Abstract
The foreign body response (FBR) begins with injury acquired during implantation of a biomaterial (BM) and is detrimental due to the eventual encapsulation of the implant. Fusion of macrophages to form foreign body giant cells (FBGC), a hallmark of the FBR, is the consequence of a multistep mechanism induced by interleukin (IL)-4 that includes the acquisition of a fusion competent state and subsequent cytoskeletal rearrangements. However, the precise mechanism, regulation, and interplay among molecular mediators to generate FBGCs are insufficiently understood. Seeking novel mediators of fusion that might be regulated at the post-transcriptional level, we examined the role of microRNAs (miRs) in this process. A miR microarray was screened and identified miR-223 as a negative regulator of macrophage fusion. In addition, transfection of primary macrophages with a mir-223 mimic attenuated IL-4-induced fusion. Furthermore, miR-223 KO mice and mir-223 deficient cells displayed increased fusion in vivo and in vitro, respectively. Finally, we developed a method for in vivo delivery of miR-223 mimic utilizing PLGA nanoparticles, which inhibited FBGC formation in a biomaterial implant model. Our results identify miR-223 as a negative regulator of fusion and demonstrate miR-223 mimic-loaded nanoparticles as a therapeutic inhibitor of macrophage fusion.
Keywords: nanoparticle, microRNA, biomaterial, macrophage fusion, foreign body giant cell
Introduction
Implantation of biomaterials and devices in vascularized tissue induces a cascade of events known as the foreign body response (FBR). Part of the initial response involves the recruitment of macrophages that can undergo fusion and form foreign body giant cells (FBGC), which can cause direct degradation of the implant leading to its malfunction [1–3]. Specifically, it has been shown that FBGCs secrete reactive oxygen species, degradative enzymes, and create an acidic microenvironment at the implant interface.
Despite extensive studies directed at limiting the FBR in vivo, implant malfunction due to FBGC and collagenous capsule formation remains an insurmountable obstacle in many applications. Recent studies focus on creating novel materials for implants, various polymer coatings, or change to the physical structure of the implant to circumvent this issue. For example, in attempt to create novel biopolymers with increased in vivo compatibility, Chandorkar et al fabricated a biodegradable salicylic acid releasing polyester that, when implanted subcutaneous (SC) in mice, reduces the FBR compared to a poly(lactic-co-glycolic acid) (PLGA) polymer [4]. Likewise, Udpa et al utilized chitosan coatings on propylene mesh in a rat abdominal wall model of hernia repair to compare the biocompatibility to currently available commercial meshes [5]. Chitosan coated mesh was associated with a decrease in inflammatory response and in overall fibrosis around the implant. Coating of glucose sensors with a polymer that quickly dissolves upon tissue implantation, allowing for release of a slower degrading polymer containing masitinib microparticles, has proven effective in limiting capsule thickness in the mouse SC FBR model [6]. Wang et al fabricated a novel polymer coating comprised of PLGA microspheres dispersed in polyvinyl alcohol (PVA) hydrogels [7]. When used to coat glucose sensors, this material allowed for slow release of dexamethasone from the microsphere while allowing for rapid diffusion of glucose and other sensing molecules, effectively reducing the FBR. Despite advancements in material coatings, in vivo attenuation of the FBR remains an unmet challenge.
Extensive changes in the transcription of several fusion mediators have been observed during in vitro and in vivo macrophage fusion (Reviewed in [8, 9]). While many of the fusion mediators are known to be transcriptionally regulated, some are mediated by yet unknown processes. For example, the janus kinase (JAK) and signal transducer and activator of transcription (JAK/STAT) pathway is well known to induce transcription of the essential fusion mediator epithelial-cadherin (ECAD) [10, 11]. Likewise, DNAX-activating protein of molecular mass 12 kilodaltons (DAP-12) was shown to mediate transcriptional programming of IL-4 induced macrophages through TREM2 and SYK signaling [12]. Other fusion mediators –including monocyte chemotactic protein-1 (MCP-1), Ras-related C3 botulinum toxin substrate 1 (Rac-1), and tumor necrosis factor (TNF)— are regulated by unknown mechanisms. One possibility is that microRNAs (miRs) play a role in regulating expression of these transcripts. Support for this possibility was recently reported in a study were human monocyte-derived macrophages treated with IL-4/IL-13 displayed increased miR-142-5p and decreased miR-130-3p [13]. In the activation model used in this study, miR-142-5p regulated the phosphorylation of STAT6.
MiRs, small non-coding RNAs that regulate gene expression at the post-transcriptional level, have been linked to regulation of macrophages in many inflammatory settings and cell fusion scenarios [14]. In tightly regulated processes, homologous cell fusion occurs under both physiological and pathological conditions. Examples include myoblasts in skeletal muscle formation, and cells of the monocytic lineage in osteoclast and FBGC formation [9, 15–18]. MiRs have been found to extensively regulate macrophage functions. For example, miR-223 has been shown to be essential for macrophage differentiation [14]. However, less attention has been given to macrophage fusion during the FBR. Specifically, a single study has reported miR-7a-1 as a direct regulator of DC-STAMP during IL-4 induced macrophage fusion in vitro. [19]. There is also support for miR regulation of other fusion mediators but not in the context of FBGC formation. For example, MCP-1 has been shown to be regulated by miRs in scenarios ranging from adipose tissue production, rheumatoid arthritis, and proliferation and apoptosis of human umbilical vein endothelial cells [20–22]. Rac-1, an essential mediator of shape change and cytoskeletal rearrangements during macrophage fusion, has been shown to be regulated by miRs in various types of cancer. Specifically, Rac-1 levels are negatively regulated by miR-146a in prostate cancer, miR-142 in hepatocellular carcinoma, miR-101 in papillary thyroid cancer, and miR-137 in glioblastomas [23–26].
Mir-based approaches to treat various conditions have been described [27–30]. Moreover, nanoparticles are promising carriers for microRNA mimics and antagomiRs, as they can protect and deliver oligonucleotides in vivo. For example, Babar et al. used PLGA nanoparticles to inhibit an anti-miR-155 construct in lymphoma, illustrating the role of nanoparticles in enhancing stability and controlling release over time, which was possible because of the high loading capacity of the nanoparticles[31]. Devalliere et al demonstrated that microRNA mimic can be loaded into PLGA nanoparticles and that these nanoparticles loaded with microRNA-132 provided sustained angiogenic activities in vivo [32]. Use of nanoparticles to deliver miRs circumvents many of the challenges to miR delivery in the clinical setting, providing protection of the miR until it has reached the target cell, targeted biodistribution, and decreases chances of off target affects [33].
Given previous studies indicating miR regulation of fusion mediators, and miR involvement in other fusion scenarios, we investigated miR mediation of macrophage fusion during the FBR. Through miR microarrays, we compared macrophages that were cultured in the presence or absence of IL-4, and identified miR-223 as an essential negative regulator of fusion. We demonstrated, through in vitro transfection with a miR-223 mimic, that over expression of this miR lead to a reduction in fusion. Likewise, in vitro and in vivo fusion assays with miR-223 KO cells and mice, respectively, indicated that the absence of this miR allows for amplification of fusion. Thus, we developed a nanoparticle-based strategy to deliver miR-223 mimic in vivo to attenuate fusion during the FBR. We fabricated miR-223 mimic-loaded nanoparticles and delivered these particles following biomaterial implantation to successfully attenuate FBGC formation in vivo.
Results
Identification of fusion associated miRs
To determine miRs involved in the regulation of macrophage fusion during the FBR, a series of miR microarrays were used to identify differentially expressed miRs following IL-4 stimulation. First, as described previously, bone marrow derived monocytes (BMDM) were isolated from 3–6 month old C57B6 WT mice [34]. The onset of BMDM fusion in vitro is routinely observed at 48 hours, but induction of molecules regulating fusion can be observed as early as 24 hours [35]. Thus, 24 hours after stimulation, miRNA was isolated to compare miR expression from two conditions: cells treated with IL-4 (fusion-induced) and not treated (control). Analysis identified miRs with the most fold change, and allowed for creation of a list of candidate miRs to explore further (Figure 1A). Additional miR microarray screens were performed to compare non-induced and fusion-induced samples at 48 hours (SI Figure 1).
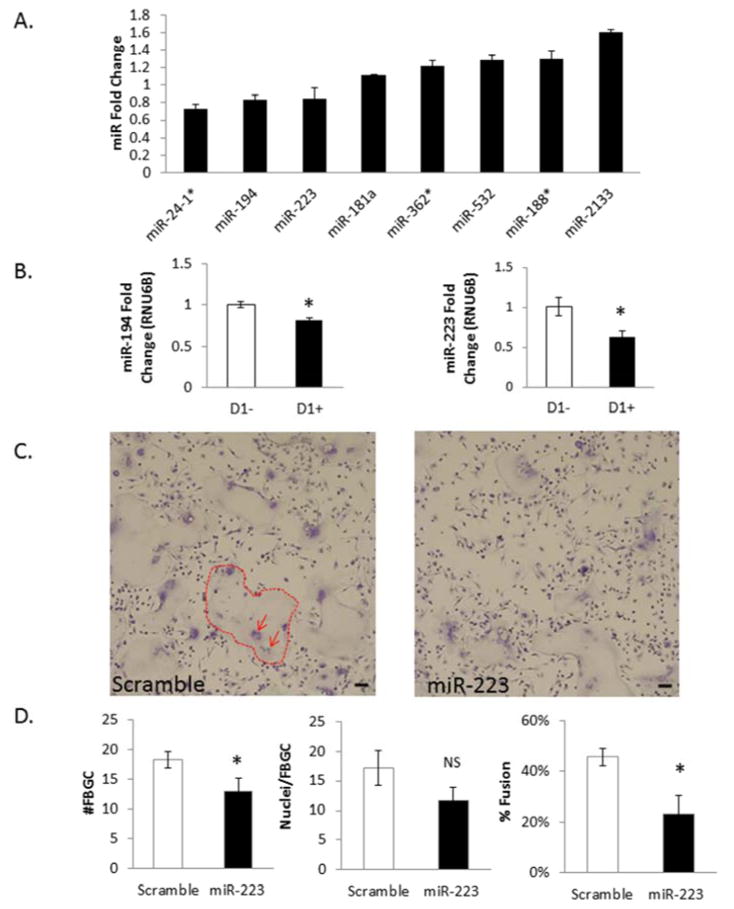
Figure 1. miR-223 negatively regulates FBGC formation.
24 Hour microRNA microarray comparing samples treated and not treated with IL-4 for 24hours. A fold change greater than 1 indicates the miR is induced following IL-4 stimulation while a fold change less than 1 indicates the miR is decreased following IL-4 stimulation. Potential candidate miRs are shown based on P-Value stringency of a two tailed, unpaired TTEST of raw data (A). miR-194 and miR-223 qPCR validation of microarray fold changes (B). 50 nM miR-223 was transfected into C57/BL6 BMDM for twelve hours and cells treated with IL-4 for 5 days. At day 5 cells were stained with a combination of Giemsa and May Grunwald. Dotted lines outline example FBGC and arrows point to clumps of nuclei within FBGC (C). Total number for FBGC, nuclei/FBGC, and percent fusion were assayed as described above (D). * indicates p-value < 0.05, scale bar=50um, N=3.
qRT-PCR validation of candidate miRs
Once candidate miRs were identified via microarray analysis, qRT-PCR was performed to verify that they were differentially expressed during macrophage fusion in vitro. Aliquots of the original miRNAs isolated for the 24hour miR microarray were used to assess miR expression changes. Initial attention was given to the 24 hour time point as future attempts to inhibit the FBR in vivo would be more successful if the process was inhibited as early as possible. Based on microarray fold change and known interactions of miRs in other fusion or macrophage related processes, a list of candidates were chosen for q-RT-PCR analysis. Two miRs—miR-194 and miR-223—displayed reproducible fold changes that were consistent with the microarray data (Figure 1B).
miR-223 mimic transfected cells exhibit decreased fusion in vitro
Based on the decrease in miR-223 levels in cells treated with IL-4, transfection experiments were performed to determine if the miR was biologically relevant to fusion. Specifically, BMDM were transfected with miR-223 mimic and treated with IL-4 in order to evaluate changes in fusion. Prior to this study, transfection efficiency was evaluated with a Cy3 labeled scramble miR mimic (SI Figure 2A). Additionally, miRNA was isolated from wells transfected with mir-223 mimic to confirm an increase in mir-223 levels (SI Figure 2B). Analysis showed that efficient transfection was achieved at concentrations of mir mimic as low as 30nM.
At day 5 post-transfection, cells were fixed and stained via a combination of Wright-Giemsa and May-Grunwald to visualize FBGCs and quantify fusion as described previously [35]. Briefly, 10 low power images per well and three wells per transfection condition (miR-223 mimic, miR-scramble) were analyzed [35]. Number of FBGC, number of nuclei per FBGC, and number of single cells were counted to determine the fusion index [34–38]. In vitro fusion experiments typically range from five to ten days, with the fusion index reaching as high as 80% at day 10. Experiment was stopped at day 5 to ensure cell health and optimal miR-223 transfection levels. At day five, a reduction in fusion in the miR-223 transfected WT cells was observed, correlating with a decrease in total number of FBGC per field. No change in number of nuclei per FBGC was detected (Figure 1C, D). Transfection experiments were also performed with miR-194 mimic, but no changes in fusion were observed (data not shown), suggesting the specificity of miR-223 to altering FBGC formation. To determine if ablation of microRNAs within macrophages would influence macrophage fusion during the FBR, a fusion assay was performed to compare Drosha KO BMDM to WT BMDM (data not shown). The RNase enzyme Drosha is required for canonical biogenesis and processing of the primary microRNA to be cleaved into the precursor microRNA before being exported to the cell cytoplasm [39].
miR-223 KO cells display increased fusion in vitro and in vivo
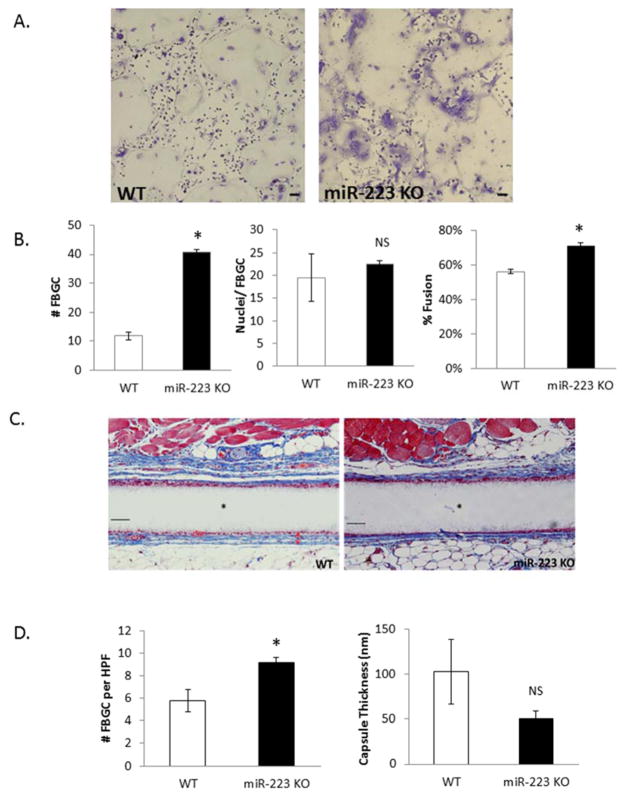
To confirm the role of miR-223 in regulating FBGC formation during the FBR, experiments were performed to determine the effect of miR-223 deficiency in this process. BMDM were isolated from MiR-223 KO mice and an IL-4-induced fusion assay was performed in vitro as described above. MiR-223 KO cells displayed increased fusion when compared to WT cells, correlating with an increase in the number of FBGC per field (Figure 2A, B). Based on miR-223’s influence on fusion in vitro, an in vivo characterization of FBGC formation in miR-223 KO animals was performed. Mice were implanted with Millipore filters SC for four weeks as described previously [40]. Total number of FBGC and capsule thickness were then analyzed to reveal an increase in number of FBGC while no changes were seen in capsule thickness (Figure 2C,D).
Figure 2. miR-223 ablation results in increased FBGC formation.
BMDM derived from WT or miR-223 KO animals were treated with IL-4 for 6 days and then stained with Giemsa and May Grunwald (A). Number of FBGC per field of view, nuclei/FBGC, and percent fusion were assayed (B). WT and miR-223 KO mice were implanted SC with Millipore filters for four weeks. Implants were removed, stained with Masson’s Trichrome to visualize FGBC and capsule (C). Number of FBGC per high power field and capsule thickness were quantified using Image J software (D). Scale bar = 50um,* indicates p-value <0.05, N=3.
Consistent with the observation that mir-223 levels need to be reduced during macrophage fusion, we observed that macrophages deficient in fusion failed to regulate mir-223. Specifically, we analyzed mir-223 levels in IL-4-treated MCP-1 KO macrophages, which display compromised fusion (SI Figure 3). Unlike in their wild counterparts, mir-223 levels remained constant in these cells suggesting a putative role for MCP-1 in the regulation of this mir.
Fabrication and characterization of miR-223 mimic nanoparticles
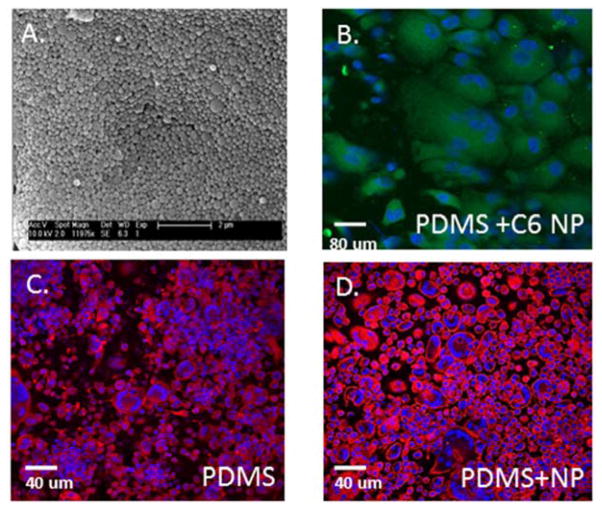
Based on observations that miR-223 negatively regulates FBGC formation in vitro and in vivo, we developed an in vivo nanoparticle delivery strategy for miR-223. PLGA nanoparticles were fabricated using the double emulsion solvent evaporation method as described previously [41]. Each batch of nanoparticles was visualized using SEM and DLS was performed to determine the morphology and size of particles (Figure 3A): nanoparticles were generally spherical, with an average diameter of 400 nm. A pilot study was performed comparing FBGC formation on polydimethylsiloxane (PDMS) implanted into the peritoneal cavity (IP) of WT mice that were subsequently treated or not treated with PLGA nanoparticles. Specifically, injection of empty PLGA nanoparticles 24 hr following IP implantation of PDMS discs did not alter FBGC formation (Figure 3C–D). In addition, injection of C6 labeled nanoparticles demonstrated extensive uptake by implant-adherent macrophages at day 5, demonstrating the efficiency of the nanoparticles to target implant-adherent cells.
Figure 3. PLGA nanoparticles do not alter FBGC formation in IP implant model.
Scanning electron micrographs of empty PLGA nanoparticles (A) and confocal of C6 labeled nanoparticles on surface of PDMS implant (B) reveal morphology and in vivo distribution of nanoparticles following IP implantation of PDMS discs for 5 days. Dapi and Phalloidin stain of PDMS implant (C) and PDMS implant following injection of empty PLGA nanoparticles (D) demonstrate no change in FBGC formation.
IP delivery of miR-223 mimic nanoparticles attenuates fusion at implant site
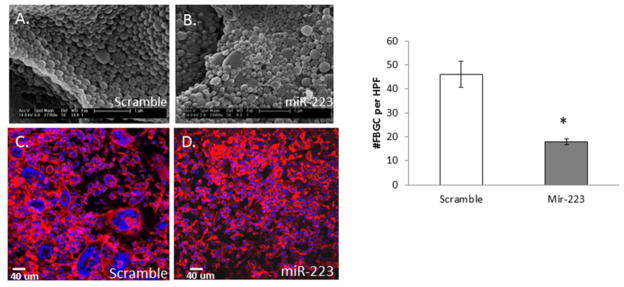
Nanoparticles encapsulating miR-223 mimic or miR scramble, as well as the empty particles, were analyzed for amount of nucleic acid loading per particle so that an optimal delivery amount could be determined. To determine the efficacy of our miR-223 loaded nanoparticles at decreasing fusion in vivo, PDMS discs were implanted IP into WT mice. We have previously shown extensive uptake of microparticles by cells adherent to biomaterials in this model [37]. After 24 hours, nanoparticles containing 3,000 pmol of either miR-223 mimic or scrambled miR were injected into the peritoneal space. The 24 hour delay allowed for the first layer of cells to adhere to the implant prior to introduction of the nanoparticles. Implants were excised at day 5, fixed, stained for Dapi and Rhodamine-Phalloidin to visualize cells, and analyzed by confocal microscopy and Volocity software. Quantitative analysis showed that the formation of FBGC was decreased in animals that received miR-223 mimic (Figure 4).
Figure 4. miR-223 mimic loaded nanoparticles attenuate FBGC formation in vivo.
SEM of miR scramble control (A) or miR-223 mimic (B) loaded nanoparticles reveals consistent size and morphology. Dapi and Rhodamine- Phalloidin stain of PDMS implants following injection of scramble (C) or miR-223 mimic (D) nanoparticles demonstrate a reduction of FBGC following nanoparticle delivery and implantation for five days. Quantification of total number of FBGC per implant indicates a decrease in fusion following delivery of miR-223 mimic loaded nanoparticles (D). * indicates p-value < 0.05, N=3.
Discussion
FBGC formation remains a significant complication despite extensive research into the FBR. Most attempts at modification of the FBR focus on implant design such as surface chemistry. Despite widespread involvement of miRs in osteoclast formation, little is known about miR regulation of FBGC formation. A single study demonstrating the role of microRNAs introduces the idea, but still leaves many questions [19]. Specifically, Sissons et al utilized Dicer KO cells to demonstrate the importance of miRs in regulating macrophage fusion. However, this study involved very low rates of fusion (below 50%) and low number of nuclei per FBGCs suggesting that the findings were not robust. In contrast, most studies report fusion indexes that exceed 70 percent and extensive multinucleation [34]. Furthermore, the same study identified miR-7a as a direct regulator of DC-STAMP to control macrophage fusion in vitro, but did not describe a mechanism of miR regulation of the fusion pathways or demonstrate the importance of this miR to macrophage fusion in vivo [19]. Nevertheless, the findings in Dicer KO cells regarding the negative regulation of fusion by mirs are consistent with our findings with mir-223. Furthermore, we have evaluated IL-4-induced fusion in BDMD from Drosha KO and observed increased fusion (unpublished observation). Interestingly, in a recent publication of IL-4-regulated miRs in macrophages, changes in either mir-223 or mir7-a were not reported [13]. We believe that this is due to the fact that the analysis was performed in macrophages plated on tissue culture plastic, which was previously shown to be non-permissive for fusion [8, 9]. Therefore, the effects of IL-4 were limited to activation and did not lead to engagement of the fusion relevant pathways.
Our experimental approach allowed us to identify miR-223 as a negative regulator of macrophage fusion during the FBR in both in vitro and in vivo. Specifically, miRNA microarrays and qRT-PCR comparing miR expression demonstrated reduction in miR-223 levels following 24 hours of IL-4 stimulation. In vitro fusion assays demonstrate the role of miR-223 in limiting fusion, with miR-223 mimic greatly reducing fusion. In addition, in vitro fusion assays with miR-223 KO cells demonstrate that when miR-223 is completely removed, fusion is significantly increased. Furthermore, SC implantation of Millipore filters into miR-223 KO mice reveal the same trend, supporting the role of miR-223 in vivo.
MiR-223 is an important regulator of cells of the hematopoietic lineage so it is not surprising that it plays a role in macrophage fusion. In fact, miR-223 has already been shown to be essential for osteoclast fusion [42, 43]. Sugatani et al. demonstrate that RAW cells treated with RANKL to induce osteoclastogenesis in the presence of an siRNA to knockdown miR-223 had a significant increase in fusion, as indicated by an increase in TRAP+ cells. In contrast, cells that were treated with a pre-miR-223 were unable to undergo fusion [43].
Although it is yet unclear how IL-4 levels result in a decrease in miR-223 during fusion, current findings provide some insight. Specifically, studies indicate that miR-223 is generally controlled by at least three transcription factors: NFI-A, C/EBPα and PU.1. C/EBPα and PU.1 positively regulate miR-223 while NFI-A negatively regulates miR-223 by competitively binding at the C/EBPα site [44]. It should be also noted that in vivo microRNA regulation of targets in the FBR are likely complex. Thus, it is possible that other miRs play a significant role during the FBR. One possibility is that miRs could be acting to repress regulators of fusion until the appropriate signals are propagated. For example, several miRs have been linked to fusion mediator MCP-1. Kawano et al identified miR-124a as a major mediator of MCP-1 secretion during rheumatoid arthritis [22]. In addition, reviews outline the multitude of miRs found in the wound environment, which mediate inflammation and repair [45, 46]. Nevertheless, identification of miR-223 regulation of fusion in vitro and in vivo during the FBR adds to our expanding understanding of the process of biomaterial-macrophage interactions. Moreover, miR-223 and other miR regulators of fusion could prove to be essential targets in attenuating the FBR in vivo.
Many issues arise when delivering miRs in vivo, including administration of the miRNA, cellular entry, poor in vivo stability due to degradation by ribonucleases, and off-target effects of the miR. Our approach with PLGA nanoparticles as a vehicle for delivery of miRs during the in vivo FBR combats many of these issues. First, the IP implantation model of the FBR allows for direct injection of the miR loaded nanoparticles in the immediate vicinity of the implant. Previous studies have demonstrated that the majority of adherent cells 24–48 hours after IP implantation are macrophages [40]. Thus, injecting nanoparticles 24 hours post implantation ensures that the majority of the implant-adherent cells that will be targeted by the nanoparticles are macrophages. Moreover, the phagocytic nature of macrophages provides a distinct advantage by enhancing the uptake of the nanoparticles. Finally, the nanoparticles protect the miR from degradation until they have been taken by cells, allowing for controlled release over time [33].
Previous work has established a robust model of FBGC formation via implantation of PDMS disks into the peritoneal cavity, which was used here to evaluate the functional relevance of the miR: target interaction during fusion in vivo [40]. Because of their biocompatibility, we sought to use PLGA nanoparticles as a vehicle for miR-223 delivery in vivo in a mouse IP model of the FBR. Our experimental approach has allowed for successful fabrication of miR-223 mimic nanoparticles with consistent morphology and size. Furthermore, injection of the miR-223 mimic nanoparticles resulted in a significant decrease of macrophage fusion during the FBR. It should be noted that our study is restricted to the IP model of FBR and should be further tested in other scenarios including SC implantation. Together, these data demonstrate the importance of miR-regulated pathways in the control if macrophage fusion and suggest that delivery of miRs via nanoparticles could be used to attenuate FBGC formation in other models.
Materials and methods
Recombinant human (rh) M-CSF, recombinant mouse (rm) IL-4, and rh FLT-3were purchased from R&D Systems (Minneapolis, MN). Vectastain ABC kit for immunohistochemistry and Vectashield mounting medium was purchased from Vector Laboratories (Burlingame, CA). May-Grunwald and Wright-Giemsa stains were obtained from Sigma (St. Louis, Missouri). Filters (0.45-μm pore size, mixed cellulose ester) were acquired from Millipore (Billerica, MA) GeneChip miRNA 2.0 Arrays and mouse Gene 1.0st Arrays were purchased from Affymetrix (Santa Clara, California). PLGA was obtained from Durect Corporation (Pelham, Alabama), spermidine and Coumarin 6 from Sigma-Aldrich (St. Louis, Missouri). A custom miRIDIAN mimic for mmu-miR-223-3p was purchased from Thermo Fisher Scientific Biosciences Inc. (Chicago, IL) and miRIDIAN mimic negative control #1 was purchased from GE Healthcare Dharmacon, Inc. (Lafayette, Colorado). Polydimethylsiloxane (PDMS) was purchased from Invotec International (Jacksonville, Florida) and was cut into 4mm diameter and 1mm thick discs using a biopsy punch from Acuderm (Fort Lauderdale, Florida).
BMDM isolation and in vitro fusion assay
Murine macrophages were expanded from the bone marrow of Mir-223-KO and WT mice as described previously [40]. Briefly, marrow was flushed from the femurs of mice and collected in Iscove’s modified Dulbecco’s medium (Life Technologies, Inc., Grand Island, NY) supplemented with 10% fetal bovine serum and penicillin / streptomycin. Subsequently, the mononuclear cell fraction was collected by centrifugation over Lympholyte-M (Cedarlane Labs, Burnlington NC) according to supplier’s instructions. These pre-mononuclear cells were expanded and differentiated into macrophages by growth on non-tissue culture treated polystyrene plates (BD, Franklin Lakes, NJ) in the presence of 1.5 ng/ml macrophage-colony stimulating factor (huM-CSF) (R&D Systems, Minneapolis, MN) and 100 ng/ml fetal liver tyrosine kinase 3 ligand (huflt-3) (R&D Systems, Minneapolis, MN) [34, 35, 38, 47]. Fluorescence activated cell sorting for the macrophage marker, F4/80, has been used in the past to ensure that the isolation procedure resulted in a pure and homogeneous population [37]. After ten days of treatment with FLT-3 and M-CSF, mature macrophages were harvested and treated with 10ng/ml IL-4 to induce fusion [34, 35, 38]. Cells were fed on day 5 and collected by scraping on day 10. Mature macropahges were plated at 5 × 105 cells/well in 24-well non-tissue culture treated polystyrene plates (BD) in media containing 10 ng/ml rm IL-4. Numerous studies have demonstrated robust macrophage fusion utilizing this protocol [36, 40, 48].
miRNA microarrays
Expanded macrophages (day 10 after isolation), were plated on non-tissue treated plastic and total RNA enriched for miRNA was isolated from two conditions: cells treated with IL-4 (fusion-induced) and not treated (control) at 24 and 48 hr. Isolated miRNA was then assayed for quality and quantity, and analyzed by Affymetrix Gene Chip miRNA 2.0 array technology at the Yale Keck Facility. miR microarrays were performed in triplicate to allow for comparison of global miR expression changes under non-fusion (control) and fusion-induced (IL-4 treatment). Analysis of the microarray data was performed to determine abundance of miRs and significant fold change of miRs between non-IL-4 treated and IL-4 treated samples. Additionally, analysis of microarray data was performed using Partek software to identify differentially expressed microRNAs.
Cell Transfection in vitro
Cells were transfected using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) and Optimem-I (Gibco) as suggested by manufacturer. BMDM, isolated as described previously, were plated in 6 well non-coated petri plastic dishes (for miRNA isolation) or 24 well non-coated petri plastic dishes (for fusion assay) and transfected with 50nM of miRVana miR-223- mimic, miRVana miRNA mimic scramble NEGCTRL2, or Cy3 labeled pre-miR negative control from Life Technologies (Grand Island, NY). Following twelve hours of transfection, cells were washed with 1xPBS and fresh media with IL-4 was added to induce fusion.
qPCR Analysis miRNA & mRNA
miRNA or mRNA was extracted from Millipore filter implants in vivo or BMDM plated on petri plastic in vitro with the miRNeasy or mRNeasy Mini Kit from Qiagen (Valecia, CA). 1μg of total RNA or 2μg miRNA per sample was translated into single-stranded cDNA using the Superscript II cDNA synthesis kit and Oligo-dT both from Invitrogen (Carlsbad, CA), according to supplier’s instructions for mRNA, or using miScript II RT Kit from Qiagen (Valecia, CA). Gene expression levels were determined using the IQ5 multi-color real-time PCR and SYBR green supermix from Bio-Rad Laboratories, Inc. (Hercules, CA) for mRNA and miScript SYBR Green qPCR kit from Qiagen (Valecia, CA). The amplification of each sample was performed in triplicate. For mRNA, data was normalized to the levels of the housekeeping gene β-actin and displayed normal distribution. For miRNA samples, primers from Qiagen were used: mmu-miR-223-3p, mmu-miR-194-3p, and hs-RNU6-2 primer assays were used. Fold-change was determined in comparison to non-IL4-treated WT samples at 24 h.
Implantation of BMs
All procedures were performed in accordance with the regulations adopted by the National Institutes of Health and approved by the Animal Care and Use Committee of Yale University. For characterization of miR-223 KO FBR in vivo, a total of 5 miR-223 KO and 5 WT mice were used per time point. Each mouse received one 25 mm2 Millipore filter SC implantation and incision was closed with sterile clips. As we described previously, this material was chosen because it preserves the tissue-implant interface following processing and sectioning and allows for accurate quantification of FBGC formation [40]. Implants were excised at 2 and 4 weeks after implantation. IP implantations of PDMS for in vivo nanoparticle delivery were performed as described previously [40]. Because implants are analyzed by confocal microscopy and sectioning is not required, PDMS is a suitable biomaterial choice for this experiment. A total of 20 WT mice were used for in vivo experiments. Mice were age-matched (3–4 months of age) in all experiments. For implantation of PDMS, each mouse received one 4mm diameter PDMS disc in the peritoneum through a 0.7 cm incision in the skin and peritoneum. The peritoneum was closed with sterile sutures (nylon, size 8.0) and the skin was closed with sterile clips. Nanoparticles (empty, C6, miR-scramble, miR-223 mimic) were resuspended in 1X PBS and injected into the peritoneum at 24 hours post-implantation. Implants were excised at 5 days after implantation. For each type of nanoparticle (empty, C6, miR-scramble and miR-223 mimic) 5 mice were used (n=5). Biomaterials and nanoparticles were analyzed for endotoxin content via the Limulus Amebocyte Lysate prior to implantation as described previously [34, 49]. Specifically, endotoxin content of nanoparticles was 0.065 EU/mg for nanoparticles and negligible for solid implants.
Immnunohistochemistry and Microscopy
Millipore filter implants excised from miR-223 KO and WT mice were processed and sectioned as described previously [40]. For each genotype, 5 mice were implanted and 5 images were taken per implant to analyze FBGC formation in vivo. Number of FBGCs and capsule thickness was determined in sections stained with Mason’s Trichrome. PDMS discs were excised and fixed with 4% PFA overnight and stained with 4,6-diamidino-2-phenylindole (DAPI) (1:1000 dilution; Sigma) and rhodamine-phalloidin (1:1000 dilution; Molecular Probes) and analyzed by confocal microscopy using a Leica TCS SP5 Spectral confocal microscope and Volocity software (Perkin Elmer). Nanoparticles were characterized for morphology and size by visualizing particles via SEM using an XL-30 ESEM-FEG SEM (FEI, Hillsboro, Oregon) and analyzing particle images with Image J software (National Institutes of Health).
Fabrication and characterization of nanoparticles
Nanoparticles were created using a double emulsion solvent evaporation technique as described previously [50]. One hundred (100) nmol of nucleic acid, miR-223 mimic or miR-mimic scramble miRIDIAN mimic negative control #1, was resuspended in 1X TE buffer and mixed with spermidine at an N to P ratio of 8. Nucleic acid mixtures were then added dropwise under vortex to 100 mg PLGA dissolved in 1 mL dichloromethane. The resultant mixture was then sonicated 3 × 10 s. This first emulsion was added dropwise under vortex to a solution of 2 mL 5% PVA and sonicated 3 × 10 s. This second emulsion was then added to 50 mL 0.3% PVA solution and stirred for 3 hours at RT. Nanoparticles were dissolved in dichloromethane for 2 hours at RT and then added to TE buffer, vortexed, and centrifuged to allow nucleic acid to be extracted into the aqueous phase. The amount of nucleic acid present in the aqueous phase was then measured with nanodrop (Thermo Scientific).
Coumarin 6 nanoparticles were fabricated using a single emulsion procedure, and were used to detect the delivery of particles to implant-adherent cells. 0.2 mg Coumarin 6 was dissolved in 1 mL dicholormethane with 100 mg PLGA. The C6/PLGA solution was added dropwise to 2 mL 5% PVA under vortex. The resultant solution was sonicated 3 × 10s, added to 50 mL 0.3% PVA solution and stirred for 3 hours at RT. Nanoparticles were washed 2 X with DI water and collected via centrifugation. Particles were freeze-dried and stored frozen until use. Nanoparticles were dissolved in dichloromethane for 2 hours at RT and then added to TE buffer, vortexed, and centrifuged to allow nucleic acid to be extracted into the aqueous phase. The amount of nucleic acid present in the aqueous phase was then measured with nanodrop (Thermo Scientific). Average loading efficiency and total loading for the miR-223 nanoparticles were 8.5% and 131 pmol of miR-223/mg of nanoparticle, respectively. Likewise, average loading efficiency and total loading for the miR scramble nanoparticles were 9.3% and 60 pmol of miR/mg of nanoparticle, respectively. For both the miR-223 mimic and miR scramble nanoparticles, 3,000 pmol of total miR was injected IP per animal.
Statistical Analysis
Data are expressed as the mean ± the standard error of the mean. The student t-test and ANOVA were used to assess statistical significance. p-values ≤ 0.05 were considered significant.
Supplementary Material
Figure S1: 48 Hour miR microarray
microRNA microarray comparing samples treated and not treated with IL-4 for 48 hours. A fold change greater than 1 indicates the miR is induced following IL-4 stimulation while a fold change less than 1 indicates the miR is decreased following IL-4 stimulation. Potential candidate miRs are shown based on P-Value stringency of a two tailed, unpaired TTEST of raw data. P-Value of ≤ 0.05
Figure S2 In vitro transfection of miR-223 mimic
A. Cells transfected with 50nM Cy3 labelled miR scramble demonstrate cells in which scrambled miR has successfully entered cell. Left panel is bright field microscopy and right panel is immunofluorescence. B.miRNA qPCR demonstrates miR-223 induced expression at 50nM concentrations of miR-223. Scale bar=50um, p-value indicates <0.05, N=3.
Figure S3: Absence of IL-4 induced inhibition of miR-223 in MCP-1 KO cells
qPCR showing miR-223 transcript levels following IL-4 induction for 24 hours in WT cells (A.) or MCP-1 KO cells (B.) * indicates p-value < 0.05, N=3
Acknowledgments
Funding Sources
This work was funded by National Institutes of Health Grants (GM-072194), CRISP (NSF MRSEC DMR 1119826), NSF-GRFP (L.B.M DGE-1122492.), and National Institutes of Health Vascular Research Training Grants (T32HL007950-12; A.J.S) and (T32GM007223; L.B.M)
We thank Nina Kristofik, Jagannath Padmanabhan, Young-Eun Seo, and Jenny Jumai’an for technical assistance and Drs. Jun Lu and Agnes Vignery for helpful discussions. Yale Institute for Nanoscience and Quantum Engineering and CRISP provided partial support for facilities used for this work.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Anderson JM, Rodriguez A, Chang DT. Foreign body reaction to biomaterials. Semin Immunol. 2008;20(2):86–100. doi: 10.1016/j.smim.2007.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dadsetan M, et al. In vivo biocompatibility and biodegradation of poly(ethylene carbonate) J Control Release. 2003;93(3):259–70. doi: 10.1016/j.jconrel.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 3.Ratner BD. Reducing capsular thickness and enhancing angiogenesis around implant drug release systems. J Control Release. 2002;78(1–3):211–8. doi: 10.1016/s0168-3659(01)00502-8. [DOI] [PubMed] [Google Scholar]
- 4.Chandorkar Y, et al. Long-term sustained release of salicylic Acid from cross-linked biodegradable polyester induces a reduced foreign body response in mice. Biomacromolecules. 2015;16(2):636–49. doi: 10.1021/bm5017282. [DOI] [PubMed] [Google Scholar]
- 5.Udpa N, et al. Effects of chitosan coatings on polypropylene mesh for implantation in a rat abdominal wall model. Tissue engineering Part A. 2013;19(23–24):2713–23. doi: 10.1089/ten.tea.2012.0739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Avula MN, et al. Modulation of the foreign body response to implanted sensor models through device-based delivery of the tyrosine kinase inhibitor, masitinib. Biomaterials. 2013;34(38):9737–46. doi: 10.1016/j.biomaterials.2013.08.090. [DOI] [PubMed] [Google Scholar]
- 7.Wang Y, Papadimitrakopoulos F, Burgess DJ. Polymeric “smart” coatings to prevent foreign body response to implantable biosensors. J Control Release. 2013;169(3):341–7. doi: 10.1016/j.jconrel.2012.12.028. [DOI] [PubMed] [Google Scholar]
- 8.Moore LB, et al. Loss of monocyte chemoattractant protein-1 alters macrophage polarization and reduces NFkappaB activation in the foreign body response. Acta Biomater. 2015;11:37–47. doi: 10.1016/j.actbio.2014.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Helming L, Gordon S. Molecular mediators of macrophage fusion. Trends Cell Biol. 2009;19(10):514–22. doi: 10.1016/j.tcb.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 10.Moreno JL, et al. IL-4 promotes the formation of multinucleated giant cells from macrophage precursors by a STAT6-dependent, homotypic mechanism: contribution of E-cadherin. J Leukoc Biol. 2007;82(6):1542–53. doi: 10.1189/jlb.0107058. [DOI] [PubMed] [Google Scholar]
- 11.Van den Bossche J, et al. Alternatively activated macrophages engage in homotypic and heterotypic interactions through IL-4 and polyamine-induced E-cadherin/catenin complexes. Blood. 2009;114(21):4664–74. doi: 10.1182/blood-2009-05-221598. [DOI] [PubMed] [Google Scholar]
- 12.Helming L, et al. Essential role of DAP12 signaling in macrophage programming into a fusion-competent state. Sci Signal. 2008;1(43):ra11. doi: 10.1126/scisignal.1159665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Su S, et al. miR-142–5p and miR-130a-3p are regulated by IL-4 and IL-13 and control profibrogenic macrophage program. Nat Commun. 2015;6:8523. doi: 10.1038/ncomms9523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li T, et al. MicroRNAs modulate the noncanonical transcription factor NF-kappaB pathway by regulating expression of the kinase IKKalpha during macrophage differentiation. Nat Immunol. 2010;11(9):799–805. doi: 10.1038/ni.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brodbeck WG, Anderson JM. Giant cell formation and function. Curr Opin Hematol. 2009;16(1):53–7. doi: 10.1097/MOH.0b013e32831ac52e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Helming L, Gordon S. The molecular basis of macrophage fusion. Immunobiology. 2007;212(9–10):785–93. doi: 10.1016/j.imbio.2007.09.012. [DOI] [PubMed] [Google Scholar]
- 17.Anderson JM. Multinucleated giant cells. Curr Opin Hematol. 2000;7(1):40–7. doi: 10.1097/00062752-200001000-00008. [DOI] [PubMed] [Google Scholar]
- 18.McNally AK, Anderson JM. Macrophage fusion and multinucleated giant cells of inflammation. Adv Exp Med Biol. 2011;713:97–111. doi: 10.1007/978-94-007-0763-4_7. [DOI] [PubMed] [Google Scholar]
- 19.Sissons JR, et al. Cutting edge: microRNA regulation of macrophage fusion into multinucleated giant cells. J Immunol. 2012;189(1):23–7. doi: 10.4049/jimmunol.1102477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arner E, et al. Adipose tissue microRNAs as regulators of CCL2 production in human obesity. Diabetes. 2012;61(8):1986–93. doi: 10.2337/db11-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu D, et al. MicroRNA-495 regulates the proliferation and apoptosis of human umbilical vein endothelial cells by targeting chemokine CCL2. Thromb Res. 2015;135(1):146–54. doi: 10.1016/j.thromres.2014.10.027. [DOI] [PubMed] [Google Scholar]
- 22.Kawano S, Nakamachi Y. miR-124a as a key regulator of proliferation and MCP-1 secretion in synoviocytes from patients with rheumatoid arthritis. Ann Rheum Dis. 2011;70(Suppl 1):i88–91. doi: 10.1136/ard.2010.138669. [DOI] [PubMed] [Google Scholar]
- 23.Sun Q, et al. miR-146a functions as a tumor suppressor in prostate cancer by targeting Rac1. Prostate. 2014;74(16):1613–21. doi: 10.1002/pros.22878. [DOI] [PubMed] [Google Scholar]
- 24.Wu L, et al. MicroRNA-142-3p, a new regulator of RAC1, suppresses the migration and invasion of hepatocellular carcinoma cells. FEBS letters. 2011;585(9):1322–30. doi: 10.1016/j.febslet.2011.03.067. [DOI] [PubMed] [Google Scholar]
- 25.Lin X, et al. miR-101 inhibits cell proliferation by targeting Rac1 in papillary thyroid carcinoma. Biomed Rep. 2014;2(1):122–126. doi: 10.3892/br.2013.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun G, et al. Overexpressed miRNA-137 inhibits human glioma cells growth by targeting Rac1. Cancer Biother Radiopharm. 2013;28(4):327–34. doi: 10.1089/cbr.2012.1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krutzfeldt J, et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438(7068):685–9. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 28.Sazani P, et al. Systemically delivered antisense oligomers upregulate gene expression in mouse tissues. Nat Biotechnol. 2002;20(12):1228–33. doi: 10.1038/nbt759. [DOI] [PubMed] [Google Scholar]
- 29.Elmen J, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452(7189):896–9. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 30.Esau CC. Inhibition of microRNA with antisense oligonucleotides. Methods. 2008;44(1):55–60. doi: 10.1016/j.ymeth.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 31.Babar IA, et al. Nanoparticle-based therapy in an in vivo microRNA-155 (miR-155)-dependent mouse model of lymphoma. Proc Natl Acad Sci U S A. 2012;109(26):E1695–704. doi: 10.1073/pnas.1201516109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Devalliere J, et al. Sustained delivery of proangiogenic microRNA-132 by nanoparticle transfection improves endothelial cell transplantation. FASEB J. 2014;28(2):908–22. doi: 10.1096/fj.13-238527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Y, Wang Z, Gemeinhart RA. Progress in microRNA delivery. J Control Release. 2013;172(3):962–74. doi: 10.1016/j.jconrel.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kyriakides TR, et al. The CC chemokine ligand, CCL2/MCP1, participates in macrophage fusion and foreign body giant cell formation. Am J Pathol. 2004;165(6):2157–66. doi: 10.1016/S0002-9440(10)63265-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.MacLauchlan S, et al. Macrophage fusion, giant cell formation, and the foreign body response require matrix metalloproteinase 9. J Leukoc Biol. 2009;85(4):617–26. doi: 10.1189/jlb.1008588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jay SM, et al. Foreign body giant cell formation is preceded by lamellipodia formation and can be attenuated by inhibition of Rac1 activation. Am J Pathol. 2007;171(2):632–40. doi: 10.2353/ajpath.2007.061213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jay SM, et al. Macrophage fusion leading to foreign body giant cell formation persists under phagocytic stimulation by microspheres in vitro and in vivo in mouse models. J Biomed Mater Res A. 2010;93(1):189–99. doi: 10.1002/jbm.a.32513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McNally AK, Anderson JM. Interleukin-4 induces foreign body giant cells from human monocytes/macrophages. Differential lymphokine regulation of macrophage fusion leads to morphological variants of multinucleated giant cells. Am J Pathol. 1995;147(5):1487–99. [PMC free article] [PubMed] [Google Scholar]
- 39.Murchison EP, Hannon GJ. miRNAs on the move: miRNA biogenesis and the RNAi machinery. Curr Opin Cell Biol. 2004;16(3):223–9. doi: 10.1016/j.ceb.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 40.Skokos EA, et al. Lack of TNF-alpha-induced MMP-9 production and abnormal E-cadherin redistribution associated with compromised fusion in MCP-1-null macrophages. Am J Pathol. 2011;178(5):2311–21. doi: 10.1016/j.ajpath.2011.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Steinbach JM, et al. Polymer nanoparticles encapsulating siRNA for treatment of HSV-2 genital infection. J Control Release. 2012;162(1):102–10. doi: 10.1016/j.jconrel.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kagiya T, Nakamura S. Expression profiling of microRNAs in RAW264.7 cells treated with a combination of tumor necrosis factor alpha and RANKL during osteoclast differentiation. J Periodontal Res. 2013;48(3):373–85. doi: 10.1111/jre.12017. [DOI] [PubMed] [Google Scholar]
- 43.Sugatani T, Hruska KA. MicroRNA-223 is a key factor in osteoclast differentiation. J Cell Biochem. 2007;101(4):996–9. doi: 10.1002/jcb.21335. [DOI] [PubMed] [Google Scholar]
- 44.Agatheeswaran S, et al. BCR-ABL mediated repression of miR-223 results in the activation of MEF2C and PTBP2 in chronic myeloid leukemia. Leukemia. 2013;27(7):1578–80. doi: 10.1038/leu.2012.339. [DOI] [PubMed] [Google Scholar]
- 45.Roy S, Sen CK. MiRNA in innate immune responses: novel players in wound inflammation. Physiol Genomics. 2011;43(10):557–65. doi: 10.1152/physiolgenomics.00160.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rutnam ZJ, Wight TN, Yang BB. miRNAs regulate expression and function of extracellular matrix molecules. Matrix Biol. 2013;32(2):74–85. doi: 10.1016/j.matbio.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kyriakides TR, Bornstein P. Matricellular proteins as modulators of wound healing and the foreign body response. Thromb Haemost. 2003;90(6):986–92. doi: 10.1160/TH03-06-0399. [DOI] [PubMed] [Google Scholar]
- 48.McNally AK, Anderson JM. Beta1 and beta2 integrins mediate adhesion during macrophage fusion and multinucleated foreign body giant cell formation. Am J Pathol. 2002;160(2):621–30. doi: 10.1016/s0002-9440(10)64882-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Look M, et al. The nanomaterial-dependent modulation of dendritic cells and its potential influence on therapeutic immunosuppression in lupus. Biomaterials. 2014;35(3):1089–95. doi: 10.1016/j.biomaterials.2013.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cheng CJ, Saltzman WM. Polymer nanoparticle-mediated delivery of microRNA inhibition and alternative splicing. Mol Pharm. 2012;9(5):1481–8. doi: 10.1021/mp300081s. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: 48 Hour miR microarray
microRNA microarray comparing samples treated and not treated with IL-4 for 48 hours. A fold change greater than 1 indicates the miR is induced following IL-4 stimulation while a fold change less than 1 indicates the miR is decreased following IL-4 stimulation. Potential candidate miRs are shown based on P-Value stringency of a two tailed, unpaired TTEST of raw data. P-Value of ≤ 0.05
Figure S2 In vitro transfection of miR-223 mimic
A. Cells transfected with 50nM Cy3 labelled miR scramble demonstrate cells in which scrambled miR has successfully entered cell. Left panel is bright field microscopy and right panel is immunofluorescence. B.miRNA qPCR demonstrates miR-223 induced expression at 50nM concentrations of miR-223. Scale bar=50um, p-value indicates <0.05, N=3.
Figure S3: Absence of IL-4 induced inhibition of miR-223 in MCP-1 KO cells
qPCR showing miR-223 transcript levels following IL-4 induction for 24 hours in WT cells (A.) or MCP-1 KO cells (B.) * indicates p-value < 0.05, N=3