Abstract
Characterization by partial 16S rRNA gene sequencing, ribotyping, and green fluorescent protein-based nisin bioassay revealed that 6 of 20 human milk samples contained nisin-producing Lactococcus lactis bacteria. This suggests that the history of humans consuming nisin is older than the tradition of consuming fermented milk products.
One of the most studied bacteriocins, nisin is naturally produced by Lactococcus lactis. Both nisin variants A and Z, with a difference of an amino acid (1), are approved for use in foodstuffs by food additive legislating bodies in the United States (Food and Drug Administration), the United Kingdom, and the European Union (16). Nisin is employed in the dairy industry to inhibit Clostridium botulinum and Bacillus cereus, due to its capacity to prevent spore germination (7, 20). Nisin was first identified in fermented cow milk (13). Since then, it has been isolated from various milk and dairy products (8, 16) as well as from plant material (5, 16) and river water (21). In this study, human milk was screened for bacteria to reveal antibacterial activity caused by nisin producers.
Early lactational (within 80 days of birth) human milk samples (n = 20) were collected in southern Finland from healthy first-time deliverers and from mothers with several children. Two milk samples were received within a month from one donor. The donors were requested to collect milk in sterile test tubes with minimal skin contact. The fresh milk was screened within hours of delivery for bacteria with antibacterial properties by the agar diffusion test (17). Samples were spotted on nonselective Luria-Bertani agar (14) overlaid with 100 μl (at an optical density at 600 nm of 1.6) of Micrococcus luteus A1 NCIMB86166 (National Collection of Industrial and Marine Bacteria), a strain sensitive to many antibacterial substances. Plates covered with M. luteus were allowed to dry with the lid open before 30-μl milk sampling and were incubated overnight at 37°C in an aerobic atmosphere. Seven samples from six different mothers contained strains with strong antibacterial activity. Of the 38 isolated strains, 20 colonies produced a clear inhibition zone around M. luteus and were isolated on plates of M17 agar (Oxoid Ltd., Basingstoke, England) with 0.5% (wt/vol) glucose (M17G) and grown overnight at 30°C.
The 20 isolated inhibitory strains from human milk were characterized by partial 16S rRNA gene sequencing. A region of the 16S rRNA gene was amplified by 29 cycles of PCR (consisting of 30 s at 94°C, 60 s at 55°C, and 90 s at 72°C, with a final 120-s extension step at 72°C) with purified chromosomal DNA (18) from the strains as template and using universal primers pA (5′ AGA GTT TGA TCC TGG CTC AG 3′) and pE′ (5′ CCG TCA ATT CCT TTG AGT TT 3′) (3). The amplified 900-bp fragments were harvested from low-melting-point gel LM-3 (Pronadisa, Madrid, Spain), purified with chloroform-propanol (14), and sequenced with an Autoread sequencing kit with an ALF DNA sequencer (Pharmacia, Piscataway, N.J.), all performed by the DNA Synthesis and Sequencing Laboratory (Institute of Biotechnology, Helsinki, Finland). The sequences obtained were compared against the National Center for Biotechnology Information genome BLAST library (version 2.2.8, accessed 25 March 2004; http://www.ncbi.nlm.nih.gov/BLAST/). The sequence homology (>99%) to known sequences of the L. lactis subsp. lactis 16S rRNA gene confirmed that all strains analyzed represented this species. One L. lactis strain per milk sample was selected for continued characterization.
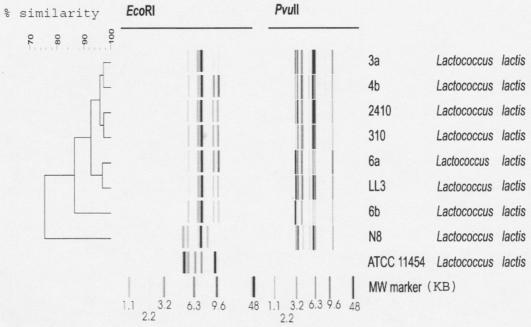
The seven L. lactis strains (LL3, 310, 2410, 3A, 4B, 6A, and 6B) were further characterized by the RiboPrinter and compared to the riboprint of L. lactis strains ATCC 11454 (15) and N8 (4) producing nisin A or nisin Z, the two natural nisin variants. Bacteria were grown on M17G plates (Oxoid Ltd.) overnight at 30°C and then processed as described previously (10) with a lactic agent (DuPont Qualicon, Wilmington, Del.) by using an automated ribotyper (RiboPrinter microbial characterization system; DuPont Qualicon) with the 2000 RiboPrinter system data analysis program. Ribopatterns were exported as TIFF files and analyzed by Bionumerics (Applied Maths BVBA, Sint-Martens-Latem, Belgium). This analysis showed that the ribopatterns isolated from human milk form a homologous cluster when cleaved with EcoRI and PvuII enzymes. The human L. lactis strains clearly differ from the nisin A and nisin Z strains isolated from cow milk. When fragments obtained by the two enzymes were combined and analyzed with a UPGMA (unweighted pair-group mean arithmetic method) dendrogram using the Dice coefficients, a major cluster emerged. Human milk contained L. lactis strains having a >90% similarity (Fig. 1).
FIG. 1.
Human milk-derived L. lactis subsp. lactis strain fragments, obtained by EcoRI and PvuII enzyme treatments, were combined and analyzed with a UPGMA (unweighted pair group method with arithmetic average) dendrogram (Applied Maths BVBA).
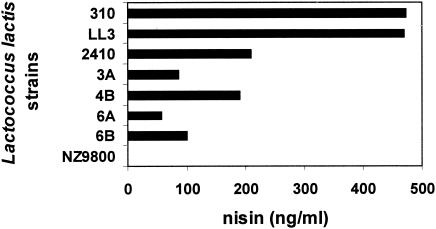
To determine whether the antibacterial activity of the L. lactis strains identified in this study was due to nisin, a green fluorescent protein (GFP)-based nisin bioassay (12) was employed. In this bioassay, nisin was identified and quantified with fluorescence correlated with the nisin concentration in the samples. For this, the strains were grown in M17G broth containing 5 μg of erythromycin/ml, 0.1% Tween 80, and LAC240 as an indicator strain overnight at 30°C. Fluorescence (excitation, 485 nm; emission, 538 nm) was detected in terms of relative fluorescence units with a Fluoroskan Ascent 374 scanning fluorometer (Labsystems, Helsinki, Finland), computer-linked with Ascent version 1.2 software (Labsystems). Viable count for all strains was measured on M17G plates (48 h, 30°C) before quantifying nisin production. The result (Fig. 2) revealed that 6 out of 20 strains produced nisin; therefore, 30% of healthy human donors deliver human milk naturally containing nisin-producing bacteria, with a bacterial count between 2 × 102 to 8.7 × 104 CFU/ml (average, 1.4 × 103 CFU/ml). A nisin producer strain was isolated twice (L. lactis strains 310 and 2410) at monthly intervals in milk samples from the same donor. Lactic acid bacterial species have previously been found in human milk (6, 19), but only one L. lactis strain producing nisin has previously been isolated (6). A reason for this could be that the plating technique and plates used in previous studies led to a lower frequency of isolating nisin-producing L. lactis strains. The screening method in this work revealed higher numbers of bacteria with antibacterial activity. L. lactis LL3 produced the highest nisin titer (Fig. 2). This strain also produced nisin in human milk (3.75 μg/ml) and in infant formula (5 μg/ml) in overnight incubation in quantities that can inhibit many pathogens (16). These nisin values are fluorescence equivalents on a standard curve.
FIG. 2.
Nisin production in M17G broth (Oxoid Ltd.) by overnight cultures of L. lactis strains 310, LL3, 2410, 3A, 4B, 6A, and 6B, of human origin, and the negative control NZ9800 according to the GFP bioassay (12).
Three natural variants of nisin have been described (1, 21). To analyze which variant the human milk isolate is, the nisin structural gene of L. lactis LL3 was amplified by PCR using primers O423 (5′ ATC TGA ATC GAT GGA TCC TGA TCA TAG AGA TAG GTT TAT TGA GTC TTA GAC ATA CT 3′) and O424 (5′ ATC TGA GTC GAC GGA TCC TGA TCA ATC GAT CGG TTG AGC TTT AAA TGA ACT TTT TAT CAT 3′). It was then cloned into pBluescript II vector (Stratagene, La Jolla, Calif.) for sequencing, essentially as described previously (4). Sequencing of its nisin structural gene revealed it to be a nisin A producer.
Nisin-producing L. lactis strain LL3 was examined for its ability to metabolize lactose, an ingredient of infant formula, and other carbohydrates. The bacterial mass from strain LL3 grown on an M17G plate overnight at 30°C was prepared according to the 1999 Biolog manual (Biolog Inc., Hayward, Calif.): cells were scraped into GN/GP buffer (Biolog Inc.) to achieve a 20% turbidometric solution (Microlog system; Biolog Inc.). A volume of 150 μl of bacterial solution was added to a GP2 microtiter plate (Biolog Inc.) and incubated covered for 4 and 20 h at 30°C before reading the results with Biolog (Microlog system; Biolog Inc.). L. lactis LL3 was able to metabolize the following carbohydrates in 20 and 48 h of incubation: N-acetyl-d-glucosamine, N-acetyl-d-mannosamine, amygdalin, arbutin, cellobiose, fructose, d-galactose, gentiobiose, α-d-glucose, α-d-lactose, lactulose, d-mannose, β-methyl d-galactoside, β-methyl d-glucoside, palatinose, salicin, sucrose, d-trehalose, acetic acid, and glycerol. As L. lactis LL3 could metabolize lactose, it could, when used as a protective culture in infant formula, use lactose for its growth and nisin production. L. lactis LL3 has been deposited in the Deutsche Sammlung von Mikroorganismen und Zellkulturen as L. lactis DSM14456.
Our finding that approximately 30% of human milk contains nisin-producing bacteria suggests that humans may have a long history of consuming nisin-producing bacteria. However, little is known about the potential effects of ingesting nisin-producing bacteria. Nisin-producing L. lactis may protect mothers (mastitis) and infants (toxication) from pathogenic skin flora, such as Staphylococcus aureus (11). The effect of nisin producers on the human gastrointestinal tract and its microbiota remains unclear. Nisin producers survive passage through the intestine (2, 9), but it is not known if nisin is produced in the intestine. Clearly, more study is needed to answer these questions.
REFERENCES
- 1.De Vos, W. M., J. W. Mulders, R. J. Siezen, J. Hugenholtz, and O. P. Kuipers. 1993. Properties of nisin Z and distribution of its gene, nisZ, in Lactococcus lactis. Appl. Environ. Microbiol. 59:213-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Drouault, S., G. Corthier, S. D. Ehrlich, and P. Renault. 1999. Survival, physiology, and lysis of Lactococcus lactis in the digestive tract. Appl. Environ. Microbiol. 65:4881-4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Edwards, U., T. Rogall, H. Blocker, M. Emde, and E. C. Böttger. 1989. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 17:7843-7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Graeffe, T., H. Rintala, L. Paulin, and P. E. J. Saris. 1991. A natural nisin variant, p. 260-268. In G. Jung and H.-G. Sahl (ed.), Nisin and novel lantibiotics. ESCOM Science Publishers, Leiden, The Netherlands.
- 5.Harris, L. J., H. P. Fleming, and T. R. Klaenhammer. 1992. Characterization of two nisin-producing Lactococcus lactis subsp. lactis strains isolated from a commercial sauerkraut fermentation. Appl. Environ. Microbiol. 58:1477-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heikkilä, M. P., and P. E. J. Saris. 2003. Inhibition of Staphylococcus aureus by the commensal bacteria of human milk. J. Appl. Microbiol. 95:471-478. [DOI] [PubMed] [Google Scholar]
- 7.Hurst, A., and D. G. Hoover. 1993. Nisin, p. 369-394. In P. M. Davidson and A. L. Branen (ed.), Antimicrobials in foods, 2nd ed. Marcel Dekker Inc., New York, N.Y.
- 8.Klijn, N., A. H. Weerkamp, and W. M. de Vos. 1995. Detection and characterization of lactose-utilizing Lactococcus spp. in natural ecosystems. Appl. Environ. Microbiol. 61:788-792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klijn, N., A. H. Weerkamp, and W. M. de Vos. 1995. Genetic marking of Lactococcus lactis shows its survival in the human gastrointestinal tract. Appl. Environ. Microbiol. 61:2771-2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pirttijärvi, T. S. M., M. A. Andersson, A. C. Scoging, and M. S. Salkinoja-Salonen. 1999. Evaluation of methods for recognizing strains of the Bacillus cereus group with food poisoning potential among industrial and environmental contaminants. Syst. Appl. Microbiol. 22:133-144. [DOI] [PubMed] [Google Scholar]
- 11.Pittard, W. B., III, K. M. Geddes, S. Brown, S. Mintz, and T. C. Hulsey. 1991. Bacterial contamination of human milk: container type and method of expression. Am. J. Perinatol. 8:25-27. [DOI] [PubMed] [Google Scholar]
- 12.Reunanen, J., and P. E. J. Saris. 2003. Microplate assay for nisin in foods, based on nisin-induced green fluorescent protein fluorescence. Appl. Environ. Microbiol. 69:4214-4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rogers, L. A. 1928. The inhibiting effect of Streptococcus lactis on Lactobacillus bulgaricus. J. Bacteriol. 16:321-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 15.Schleifer, K. H., J. Kraus, C. Dvorak, R. Klipper-Bälz, M. D. Collins, and W. Fischer. 1985. Transfer of Streptococcus lactis and related streptococci to the genus Lactococcus gen. nov. Syst. Appl. Microbiol. 6:183-195. [Google Scholar]
- 16.Thomas, L. V., M. R. Clarkson, and J. Delves-Broughton. 2000. Nisin, p. 463-524. In A. S. Naidu (ed.), Natural food antimicrobial systems. CRC Press, Boca Raton, Fla.
- 17.Tramer, J., and J. J. Fowler. 1964. Estimation of nisin in foods. J. Sci. Food Agric. 15:522-528. [Google Scholar]
- 18.Van der Meer, J. R., J. Polman, M. M. Beerthuyzen, R. J. Siezen, O. P. Kuipers, and W. M. de Vos. 1993. Characterization of the Lactococcus lactis nisin A operon genes nisP, encoding a subtilisin-like serine protease involved in precursor processing, and nisR, encoding a regulatory protein involved in nisin biosynthesis. J. Bacteriol. 175:2578-2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.West, P. A., J. H. Hewitt, and O. M. Murphy. 1979. The influence of methods of collection and storage on the bacteriology of human milk. J. Appl. Bacteriol. 46:269-277. [DOI] [PubMed] [Google Scholar]
- 20.Zapico, P., M. Medina, P. Gaya, and M. Nuñez. 1998. Synergistic effect of nisin and the lactoperoxidase system on Listeria monocytogenes in skim milk. Int. J. Food Microbiol. 40:35-42. [DOI] [PubMed] [Google Scholar]
- 21.Zendo, T., M. Fukao, K. Ueda, T. Higuchi, J. Nakayama, and K. Sonomoto. 2003. Identification of the lantibiotic nisin Q, a new natural nisin variant produced by Lactococcus lactis 61-14 isolated from a river in Japan. Biosci. Biotechnol. Biochem. 67:1616-1619. [DOI] [PubMed] [Google Scholar]