetic diseases, cancer, and drug resistance) improvements and plays a key role in our understanding of evolution. In S. cerevisiae, genetic instability is associated with a high rate of loss of heterozygosity (LOH), chromosome size changes, and the appearance of spontaneous recessive homozygous mutants (14). Although the genetic instability of yeasts has been analyzed in detail in recent years (1, 4, 5, 9, 19, 24, 26), little is known about the causes of this instability or the suppression mechanisms in genetically stable cells (reviewed by Kolodner et al. [14]). More than 50 genes (many of them involved in DNA recombination, S-phase checkpoints, and telomere maintenance) have been implicated in the maintenance of S. cerevisiae genome stability (14). The inactivation of S-phase checkpoints and recombination, mismatch repair, and telomere maintenance defects are among the known causes of genetic rearrangements (14).
Most commercial wine yeasts are naturally occurring strains of S. cerevisiae isolated from wines and spontaneously fermenting musts. The phenotype of these wild yeasts usually is more variable than that of domesticated laboratory yeast strains, which have necessarily been selected for genetic stability to obtain reproducible research results. In wine yeasts, karyotype changes have been reported during vegetative growth (3, 15, 21), as have genetic changes that modify the yeast's metabolic properties (24). Genetic instability may alter useful properties of industrial yeasts, resulting in problems in biotechnological processes or lower quality of products such as bread, pastry, beer, or wine. For example, loss of the killer phenotype may result in protracted wine fermentation (22). Thus, obtaining genetically stable yeasts from economically interesting industrial and natural yeast strains is of economic importance. Moreover, the mechanisms responsible for genetic instability may help drive the evolution of genetic variability in natural yeast populations and could be used for controlled in vivo manipulation of the yeast genome.
Naturally occurring strains of S. cerevisiae usually are very sensitive to cycloheximide (CYH) and are inhibited by 1 to 1.5 μg/ml. The frequency of spontaneous CYH-resistant (CYHR) mutants is <2 × 10−5 (6, 23). Of the known CYHR mutations, the best characterized occur at the cyh2 gene, which encodes the 60S ribosomal subunit L28 protein. This mutation changes amino acid Gln (Q) 37 to Glu (E) (13, 33). The CYHR phenotype has been described as recessive (30), semidominant (6, 11, 18, 32), and dominant (27).
We analyzed the genetic instability of heterozygous cyh2R/cyh2S hybrids from naturally occurring wine yeasts. We chose the CYHR phenotype because (i) it is easy to obtain spontaneous CYHR mutants from diploid wine yeasts without altering their physiological and technological properties (23); (ii) by using the CYHR and killer K2 phenotypes, which are common in wine yeasts, heterozygous hybrids of homothallic yeasts can be obtained without the need for crosses with laboratory strains, thereby preserving the yeast's natural properties such as the genome instability itself (27); and (iii) it is easy to detect the homozygous yeasts (cyh2R/cyh2R or cyh2S/cyh2S) that originate from the heterozygous hybrids (cyh2R/cyh2S).
Our objectives in this study were to characterize and quantify the genetic instability in natural wine yeasts and hybrids synthesized in the laboratory due to LOH at the cyh2 locus. We show that the heterozygous yeast populations, when grown under nonselective laboratory conditions, can change their phenotype more rapidly than was previously known. By understanding this phenomenon, the sudden loss of interesting properties of industrial yeasts can be avoided and our knowledge of how these organisms change and evolve in industrial and field settings can be increased.
MATERIALS AND METHODS
Yeast strains, culture media, and phenotype tests.
S. cerevisiae wine yeast strains were isolated from several Spanish wineries. All of the strains are prototrophic, homothallic, and homozygous for CYH sensitivity (cyhS/cyhS) and sporulate on appropriate media. EX33 is a virus-free killer-sensitive yeast. The rest (EX1, EX34, EX47, EX70, EX73, EX85, EX88, and EX93) are killer K2. The yeasts were sampled, isolated, and identified as previously described (29). 88P1A and 85P4D are cyhS/cyhS killer K2 spore clones from EX88 and EX85, respectively. 73R and 85R are spontaneous CYHR killer-sensitive mutants, which have lost the ScV-M2 virus, derived from EX73 and EX85, respectively. Our study focused on these two mutants because they are resistant to high concentrations of CYH (≥50 μg/ml) and thus are easy to work with and because the mutation is thought to be located in the cyh2 gene (30). Isolation of spontaneous CYHR mutants and MIC determination were performed as described by Pérez et al. (23). 73R11D, 85R4A, 85R6B, and 85R23D are cyhR/cyhR killer-sensitive spore clones from 73R and 85R, respectively. Laboratory yeast strains (Table 1) were used for genetic mapping and the killer assay. Standard culture media were used for yeast growth and phenotype tests in the genetic mapping (10). YEPD+CYH is yeast extract-peptone-dextrose (YEPD) agar supplemented with CYH to the desired final concentration (2 μg/ml unless a different concentration is given). Minimal medium for auxotroph analysis was Difco yeast nitrogen base (without amino acids, with ammonium sulfate; Difco, Detroit, Mich.).
TABLE 1.
S. cerevisiae yeast strains used for genetic mapping and killer phenotype assay
| Strain | Origin | Genotype |
|---|---|---|
| F573 | C. R. Vázquez de Aldanaa (from G. R. Fink) | MATaspo11 ura3 ade1 his1 leu2 lys7 met3 trp5 |
| F570 | C. R. Vázquez de Aldana (from G. R. Fink) | MATα spo11 ura3 ade6 arg4 aro7 asp5 met14 lys2 pet17 trp1 |
| F568 | C. R. Vázquez de Aldana (from G. R. Fink) | MATα spo11 ura3 can1 cyh2 ade2 his7 hom3 tyr1 |
| F572 | C. R. Vázquez de Aldana (from G. R. Fink) | MATα spo11 ura3 his2 leu1 lys1 met4 pet8 |
| F4 | A. G. Hinnebuschb | MATα leu2-3 leu2-112 ura3-52 ino1 GCN2 [HIS4::lacZ ura3-52] [k1+] |
| F1 | A. G. Hinnebusch | MATaino1 can1 ura3-52 [k1+] |
| F19 | A. G. Hinnebusch | MATaura3-52 leu2 [HIS4::lacZ ura3-52] GCN3 gcn2::LEU2 gcn1-1 [k1+] |
| F15 | A. G. Hinnebusch | MATaura3 his3 trp1 [k1+] |
| 5 × 47 | J. C. Ribasc (from R. B. Wickner) | MATα/MATaho/ho [k10] [k20] |
| F166 | J. C. Ribas (from R. B. Wickner) | MATα leu1 kar1 LA-HNB M1 [k1+] |
C. R. Vázquez de Aldana, Departamento de Microbiologia y Genética, Universidad de Salamanca, Salamanca, Spain.
A. G. Hinnebusch, Laboratory of Gene Regulation and Development, National Institute of Child Health and Human Development, National Institutes of Health, Bethesda, Md.
J. C. Ribas, Departamento de Microbiologia y Genética, Universidad de Salamanca, Salamanca, Spain.
Standard yeast genetic procedures were used for sporulation of cultures and dissection of asci (12). Cells were grown on YEPD plates for 2 days at 30°C, transferred to sporulation plates (1% potassium acetate, 0.1% Bacto Yeast Extract, 0.05% glucose, 2% Bacto Agar) and incubated for 7 to 20 days at 25°C until more than 50% of the cells had sporulated. Twenty-four asci from each parental yeast were dissected on YEPD plates and incubated for 5 days at 30°C, at which time the percentage of viable spores was determined. The homothallic spore clones were tested for phenotypic segregation by replica plating on the appropriate media. The phenotype test was performed only if spore viability was >95%. The assay for killer activity was performed with low-pH (pH 4) blue plates (4MB) (12) seeded with 100 μl of a 48-h culture of a sensitive strain (EX33 or 5x47). Strains being tested for killer activity were loaded (4 μl of a 48-h culture to produce a patch approximately 5 mm in diameter) or replica plated onto the seeded 4MB plates and incubated for 4 days at 20°C. Killer strains produce a clear halo as result of killing the seeded sensitive yeasts.
Detection of genetic instability.
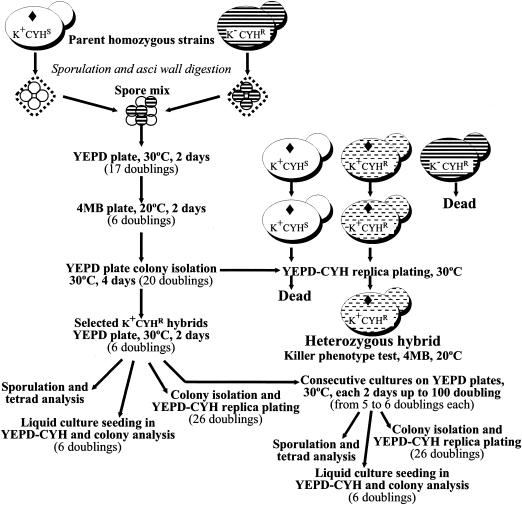
Standard genetic crosses were performed to map the cyhR mutations (10). Hybridization of natural homothallic wine strains was performed as described by Ramírez et al. (27). All of the hybrids from each cross are isogenic because the parent strains are spore clones from homothallic wine yeasts. Genetic instability was assayed by measuring LOH in the populations of newly obtained heterozygous cyhR/cyhS hybrids before and after manipulation (Fig. 1). All of the hybrids were replicated and grown at the same time on the same YEPD plate (side by side) for each transfer, so all grew under the same nonselective conditions (pH 6, 30°C; absence of CYH and killer activity). The number of population doublings for each step was determined by measuring the initial and final numbers of viable cells on YEPD plates after suitable suspension and dilution of the cultures. The starting material was hybrids selected from different crosses. By the time the hybrids were obtained, their population had already undergone 32 to 49 doublings since zygote formation, depending on when cell conjugation occurred following germination of the mixed spores. The newly obtained hybrids were cultured by serial transfers on YEPD plates at 30°C (nonselective conditions) every 24 h until the population had undergone 100 doublings (from 18 to 20 transfers). The yeast population of the hybrids (both newly obtained and after 100 doublings) was analyzed as follows.
FIG. 1.
General scheme for homothallic wine yeast hybridization, hybrid manipulations, and genetic population analyses. The number of doublings of yeasts for each step is in parentheses; calculations assume 100% cell viability. The cell types present are illustrated as schematic yeasts with relevant phenotypes identified. ⧫, ScV-M2 killer virus.
Sporulation and tetrad analysis.
Twenty-four tetrads of each hybrid were dissected with a tetrad dissection microscope (Micro Video Instruments, Inc., Avon, Mass.) on YEPD plates, incubated for 4 days at 30°C, and replica plated with sterile velvet to YEPD+CYH and YEPD (control). Heterozygous yeasts produce tetrads whose spores segregate 2CYHR:2CYHS. The 4CYHR:0CYHS or 0CYHR:4CYHS segregations belong to tetrads of homozygous resistant or sensitive yeasts that have lost heterozygosity (LOH).
Liquid culture seeding on YEPD-CYH and colony analysis.
The hybrids were inoculated (4 × 106 to 6 × 106 cells/ml from a YEPD plate culture) into sterile tubes (18-mm diameter) containing 2 ml of liquid YEPD and incubated at 30°C to saturation (for 2 days with shaking at 250 rpm, 2 × 108 to 4 × 108, about six doublings). A suitably diluted sample of each culture was spread onto a YEPD-CYH plate to obtain isolated colonies. The number of colonies of different sizes was determined. Ten to 20 colonies of each type of colony from each type of hybrid were isolated and subjected to killer phenotype and tetrad analyses.
Colony isolation and YEPD+CYH replica plating.
Colonies isolated in YEPD plates were harvested, replicated on another YEPD plate, incubated at 30°C for 2 days (roughly six doublings), replica plated to YEPD-CYH and YEPD, and incubated for 1 to 8 days at 30°C to determine CYH resistance. Ten to 20 colonies of each phenotype from each type of hybrid were harvested from the original YEPD isolation plate and subjected to killer phenotype and tetrad analysis.
In parallel with the genetic analysis, we analyzed the karyotype (pulsed-field electrophoresis) (2), the restriction pattern of mitochondrial DNA (25), and the nuclear and cellular morphology (fluorescence microscopy of 4′,6′-diamidino-2-phenylindole [DAPI]-stained yeasts) (4) of the hybrids before and after 100 doublings.
Genetic mapping, PCR, and DNA sequencing of CYHR mutants.
The cyhR mutations were mapped by analyzing crosses of single-spore clones from spontaneous CYHR mutants with a group of haploid yeast strains in which all of the S. cerevisiae chromosomes were genetically marked (Table 1). Yeast DNA was extracted as previously described (25). DNA samples for sequencing were amplified in a Pharmacia LKB-Gene ATAQ Controller (Pharmacia, Uppsala, Sweden) with the Ready-To-Go PCR Beads kit (catalog no. 27-9555-01) in accordance with the manufacturer's protocol. The primers used were F1 (5′-GAGACGCAAACGTTTTTCCTCGCA-3′; similar to a sequence located 157 to 181 bp upstream of the cyh2 start codon) and B1 (5′-GATAAAAACGTTGGGATCTGCCAC-3′; similar to a sequence located 185 to 209 bp downstream of the cyh2 gene translation stop codon [TAA] with the opposite orientation). PCR products were run in 1× Tris-acetate-EDTA electrophoresis buffer-1% agarose gels. As expected, the PCR with F1 and B1 gave a single 1,301-bp band. Purified PCR fragments were sequenced (at the Centro de Secuenciación de DNA, Facultad de Farmacia, Universidad Complutense de Madrid, Madrid, Spain) by standard methods in an ABI PRISM 377 DNA Sequencer (AME Bioscience Ltd., London, United Kingdom) by using the BigDye Terminator Cycle Sequencing ready reaction kit from PE Biosystems (Foster City, Calif.).
Virus (ScV-LA and ScV-M2) dsRNA extraction, purification, and agarose gel electrophoresis.
Rapid yeast double-stranded RNA (dsRNA) extraction and purification was performed as previously described (8). dsRNA molecules were separated in 1× TAE-1% agarose gel for 60 to 75 min.
RESULTS
Genetic stability of naturally occurring wine yeasts.
Spontaneous homozygous cyhR/cyhR mutants were recovered from half of the wine yeast strains analyzed (frequency of 1.3 × 107 to 1.4 × 105). However, continued propagation of the same wine yeast strains under laboratory conditions (20 to 24 transfers on YEPD plates at 30°C) yielded only a few spontaneous CYHR mutants for which the MIC was low. Genetic mapping and sequencing of the CYHR mutations confirmed that they mapped to the cyh2 gene on the left arm of chromosome VII. The 73R and 73R11D mutants (for which the MIC was 100 μg/ml) had a previously reported mutation, Gln37 to Glu (nucleotide C623 to G) (13, 33). However, all of the mutants analyzed for which the MIC was >100 μg/ml (500 to 1,000 μg/ml) had a previously unreported substitution at the same amino acid, Gln37 to Lys (nucleotide C623 to A).
Stability of newly obtained heterozygous cy2hR/cyh2S hybrids.
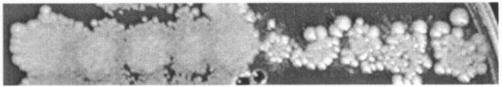
Heterozygous cyh2R/cyh2S hybrids were obtained by crossing cyh2R/cyh2R (85R6B) with cyh2S/cyh2S (88P1A and 85P4D) yeast spore clones. There were two classes of hybrids that could be distinguished following replica plating on YEPD+CYH: papilla (Pa) hybrids and lawn (Ln) hybrids (Fig. 2). The Ln hybrids grew slightly slower than the cyh2R/cyh2R parent at CYH concentrations of >50 μg/ml. Of the 334 hybrids analyzed, 149 were Ln hybrids and 185 were Pa hybrids. Each hybrid was a CYHR killer K2 clone from a single cell (Fig. 1) and was not cultured with CYH (unless indicated).
FIG. 2.
Phenotype of cyh2R/cyh2S heterozygous hybrids after replica plating on YEPD-CYH (2 μg/ml) plates. Lawn phenotype (Ln hybrids), five patches on left; papilla phenotype (Pa hybrids), five patches on right.
The 10 Pa hybrids analyzed were heterozygous (100% 2R:2S tetrad segregation). The Pa hybrids formed two main classes of single-cell colonies in YEPD+CYH, i.e., large colonies that appeared after 36 h and small colonies that appeared after 4 to 5 days and never reached the normal large size of S. cerevisiae colonies, although some of them grew to a medium size after several days. The frequency of large colonies ranged from 0.3 to 9.4% (Table 2). The large colonies originated from the cyh2R/cyh2R subpopulation present in the Pa hybrids, as deduced from the phenotypic segregation for CYH (100% 4R:0S) and mutant allele segregation (4cyh2R:0cyh2S). The papillae in the YEPD+CYH replica plates (Fig. 2) were large colonies originating from the cyh2R/cyh2R yeasts. Small colonies were the most frequent (82 to 98%) and segregated 2R:2S after tetrad dissection.
TABLE 2.
Genetic analysis of isogenic hybrids (from the cross 85R6B × 88P1A) newly obtained and after 100 doublingsa
| Hybrid | Pheno- type | Frequency (%) of colonies newly obtained (32-49 doublings)
|
Pheno- type | Frequency (%) of colonies after 100 doublings (total, 132-149)
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Liquid culture seeding in YEPD-CYH (+6 doublings)
|
Colony isolation and YEPD-CYH replicating (+26 doublings)
|
Liquid culture seeding in YEPD-CYH (+6 doublings)
|
Colony isolation and YEPD-CYH replicating (+26 doublings)
|
|||||||||||
| La colony | Me colony | Sm colony | Ln clone | Pa clone | Se clone | La colony | Me colony | Sm colony | Ln clone | Pa clone | Se clone | |||
| H7-8 | Ln, K | 29 | 0 | 71 | 50 | 50 | 0 | Ln, Se | 100 | 0 | 0 | 99 | 0 | 0.4 |
| H6-7 | Ln, K | 22 | 0 | 78 | 48 | 52 | 0 | Ln, Se | 100 | 0 | 0 | 100 | 0 | 0 |
| H1-1 | Pa, K | 9.4 | 8.3 | 82 | 0 | 100 | 0 | Pa, K | 0.9 | 1.9 | 97 | 0 | 99 | 0.3 |
| H2-1 | Pa, K | 0.3 | 1.8 | 98 | 0 | 100 | 0 | Pa, K | 0.9 | 3.4 | 96 | 0.8 | 99 | 0 |
Phenotypes: Ln, lawn; Pa, papillae (in YEPD-CYH after replica plating); K, killer, S, sensitive to K2 killer toxin. Colony sizes (single cell colony sizes in YEPD-CYH): La, large; Me, medium; Sm, small. Clone phenotypes (phenotypes after replica plating in YEPD-CYH of single-cell clones previously isolated in YEPD plate): K, killer; Ln, lawn; Pa, papillae; Se, sensitive to CYH (no growth). The small colonies in YEPD-CYH and their spore clones are killer; the large colonies and their spore clones are killer sensitive and have no ScV-M2 virus. Only two hybrids of each type are shown. Very similar results were obtained with the rest of the analyzed Pa or Ln hybrids from the 85R6B × 88P1A and 85R6B × 85P4D crosses.
Most of the tetrads from the 10 Ln hybrids analyzed also segregated 2R:2S (71 to 74%), but there were more 4R:0S tetrads (26 to 29%) than in the Pa hybrids (0.3 to 9.4%). The number of large colonies in Ln hybrids (22 to 29%) also was higher than in the Pa hybrids, which probably explains the lawn phenotype seen after replica plating on YEPD+CYH. Some of the cyh2R/cyh2S strains undergo LOH to become cyh2R/cyh2R during the 26 doublings that occur during colony isolation and replica plating analysis, and the frequency of the cyh2R/cyh2R yeasts approximately doubles (Table 2). This high LOH was not observed in Pa hybrids (presumably more genetically stable), which yielded 82 to 98% small colonies.
Changes to cyh2R/cyh2R homozygosity under nonselective conditions.
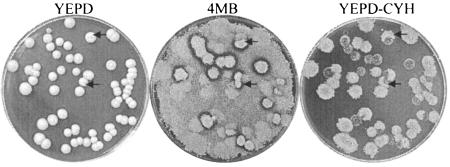
All hybrids maintained the original CYHR phenotype (lawn or papillae) after 100 doublings on YEPD with no CYH or killer activity. During this time the Ln hybrids lost the killer phenotype while Pa hybrids maintained it (Table 2). All of the cyh2R/cyh2R single-cell colonies from all of the hybrids became killer sensitive, i.e., they retained ScV-LA but lost ScV-M2, while all of the cyh2R/cyh2S and cyh2S/cyh2S colonies remained killers, i.e., they retained both ScV-LA and ScV-M2 (Fig. 3).
FIG. 3.
Replica plating on YEPD+CYH and 4MB (previously seeded with killer-sensitive strain EX33) of H6-7 Ln hybrid (after 25 doublings in YEPD at 30°C) colonies isolated in YEPD. The arrows indicate colonies with sectors of LOH in the cyh2 gene and killer phenotype loss.
More than 90% of the Ln hybrid populations became homozygous for cyh2R. The cyh2R/cyh2S yeasts all but disappeared (no small colonies or Pa clones), with only 0.4% Se clones in H7-8 after 100 doublings (Table 2). The population of heterozygous cyh2R/cyh2S single-cell clones isolated from the H6-7 and H7-8 Ln hybrids also became fully homozygous cyh2R/cyh2R after 100 doublings in YEPD (data not shown). In the H6-7 Ln hybrid, sectors of colonies that were cyh2R/cyh2R killer sensitive were detected, but no cyh2S/cyh2S sectors were found (Fig. 3). Since these yeasts are homothallic, the cyh2R/cyh2R cells could result from sporulation and mating of sister cells from the same spore clone (genomic renewal) (20). However, we detected no spores during the serial subcultures of hybrids on YEPD.
Among the Pa hybrids (Table 2), cyh2S/cyh2S cells increased for H1-1 and cyh2R/cyh2R cells increased for H2-1, while in the other hybrids (data not shown) there either was no change or the proportion of cyh2S/cyh2S yeasts decreased. This variation (always <15%) could be due to random changes in the hybrid populations that occurred during the doublings they underwent during the genetic analyses. In general, among the Pa hybrids the proportion of cyh2R/cyh2S cells decreased slightly and the proportion of cyh2S/cyh2S and/or cyh2R/cyh2R cells increased slightly.
Stability of parts of the genome other than cyh2 in Ln hybrids.
No differences were detected between the different hybrid populations in studies of pulsed-field gel electrophoresis, mitochondrial DNA restriction pattern, or nuclear and cellular morphology (data not shown). Therefore, it is unlikely that the genetic instability affects the gross organization of the cell's DNA.
DISCUSSION
On the basis of the high frequency of spontaneous cyh2R/cyh2R mutations, the genomes of some naturally occurring wine yeasts appear to be unstable. The occurrence of homozygous recessive mutants suggests that these yeasts form homozygous cyhR/cyhR strains from heterozygous cyhR/cyhS spontaneous mutants, since the frequency of simultaneous mutations in both copies of the same gene is very low (<10−12/diploid cell/division). This phenomenon is not detected in these yeasts after continued propagation under laboratory culture conditions. The original yeasts are cyhS/cyhS, so this mechanism should function during the mutant selection process in YEPD-CYH.
Heterozygous hybrids of these wild yeasts maintain the genome instability. Under nonselective conditions, about half of the hybrids (Ln hybrids) have high LOH and become homozygous cyh2R/cyh2R. The other half of the hybrids (Pa hybrids) are genetically stable. Although two classes approximately equal in frequency result from this process, the stability is not heritable as a Mendelian character. The high LOH in the Ln hybrids could explain the apparently dominant behavior of the CYH2R phenotype in some wine yeasts (27) and the two phenotypes (lawn and papillae) of the heterozygous hybrids. It also makes it possible to use a recessive selectable genetic marker (such as cyh2R) to obtain new hybrids from natural homothallic yeasts; i.e., the diploid heterozygous hybrids can easily be detected if they are genetically unstable because they frequently become homozygous for the marker (27).
The LOH rate depended upon the hybrid, the time of analysis, and the calculation method but was approximately 0.7%/diploid cell/generation on the basis of the method of Puig et al. (24). This value is 3 to 5 orders of magnitude greater than the highest LOH rates previously reported (7, 24) and greater than the estimated mitotic recombination frequency in the region of chromosome VII in which cyh2 is located (7). A similar phenomenon has been reported for spontaneous homozygous LEU1/LEU1 revertants from leu1-12/leu1-12 cells with a frequency of 0.01%/diploid cell/division (7).
Near the cyh2 gene there are many repeated sequences, e.g., retrotransposon long terminal repeats (see http://www.ncbi.nlm.nih.gov/mapview/maps.cgi?org=scerevisiae&chr=VII), a chromosomal origin of replication, and some genes involved in DNA recombination (rad54 and rad6) that could be responsible for the genetic instability and LOH we observed.
Theoretically, the cyh2R/cyh2R cells could originate from mitotic gene conversion, loss of a chromosome followed by reduplication, mitotic crossing over, or a general hyperrecombination phenotype. However, the reciprocal homozygote cyh2S/cyh2S also should occur at the same frequency because of these mechanisms, but this phenotype was never observed. To test these possibilities genetically, we made hybrids between the unstable wine yeasts and laboratory yeast strains with appropriate genetic markers, but only Pa hybrids resulted. Thus, the high genetic instability phenotype is not maintained after genetic crosses are made with domesticated laboratory yeasts. No segregation of genetic instability was observed in 16 crosses of spore clones from the parents 85R6B and 88P1A (eight spores, i.e., two tetrads, from each parent were crossed with one spore from the other parent) (data not shown). Neither did we detect any significant change in the frequency of hybrid phenotypes as the result of possible homozygosity of an instability determinant, an expected result since these wine yeasts all are homothallic and generally homozygous for their entire genomes.
The LOH mechanism appears biased toward cyh2R/cyh2R homozygosity. For example, in sectored colonies of the H6-7 Ln hybrid, sectors of cyh2R/cyh2R killer-sensitive yeasts were detected but sectors of cyh2S/cyh2S yeasts were not (Fig. 3). One possible mechanism is that in the heterozygous hybrids there is a sequence close to cyh2 at which frequent DNA double-strand breaks can occur. These breaks are repaired in the Ln hybrids, but not in Pa hybrids, by break-induced replication (14, 16, 17, 31). If the double-strand break always occurs in the chromosome carrying the cyh2S allele, then the homologous cyh2R region would serve as the template for repair events, resulting in gene conversion of cyh2S to cyh2R. Alternatively, there could be a sequence that facilitates break-induced recombination that could explain the asymmetry of the repair event. Such a sequence would be cis acting and, in our system, functional only for the chromosome carrying the cyh2S allele (17). After the initial double-strand break, both fragments could act as the invasive 3′ end in break-induced replication. In one case, both the sequence that facilitates break-induced recombination and the cyh2S allele are converted to yield a cyh2R/cyh2R strain. In another case, the resulting recombinants will be cyh2R/cyh2S and the sequence that facilitates break-induced recombination would still be located on the chromosome carrying the cyh2S allele and a new break-induced recombination event could occur. For each break-induced recombination event, the number of cyh2S alleles decreases by half, and eventually the Ln hybrid population becomes effectively cyh2R/cyh2R homozygous.
Break-induced recombination also could explain the 50:50 Ln/Pa ratio if break-induced recombination occurred just once after hybrid formation, resulting in half unstable Ln hybrids and half stable Pa hybrids. One or more of the genes involved in the break-induced replication mechanism (or its regulation) (31) and/or sequence differences between homologous chromosomes could be responsible for the observed phenotype.
We observed directly that the genetic instability was propagated through subculture of the Ln hybrids until they became fully homozygous. When four crosses were made with each of four spore clones from a tetrad of Ln hybrids after 100 doublings (cyh2R/cyh2R killer-sensitive) and the original cyh2S/cyh2S killer K2 parent, the crosses all yielded 25 to 75% single-cell clones with the genetic instability phenotype (data not shown). Thus, the cause of the genetic instability also persists in the Ln hybrids for at least 100 doublings.
As previously described (3), it is possible to obtain genetically stable heterozygous hybrids, e.g., Pa hybrids, for industrial use. However, we recommend that homozygous single-spore clones from heterozygous hybrids with the desired technological properties (27) be selected, since these homozygotes should not incur the sudden phenotypic changes that can ruin an industrial yeast strain. This process could eliminate recessive growth-retarding alleles from industrial yeast populations and improve fermentation vigor (28).
In conclusion, some natural wine yeasts and their hybrids have a genetic instability that causes asymmetrical LOH at the cyh2 locus and loss of ScV-M2 virus. The heterozygous yeast populations can change their phenotype under nonselective conditions more rapidly than previously observed without affecting the gross organization of the cell's DNA. This phenomenon may cause important, sudden phenotypic changes in industrial and pathogenic yeasts. Understanding the molecular basis of this process also could help us explain how these organisms change and evolve in industrial and field settings.
Acknowledgments
This work was funded by grant 2PR01B002 from the Extremadura Government, Spain.
REFERENCES
- 1.Adams, J., R. S. Puskas, J. Simlar, and C. M. Wilke. 1992. Adaptation and major chromosomal changes in populations of Saccharomyces cerevisiae. Curr. Genet. 22:13-19. [DOI] [PubMed] [Google Scholar]
- 2.Carle, G. F., and M. V. Olson. 1985. Separation of DNA molecules from yeast by orthogonal-field-alternation gel electrophoresis. Nucleic Acids Res. 12:5647-5664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carro, D., and B. Piña. 2001. Genetic analysis of the karyotype instability in natural wine yeast strains. Yeast 18:1-14. [DOI] [PubMed] [Google Scholar]
- 4.Cha, R. S., and N. Kleckner. 2002. ATR homolog Mec1 promotes fork progression, thus averting breaks in replication slow zones. Science 297:602-606. [DOI] [PubMed] [Google Scholar]
- 5.Codon, A. C., and T. Benítez. 1995. Variability of the physiological features and of the nuclear and mitochondrial genomes of baker's yeasts. Syst. Appl. Microbiol. 18:343-352. [Google Scholar]
- 6.Del Pozo, L., D. Abarka, M. G. Claros, and A. Jiménez. 1991. Cycloheximide resistance as a yeast cloning marker. Curr. Genet. 19:353-358. [DOI] [PubMed] [Google Scholar]
- 7.Esposito, M. S., and C. V. Bruschi. 1993. Diploid yeast cells yield homozygous spontaneous mutations. Curr. Genet. 23:430-434. [DOI] [PubMed] [Google Scholar]
- 8.Fried, H. M., and G. R. Fink. 1978. Electron microscopic heteroduplex analysis of killer double-stranded RNA species from yeast. Proc. Natl. Acad. Sci. USA 75:4224-4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ganset-Ramírez, J. M., F. Castrejon, A. Querol, D. Ramón, and T. Benítez. 1999. Genomic stability of Saccharomyces cerevisiae baker's yeasts. Syst. Appl. Microbiol. 22:329-340. [DOI] [PubMed] [Google Scholar]
- 10.Guthrie, C., and G. R. Fink. 1991. Guide to yeast genetics and molecular biology. Methods Enzymol. 194:3-57. [PubMed] [Google Scholar]
- 11.Jiménez, A., B. Littlewood, and J. Davies. 1972. Molecular mechanisms of antibiotic action on protein synthesis and membranes. Elsevier, New York, N.Y.
- 12.Kaiser, C., S. Michaelis, and A. Mitchell. 1994. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 13.Kaüfer, N. F., H. M. Fried, W. F. Schwindinger, M. Jasin, and J. R. Warner. 1983. Cycloheximide resistance in yeast: the gene and its protein. Nucleic Acids Res. 11:3123-3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kolodner, R. D., C. D. Putnam, and K. Myung. 2002. Maintenance of genome instability in Saccharomyces cerevisiae. Science 297:552-557. [DOI] [PubMed] [Google Scholar]
- 15.Longo, E., and F. Vezinhet. 1993. Chromosomal rearrangements during vegetative growth of a wild strain of Saccharomyces cerevisiae. Appl. Environ. Microbiol. 59:322-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Malkova, A., E. L. Ivanov, and J. E. Haber. 1996. Double-strand break repair in the absence of RAD51 in yeast: a possible role for break-induced DNA replication. Proc. Natl. Acad. Sci. USA 93:7131-7136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Malkova, A., L. Signon, C. B. Schaefer, M. L. Naylor, J. F. Theis, C. S. Newlon, and J. E. Haber. 2001. RAD51-independent break-induced replication to repair a broken chromosome depends on a distant enhancer site. Genes Dev. 15:1055-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McLaughlin, C. S. 1974. Ribosomes. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 19.Miklos, I., T. Varga, A. Nagy, and M. Sipiczki. 1997. Genome instability and chromosomal rearrangements in a heterothallic wine yeast. J. Basic Microbiol. 37:345-354. [DOI] [PubMed] [Google Scholar]
- 20.Mortimer, R. K., P. Romano, G. Suzzi, and M. Polsinelli. 1994. Genome renewal: a new phenomenon revealed from a genetic study of 43 strains of Saccharomyces cerevisiae derived from natural fermentation of grape musts. Yeast 10:1543-1552. [DOI] [PubMed] [Google Scholar]
- 21.Nadal, D., D. Carro, J. B. Fernández-Larrea, and B. Piña. 1999. Analysis and dynamics of the chromosomal complements of wild sparkling-wine yeast strains. Appl. Environ. Microbiol. 65:1688-1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pérez, F., M. Ramírez, and J. A. Regodón. 2001. Influence of killer strains of Saccharomyces cerevisiae on wine fermentation. Antonie van Leeuwenhoek J. Microbiol. 79:393-399. [DOI] [PubMed] [Google Scholar]
- 23.Pérez, F., J. A. Regodón, M. E. Valdés, C. De Miguel, and M. Ramírez. 2000. Cycloheximide resistance as marker for monitoring yeasts in wine fermentations. Food Microbiol. 17:119-128. [Google Scholar]
- 24.Puig, S., A. Querol, E. Barrio, and J. E. Pérez-Ortín. 2000. Mitotic recombination and genetic changes in Saccharomyces cerevisiae during wine fermentation. Appl. Environ. Microbiol. 66:2057-2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Querol, A., E. Barrio, and D. Ramón. 1992. A comparative study of different methods of yeast strain characterization. Syst. Appl. Microbiol. 15:439-446. [Google Scholar]
- 26.Rachidi, N., P. Barre, and B. Blondin. 1999. Multiple Ty-mediated chromosomal translocations lead to karyotype changes in a wine strain of Saccharomyces cerevisiae. Mol. Gen. Genet. 261:841-850. [DOI] [PubMed] [Google Scholar]
- 27.Ramírez, M., F. Pérez, and J. A. Regodón. 1998. A simple and reliable method for hybridization of homothallic wine strains of Saccharomyces cerevisiae. Appl. Environ. Microbiol. 64:5039-5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramírez, M., J. A. Regodón, F. Pérez, and J. E. Rebollo. 1999. Wine yeast fermentation vigor may be improved by elimination of recessive growth-retarding alleles. Biotechnol. Bioeng. 65:212-218. [DOI] [PubMed] [Google Scholar]
- 29.Regodón, J. A., F. Pérez, M. E. Valdés, C. De Miguel, and M. Ramírez. 1997. A simple and effective procedure for selection of wine yeast strains. Food Microbiol. 14:247-254. [Google Scholar]
- 30.Robert, S., R. S. Sikorski, and J. D. Boeke. 1991. In vitro mutagenesis and plasmid shuffling: from cloned gene to mutant yeast. Methods Enzymol. 194:302-318. [DOI] [PubMed] [Google Scholar]
- 31.Signon, L., A. Malkova, M. L. Naylor, H. Klein, and J. E. Haber. 2001. Genetic requirements for RAD51- and RAD54-independent break-induced replication repair of chromosomal double-strand break. Mol. Cell. Biol. 21:2048-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stöcklein, W., and W. Peipersberg. 1980. Altered ribosomal protein L29 in a cycloheximide-resistant strain of Saccharomyces cerevisiae. Curr. Genet. 1:177-183. [DOI] [PubMed] [Google Scholar]
- 33.Stöcklein, W., and W. Piepersberg. 1980. Binding of cycloheximide to ribosomes from wild-type and mutant strains of Saccharomyces cerevisiae. Antimicrob. Agents Chemother. 18:863-867. [DOI] [PMC free article] [PubMed] [Google Scholar]