Abstract
A horizontal, fluorophore-enhanced, repetitive extragenic palindromic-PCR (rep-PCR) DNA fingerprinting technique (HFERP) was developed and evaluated as a means to differentiate human from animal sources of Escherichia coli. Box A1R primers and PCR were used to generate 2,466 rep-PCR and 1,531 HFERP DNA fingerprints from E. coli strains isolated from fecal material from known human and 12 animal sources: dogs, cats, horses, deer, geese, ducks, chickens, turkeys, cows, pigs, goats, and sheep. HFERP DNA fingerprinting reduced within-gel grouping of DNA fingerprints and improved alignment of DNA fingerprints between gels, relative to that achieved using rep-PCR DNA fingerprinting. Jackknife analysis of the complete rep-PCR DNA fingerprint library, done using Pearson's product-moment correlation coefficient, indicated that animal and human isolates were assigned to the correct source groups with an 82.2% average rate of correct classification. However, when only unique isolates were examined, isolates from a single animal having a unique DNA fingerprint, Jackknife analysis showed that isolates were assigned to the correct source groups with a 60.5% average rate of correct classification. The percentages of correctly classified isolates were about 15 and 17% greater for rep-PCR and HFERP, respectively, when analyses were done using the curve-based Pearson's product-moment correlation coefficient, rather than the band-based Jaccard algorithm. Rarefaction analysis indicated that, despite the relatively large size of the known-source database, genetic diversity in E. coli was very great and is most likely accounting for our inability to correctly classify many environmental E. coli isolates. Our data indicate that removal of duplicate genotypes within DNA fingerprint libraries, increased database size, proper methods of statistical analysis, and correct alignment of band data within and between gels improve the accuracy of microbial source tracking methods.
Protection of humans from pathogen contamination is dependent on the purity of waters designated for recreation, drinking, and shellfish harvesting. Bacterial pathogens have been listed as major pollutants in rivers, streams, and estuaries (37). Restoration of polluted water is currently being accomplished through the development of total maximum daily loads (TMDLs). Source assessment is an important component of TMDL development in which pollutants are identified and characterized by type, magnitude, and location (38). The implementation of TMDLs has provided one of the driving forces for the development of methods to distinguish between human and animal sources of fecal pollution. Sources of fecal coliform bacteria may include runoff from feedlots and manure-amended agricultural land, wildlife, inadequate septic systems, urban runoff, and sewage discharges.
Both phenotypic and genotypic methods have been explored as means to study the ecology of fecal bacteria related to host specificity and determining potential sources of fecal bacteria found in surface water (6, 32, 34). The most widely investigated bacteria for these studies have been Escherichia coli and Enterococcus sp. strains. The use of these methods is based on the hypothesis that specific strains, or a strain's phenotypic or genetic attributes, are related to specific host animals. This hypothesis, however, has been tested in only a limited manner.
The majority of phenotypic and genotypic methodologies require the construction of known-source libraries (a host origin database) to differentiate among isolates, which are subsequently used to determine the host origin of unknown environmental isolates (34). However, in most cases, the sizes of the host origin databases are rather limited, consisting of 35 to about 500 isolates (2-4, 6, 9, 12, 13, 23-26, 31, 33, 42, 43), making broader comparisons to larger populations of E. coli and Enterococcus in the environment difficult. In addition, temporal and geographic variation in bacterial genotypes within and between animal species (7, 12, 16, 31), multiple strains within a single animal (23), and diet variation within a host animal (13) have been shown to influence the representativeness of known-source libraries. Moreover, while microbial source tracking studies done using phenotypic approaches and antibiotic resistance patterns have frequently used large known-source libraries, consisting of about 1,000 to 6,000 isolates (2, 8, 10, 15, 44-46), many of the strains examined were isolated from the same source material or sample, and thus libraries may be biased due to the presence of multiple replications (clones) of the same bacterial genotype from the same source animal.
The repetitive extragenic palindromic-PCR (rep-PCR) DNA fingerprinting technique uses the PCR and primers based on highly conserved and repetitive nucleotide sequences to amplify specific portions of the microbial genome (22, 29, 40, 41). When the PCR products are separated by agarose gel electrophoresis and visualized following staining with ethidium bromide, the resulting banding patterns produce a “fingerprint” unique to each strain. The rep-PCR technique has proven to be a valuable tool to identify and track medically and environmentally important microorganisms (5, 17, 30, 40), and it has also been recently evaluated for its use as a source-tracking tool (1, 4, 6, 20, 23). The rep-PCR DNA fingerprinting technique is relatively quick, easy, and inexpensive to perform and lends itself to high-throughput applications, making it an ideal method for microbial source-tracking studies.
Initial studies done in our laboratory indicated that rep-PCR done with Box A1R primers and E. coli yielded more consistent and complex DNA fingerprints than did studies done using REP primers (6). However, rep-PCRs done with Box, ERIC (enterobacterial repetitive intergeneric consensus), and REP primers have all been evaluated in microbial source-tracking studies (1, 4, 6, 23). Dombek et al. (6) used a minimal data set consisting of about 200 nonunique E. coli isolates and reported that 100% of chicken and cow isolates and between 78 and 90% of human, goose, duck, pig, and sheep isolates were correctly assigned to host source groups by using rep-PCR DNA fingerprinting and Box A1R primers. Similarly, Carson et al. (4) reported that rep-PCR DNA fingerprinting done using Box A1R primers produced a 96.6% average rate of correct classification for human and nonhuman E. coli isolates, and McLellan et al. (23) reported a 79.3% average rate of correct classification for E. coli analyzed using rep-PCR and REP primers.
While all these initial analyses indicated that the rep-PCR technique may be useful for determining animal sources of E. coli, these studies were done with relatively small data sets. Moreover, since rep-PCR and most other source-tracking methods require the assembly of libraries of known-source fingerprints, which is labor-intensive and time-consuming, it is very important that the fingerprint database is unbiased, has high fidelity (36), and is representative of the diversity of E. coli strains potentially present in animal hosts and in environmental samples.
rep-PCR DNA fingerprints are usually analyzed using statistical tools. Binary similarity coefficients are used to analyze data for presence and/or absence (19), and simple banding data obtained from DNA fingerprints can be analyzed using binary coefficients such as Dice or Jaccard band matching algorithms. However, more quantitative algorithms, such as Pearson's product-moment correlation coefficient, can also be applied to complex DNA banding patterns, such as those found using rep-PCR. In this case, fingerprints are analyzed as densitometric curves, taking into account both peak position and height (intensity) (11).
In this study we created a large, known-source, rep-PCR and horizontal fluorophore-enhanced rep-PCR (HFERP) DNA fingerprint database from 2,466 E. coli isolates obtained from humans and 12 animal sources (cows, pigs, sheep, goats, turkeys, chickens, ducks, geese, deer, horses, dogs, and cats) and evaluated the usefulness of this method to differentiate human from animal sources of fecal E. coli.
MATERIALS AND METHODS
Isolation of E. coli from known animal sources.
Fecal samples, representing humans and 12 animal source groups, were collected from wild and domesticated animals throughout Minnesota and western Wisconsin. Fresh fecal material was collected from individual animals as previously described (6) by swabbing the rectal or cloacal region with a Culturette7 swab transport system (BD Diagnostic Systems, Sparks, Md.), or by collecting freshly voided feces with a sterile tongue depressor. Fecal samples were placed into sterile Whirl-Pak bags (Nasco, Fort Atkinson, Wis.) and kept at 4°C until processed, usually within 6 h. Fecal material was streaked onto mFC agar plates (Difco BD Diagnostic Systems) and incubated at 44.5°C for 24 h. Characteristic blue colonies (usually six) from mFC plates were picked and evaluated using selective and differential media as previously described (6). Isolates were used for subsequent studies if growth and color responses on all media were typical for E. coli. Isolates giving atypical responses for colony color on all media or by the methylumbelliferyl-β-glucuronide reaction were further screened using API 20E test kits (bioMerieux, Inc., St. Louis, Mo.). Isolates yielding a “good” to “excellent” E. coli identification by the API 20E kit were used for DNA fingerprinting. Three E. coli colonies from each individual fecal sample were used for DNA fingerprinting and were stored at −80°C in 50% glycerol.
E. coli preparation and rep-PCR conditions.
E. coli isolates were streaked onto plate count agar (Difco BD Diagnostic Systems) and grown overnight at 37°C. Single colonies were picked with a 1-μl sterile inoculating loop (Fisher Scientific, Pittsburgh, Pa.) and suspended in 100 μl of distilled H2O in 96-well microtiter plates, and 2 μl of the resulting suspension was used as template for PCR. The rep-PCR fingerprints were obtained using the Box A1R primer (5′-CTACGGCAAGGCGACGCTGACG-3′), and PCRs were done as described previously (6, 27, 28). PCR was performed using an MJ Research PTC 100 (MJ Research, Waltham, Mass.) thermocycler according to the protocol specific for this instrument and the Box A1R primer. PCR was initiated with an incubation at 95°C for 2 min, followed by 30 cycles consisting of 94°C for 3 s, 92°C for 30 s, 50°C for 1 min, and 65°C for 8 min (27). PCRs were terminated after an extension at 65°C for 8 min, and reaction mixtures were stored at 4°C. Reaction mixtures that were not used immediately for gel electrophoresis analysis were stored at −20°C.
Electrophoresis was done at 4°C for 17 to 18 h at 70 V with constant buffer recirculation (6, 27). Gels were stained for 20 min in 0.5 μg of ethidium bromide/ml prepared in 0.5× Tris-acetate-EDTA buffer. Gel images were captured as tagged image file format files with a FOTO/Analyst Archiver electronic documentation system (Fotodyne Inc., Hartland, Wis.).
HFERP studies.
HFERP analyses were performed using a modification of the procedures of Versalovic et al. (39) as follows. Single E. coli colonies were picked with a 1-μl sterile inoculating loop (Fisher Scientific), suspended in 100 μl of 0.05 M NaOH in 96-well, low-profile PCR plates (MJ Research), heated to 95°C for 15 min, and centrifuged at 640 rpm for 10 min in a Hermle/Labnet Z383K (Edison, N.J.) centrifuge. A 2-μl aliquot of the supernatant in each well was used as template for PCR according to the protocol described above for rep-PCR. The primer consisted of a mixture of 0.09 μg of unlabeled Box A1R primer per μl and 0.03 μg of 6-FAM (6-carboxyfluorescein; Integrated DNA Technologies, Coralville, Iowa) fluorescently labeled Box A1R primer per μl. The primer mixture was used at a final concentration of 0.12 μg/25 μl of PCR mixture. A 6.6-μl aliquot of a mixture of 50 μl of Genescan-2500 ROX (6-carboxy-X-rhodamine) internal lane standard (Applied Biosystems, Foster City, Calif.) and 200 μl of nonmigrating loading dye (150 mg of Ficoll 400 per ml and 25 mg of blue dextran per ml) was added to each 25-μl PCR mixture prior to loading the PCR mixture into agarose gels; 12 μl of the resulting mixture was loaded per gel lane. DNA fragments were separated as described for rep-PCR, and HFERP images were captured using a Typhoon 8600 variable mode imager (Molecular Dynamics/Amersham Biosciences, Sunnyvale, Calif.) operating in the fluorescence acquisition mode with the following settings: green (532-nm) excitation laser, 610 BP 30 and 526 SP emission filters in the autolink mode with 580-nm beam splitter, normal sensitivity, 200-μm/pixel scan resolution, +3-mm focal plane, and 800-V power.
Computer-assisted rep-PCR fingerprint analysis.
Separated gel images (ROX-stained standards and HFERP banding patterns) were processed using ImageQuant image analysis software (Molecular Dynamics/Amersham Biosciences) and converted to 256 gray-scale tagged image file format images. Gel images were normalized and analyzed using BioNumerics v.2.5 software (Applied Maths, Sint-Martens-Latem, Belgium). rep-PCR gel lanes were normalized using the 1-kb ladder from 298 to 5,090 bp, as external reference standards, while HFERP gel lanes were normalized using the Genescan 2500 ROX internal lane standard from 287 to 14,057 bp. Band matching for rep-PCR DNA fingerprints was accomplished by using the following BioNumerics settings: minimum profiling, 5%; gray zone, 5%; minimum area, 0%; and shoulder sensitivity, 5. Band matching for HFERP DNA fingerprints was done by using 3% minimum profiling, 0% gray zone, 0% minimum area, and 0 shoulder sensitivity. DNA fingerprint similarities were calculated by using either the curve-based cosine or Pearson's product-moment correlation coefficient, with 1% optimization, or the band-based Jaccard coefficient. Dendrograms were generated using the unweighted pair group method with arithmetic means (UPGMA). The percentages of known-source isolates assigned to their correct source group were calculated by using Jackknife analysis, with maximum similarities (9).
RESULTS AND DISCUSSION
Evaluation of isolates.
Of the 2,672 E. coli strains obtained from known human and animal sources with an array of selective and differential plating media, 219 isolates gave at least one atypical result when examined by routine biochemical screening tests, the wrong color on indicator medium, or an incorrect methylumbelliferyl-β-glucuronide reaction. The biochemical characteristics of these isolates were examined further by using the API 20E system. Results of this analysis indicated that the majority of these isolates, 167, were bona fide E. coli strains, while the remainder, 52, could not be confirmed as this bacterium. The latter group was not used in rep-PCR analysis or included in the DNA fingerprint database.
Influence of duplicate E. coli strains on classification of known-source library.
Since results from several studies suggest that E. coli is genetically diverse and clonal in origin and that this may influence the usefulness of this bacterium for source-tracking studies (7), we evaluated this technology using a large library of E. coli strains obtained from humans and 12 animal sources collected throughout Minnesota and western Wisconsin (Table 1).
TABLE 1.
Animal source groups and rep-PCR DNA fingerprints generated from E. coli isolates
| Animal source group | No. of individuals sampled | Total no. of fingerprints | No. of unique fingerprintsa |
|---|---|---|---|
| Cat | 37 | 108 | 48 |
| Chicken | 86 | 231 | 144 |
| Cow | 115 | 299 | 191 |
| Deer | 64 | 179 | 96 |
| Dog | 71 | 196 | 106 |
| Duck | 42 | 122 | 81 |
| Goat | 36 | 104 | 42 |
| Goose | 73 | 200 | 135 |
| Horse | 44 | 114 | 79 |
| Human | 197 | 307 | 211 |
| Pig | 111 | 303 | 215 |
| Sheep | 37 | 101 | 61 |
| Turkey | 69 | 202 | 126 |
| Total | 982 | 2,466 | 1,535 |
Identical E. coli genotypes from each individual animal were removed.
A total of 2,466 high-quality rep-PCR DNA fingerprints were generated using the Box A1R primer and template DNA from E. coli strains obtained from the 13 human and animal sources (Table 1). About 25 to 40 PCR product bands were obtained from the E. coli isolates by rep-PCR. Jackknife analysis performed on the 2,466 DNA fingerprints from the entire known-source rep-PCR DNA fingerprint database, with Pearson's product-moment correlation coefficient, indicated that 69 to 97% of animal and human E. coli isolates were assigned to correct source groups (Table 2). This corresponds to an 82.2% average rate of correct classification for the 2,466 rep-PCR DNA fingerprints.
TABLE 2.
Total and unique E. coli isolates correctly classified into source groups by rep-PCR and HFERP DNA fingerprinting method
| Source group | % (No.) correctly classifieda
|
||||
|---|---|---|---|---|---|
| All fingerprints (n = 2,466) (rep-PCR, Pearson) | Unique fingerprints (n = 1,535)
|
||||
| rep-PCR
|
HFERP
|
||||
| Pearson | Jaccard | Pearson | Jaccard | ||
| Petb | 91.8 (279) | 61.7 (95) | 45.5 (70) | 59.1 (91) | 44.8 (69) |
| Chicken | 81.4 (188) | 59.7 (86) | 38.9 (56) | 63.2 (91) | 31.9 (46) |
| Cow | 79.6 (238) | 55.0 (104) | 47.6 (90) | 62.0 (117) | 48.2 (91) |
| Deer | 85.5 (145) | 55.2 (53) | 36.5 (35) | 62.2 (60) | 42.6 (41) |
| Waterfowlc | 81.4 (262) | 66.2 (150) | 52.8 (114) | 70.4 (152) | 56.5 (122) |
| Goat | 97.1 (101) | 66.7 (27) | 59.5 (25) | 47.6 (20) | 42.9 (18) |
| Horse | 69.3 (79) | 44.3 (35) | 34.2 (27) | 52.6 (41) | 32.1 (25) |
| Human | 78.3 (240) | 59.2 (124) | 47.4 (100) | 53.8 (113) | 45.2 (95) |
| Pig | 77.9 (236) | 63.7 (137) | 43.7 (94) | 54.4 (117) | 36.3 (78) |
| Sheep | 79.0 (80) | 7.5 (29) | 39.3 (24) | 37.7 (23) | 8.2 (5) |
| Turkey | 88.6 (179) | 73.8 (93) | 52.4 (66) | 73.0 (92) | 54.8 (69) |
| Overall | 82.2 (2,027) | 60.9 (933) | 45.8 (701) | 59.9 (917) | 43.0 (659) |
Based on Jackknife analysis with 1% optimization and maximum similarities with curve-based (Pearson's product-moment correlation coefficient) or band-based (Jaccard's coefficient) similarity calculations.
Pet group consists of cats and dogs.
Waterfowl group consists of ducks and geese.
Increasing the size of the known-source library to 2,466 isolates, however, did not necessarily lead to an increase in the ability to correctly assign strains to the correct source group. In fact, the average rate of correct classification decreased 4.2% with use of the larger library reported here, relative to what was seen with a smaller library in our previous studies (6). This may in part be due to the uncovering of increased genetic diversity among isolates, increased accumulation of errors due to gel-to-gel variation, or the presence of duplicate genotypes (DNA fingerprints) from the same individual within our original library.
Since identical DNA fingerprints from E. coli strains obtained from the same individual most likely represent isolates of clonal origin and can artificially bias subsequent analyses, we eliminated duplicate DNA fingerprints originating from E. coli strains obtained from the same individual human or source animal. Unique DNA fingerprints were defined as DNA fingerprints from E. coli isolates obtained from a single host animal whose similarity coefficients were less than 90%.
Results in Table 1 show that, of the 2,466 DNA fingerprints analyzed, 1,535 (62%) remained in the “unique” DNA fingerprint library. The influence of duplicate DNA fingerprints on the correct classification of library strains is shown in Table 2. When the 1,535 DNA fingerprints from the unique E. coli isolates were examined, Jackknife analyses indicated that only 44 to 74% of the isolates were assigned to the correct source group, with an average rate of correct classification of 60.5% (Table 2). Thus, there was a 21.7% reduction in the average rate of correct classification by using the unique DNA fingerprint library, relative to that seen with the complete library and less than we and others have previously reported with smaller libraries of E. coli strains containing duplicate DNA fingerprints from the same individual animal (4, 6, 23). Our results indicate that the clonal nature of E. coli (11, 20, 33) originating from the same source animal artificially biases the average rate of correct classification, alters the fidelity of the database, and overestimates the ability of the database to assign isolates to their correct source group.
Influence of library size on usefulness of DNA fingerprint libraries.
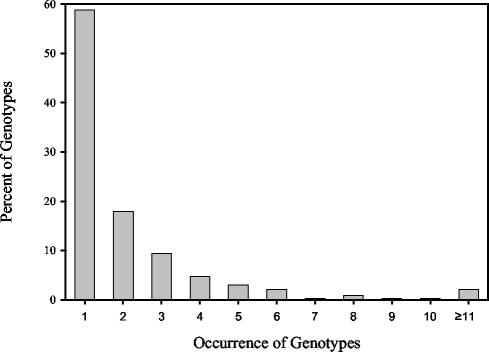
We also determined whether E. coli isolates obtained in this study were sufficient to capture the genetic diversity present within the E. coli populations sampled. E. coli isolates between animal source groups with rep-PCR DNA fingerprint similarities of 90% or greater (based on cosine coefficient, 1% optimization, and UPGMA) were assigned to the same genotype. By this definition, 657 genotypes were distinguished from the 1,535 unique E. coli isolates in the known-source database. The isolates were randomized, and a rarefaction curve was constructed by summing the number of genotypes that accumulated with the successive addition of isolates. Despite a library size of 1,535 DNA fingerprints, genetic diversity has not been saturated. This was evidenced by the apparent first-order relationship between isolate numbers (sampling effort) and accumulation of new genotypes (data not shown). Moreover, 58.75% of the genotypes from isolated strains, across all animal groups, occurred only once in the database, and a limited number occurred multiple times (Fig. 1).
FIG. 1.
Frequency of occurrence of genotypes among rep-PCR DNA fingerprints from unique E. coli isolates. Analysis was limited to the 657 genotypes identified among the 1,535 unique E. coli isolates with rep-PCR DNA fingerprint similarities of 90% or greater.
Since our rarefaction curve did not become asymptotic, our data cannot be used to predict the ultimate size that our fingerprint library needs to be. However, our data indicate that, with our present library size, each new isolate added to the library has only about a 50% chance of being new. It has been suggested that a library size of 20,000 to 40,000 isolates may be needed to capture all the genetic diversity present in E. coli (M. Samadpour, personal communication). Taken together, our data show that the use of relatively small libraries, which do not take into account the tremendous genetic diversity present in E. coli (7, 14, 23, 35) and enterococci, will make broader comparisons to larger populations of these organisms in the environment difficult.
One suggested strategy to avoid this underrepresentation problem in large regional or national libraries is to develop moderate-sized libraries for a highly confined geographical region, wherein isolates are obtained only from the animals in the study area. In this way only animals pertinent to the study site, and those likely to have an impact on the targeted watershed, need to be examined in detail. However, it is also important that in some cases animals thought to be important to or prevalent in the study site may vary over time, depending on agricultural practices and migration. Thus, a careful inventory of potential animals in the study site needs to be made prior to, and during, sampling and analysis.
HFERP DNA fingerprinting.
In our studies we noted that cluster analysis of rep-PCR DNA fingerprint data often produced groupings that were more closely related to the gels from which they originated than to the host animal from which they were isolated. We hypothesized that within-gel clustering of DNA fingerprints was in part due to intrinsic gel-to-gel variation, differential DNA migration in repeated runs of the same and different PCR samples, and the inability to correct for heat- and buffer-induced gel distortion across and between single and multiple gels. Since DNA fingerprint libraries are assembled from many different gels, this could have a major impact on the fidelity of DNA fingerprint libraries and their subsequent use for tracking sources of unknown fecal bacteria.
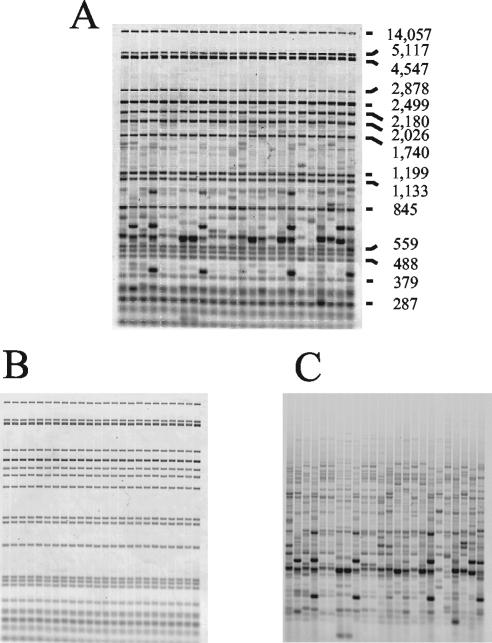
To overcome these major limitations, we developed and evaluated the use of an HFERP technique as a means to differentiate human from animal sources of fecal bacteria. In this method, alignment, correction, and normalization of fluorescently labeled, rep-PCR DNA fingerprint bands within and between gels are facilitated by the use of internal ROX-labeled molecular weight markers that are present in each lane. The technique is similar to that previously described for use with a DNA sequencer (27, 39) but instead uses a standard horizontal agarose gel and a dual-wavelength scanner. An example of an unseparated HFERP gel displaying the ROX-labeled internal lane standard and 6-FAM-labeled Box A1R DNA fingerprints is shown in Fig. 2A, and the separated gel images are shown in Fig. 2B and C. Typically, and with our E. coli strains, 12 to 20 DNA bands per strain were revealed by the HFERP technique.
FIG. 2.
Representative examples of HFERP DNA fingerprint images. Genomic DNAs from 24 E. coli strains were subjected to HFERP DNA fingerprint analysis with a mixture of unlabeled Box A1R and 6-FAM fluorescently labeled Box A1R primers. Each lane contained Genescan-2500 ROX internal lane standards and HFERP DNA fingerprints. The combined, dual-colored HFERP image (A) was captured using a Typhoon Imager and two emission filters. Values at right are sizes in base pairs. Individual images of the HFERP DNA fingerprints (B) and Genescan-2500 ROX internal lane standards (C) were acquired using one filter at a time.
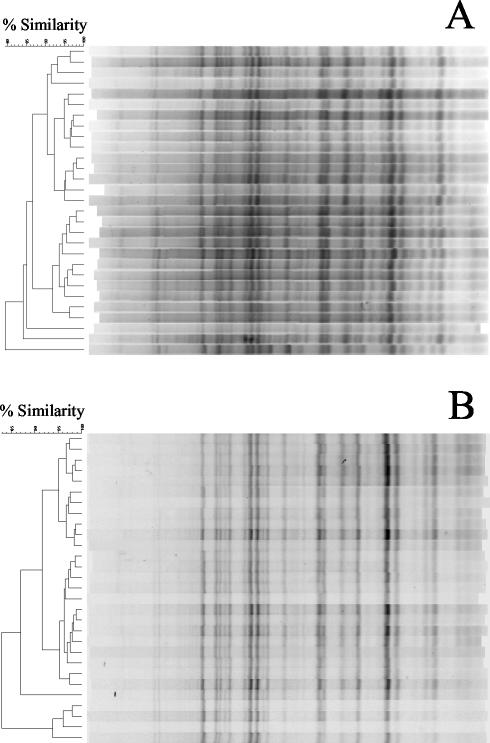
To test whether HFERP reduced within-gel groupings of DNA fingerprints, we analyzed DNA fingerprints from 40 E. coli strains obtained from dogs on two different gels by using Pearson's product-moment coefficient. Results of these studies indicated that rep-PCR DNA fingerprints from strains run on the same gel were, on average, 50% (range, 29 to 57%) more likely to be grouped together than were the same strains analyzed by the HFERP technique (data not shown). This indicates that the HFERP method considerably reduces within-gel grouping of DNA fingerprints. In addition, the HFERP method reduced alignment difficulties due to within- and between-gel variation in band migration found with rep-PCR gels (Fig. 3).
FIG. 3.
Comparison of DNA fingerprint patterns of a reference E. coli strain generated by rep-PCR and by HFERP. (A) rep-PCR DNA fingerprint patterns were assembled from 29 individual PCRs, each of which was run on a separate agarose gel. Fingerprints were generated using E. coli isolate P294 as template DNA and the Box A1R primer. (B) HFERP DNA fingerprint patterns were assembled from 29 individual PCRs, each of which was run on a separate agarose gel. Fingerprints were generated using E. coli isolate P294 as template DNA and a mixture of unlabeled Box A1R and 6-FAM fluorescently labeled Box A1R primers. Bands were aligned using Genescan-2500 ROX internal standards, which were present in each lane. Similarities were determined using the cosine algorithm of BioNumerics, and dendrograms were generated with UPGMA.
The repeatability of the rep-PCR and HFERP DNA fingerprinting methods was also examined by fingerprinting a single, reference control E. coli strain (pig isolate number 294) that was included on each gel. DNA fingerprints from 29 and 41 repetitions of E. coli control pig strain 294, each from a separate gel, were generated by the rep-PCR and HFERP methods, respectively. When analyzed with the curve-based Pearson's correlation coefficient, the rep-PCR DNA fingerprints had an average similarity of 88%, whereas the HFERP-derived DNA fingerprints had an average similarity of 92%.
Previously, Versalovic et al. (39) and Rademaker et al. (27) reported on the use of FERP, whereby polyacrylamide gel electrophoresis and automated DNA sequencers were used to separate and detect bands generated by the FERP protocol. While the more automated method presented by these authors has some advantages, the increased cost of analyses and the limited dynamic range of fragment size separation on sequencing gels did not make this technique useful in our applications. In contrast, the HFERP method described here is relatively inexpensive to perform, can be done on standard electrophoresis apparatus, has high throughput, and allows for the separation of a large range of DNA band sizes. It should be noted, however, that the intensity of HFERP bands is more variable than that of those generated by rep-PCR and that some of the gains achieved by more precise alignment of bands may be offset by more variation in band intensity. We found that this variation in intensity can be overcome by the careful mixing of all reagents in the PCR master mix and greater pipetting precision when loading gels (data not presented). Further improvements in increasing the intensity of HFERP-generated DNA fingerprints may also be obtained by varying the ratio of labeled to unlabeled primer and the final concentration of the primer mixture in PCRs. Nevertheless, our results clearly show that HFERP-derived DNA fingerprint bands are more precisely aligned than the rep-PCR bands and reduce within-gel groupings of fingerprints, which can have profound ramifications for the assembly of libraries and the analysis of unknown environmental isolates. This technology will have application to other DNA fingerprinting methods that rely on the use of PCR primers.
Assignment of E. coli isolates to source groups by using HFERP DNA fingerprints.
Of the 1,535 previously selected unique E. coli isolates from animals and humans (Table 1), 1,531 were subjected to HFERP DNA fingerprinting with a combination of fluorescently labeled and unlabeled Box A1R PCR primers. Jackknife analyses of HFERP gels done with the curve-based Pearson's correlation coefficient indicated that 38 to 73% of the isolates were assigned to the correct source group by this technique (Table 2). For the curve-based analysis, the HFERP technique had the lowest percentage of correctly classified strains in cases where the numbers of analyzed fingerprints were relatively small (for sheep, horses, and goats). The average rate of correct classification for the unique HFERP-generated DNA fingerprints was 59.9%.
In contrast, Jackknife analyses of HFERP-generated DNA fingerprints done using the band-based Jaccard analysis showed that only 8 to 56% of the E. coli isolates were assigned to the correct source group, with a 43.0% average rate of correct classification. This indicates that, for this type of data, the Pearson's product-moment correlation coefficient was superior to Jaccard's band matching algorithm for assigning known isolates to the correct source groups. Interestingly, results in Table 2 also show that, despite problems associated with within- and between-gel variation, within-gel grouping of isolates, and repeatability issues, Jackknife analysis of rep-PCR DNA fingerprints, analyzed with Pearson's correlation coefficient, indicated that 48 to 74% of the isolates were assigned to the correct source group, a 60.9% average rate of correct classification.
While band matching data obtained from DNA fingerprints can be analyzed using binary similarity coefficients, which are mostly used to analyze data for presence and/or absence (19), quantitative similarity coefficients, which require a measure of relative abundance (18), can also be applied to DNA fingerprints if they are analyzed as densitometric curves that take into account both peak position and intensity (peak height). Results of our analysis of rep-PCR DNA fingerprint data indicated that the Jaccard band-based method was not as useful in separating E. coli isolates into their correct source group as was the curve-based quantitative method. This is similar to results reported by Häne et al. (11), who demonstrated that for complex DNA fingerprints, such as those produced with the techniques we used here, a curve-based method such as Pearson's product-moment correlation coefficient more reliably identified similar or identical DNA fingerprints than did band matching formulas, such as simple matching, Dice, or Jaccard. Similarly, Louws et al. (21) reported that curve-based statistical methods worked best for analysis of complex banding profiles generated by rep-PCR, since comparison of curve data is less dependent on DNA concentration in loaded samples and is relatively insensitive to background differences in gels. More recently, Albert et al. (1) performed a statistical evaluation of rep-PCR DNA fingerprint data and reported that k-nearest neighbor classification was similar to Pearson's product-moment coefficient in its ability to correctly classify fingerprints of 584 E. coli isolates.
Groupings of fingerprint data.
In some instances, it may be sufficient to identify unknown watershed E. coli isolates to the level of larger groupings, rather than to the level of individual animal types. To determine if the HFERP-generated DNA fingerprint data from our library of unique E. coli isolates grouped well into larger categories, we assembled DNA fingerprints from pets (dogs and cats), domesticated animals (chickens, cows, goats, horses, pigs, sheep, and turkeys), wildlife (deer, ducks, and geese), and humans and used Jackknife analysis to assess the percentage of correctly classified strains. Results in Table 3 show that the HFERP DNA fingerprints, analyzed with Pearson's product-moment correlation coefficient, correctly classified about 83, 54, 71, and 59% of the isolates into the domesticated animal, human, wildlife, and pet categories, respectively. The average rate of correct classification for these groups was 74.3%. In contrast, when DNA fingerprints were analyzed with Jaccard's coefficient, the average rate of correct classification was 66.2%. As before, the least precision was found in categories having the smallest number of fingerprints, pets and humans, suggesting that there is an apparent relationship between the number of fingerprints analyzed and the percentage of correctly classified isolates.
TABLE 3.
Percentage of E. coli isolates correctly classified into domestic, human, and wildlife source groups by the HFERP DNA fingerprinting method
| Source group | No. of DNA fingerprints | % (No.) correctly classifieda
|
|
|---|---|---|---|
| Pearson | Jaccard | ||
| Domesticatedb | 855 | 83.2 (711) | 77.5 (663) |
| Human | 210 | 53.8 (113) | 45.2 (95) |
| Wildlifec | 312 | 71.4 (223) | 59.6 (186) |
| Petsd | 154 | 59.1 (91) | 44.8 (69) |
| Overall | 1,531 | 74.3 (1,138) | 66.2 (1,013) |
Done using Jackknife analysis with 1% optimization and maximum similarities with curve-based Pearson's product-moment correlation coefficient and band-based Jaccard similarity calculations.
Domesticated group includes chickens, cows, goats, horses, pigs, sheep, and turkeys.
Wildlife group includes deer, ducks, and geese.
Pet group includes dogs and cats.
In microbial source-tracking studies it may often be useful to determine if unknown isolates belong to either animal or human source groups, rather than to more specific categories. Results in Table 4 show that about 94 and 54% of E. coli isolates from animals and humans, respectively, were assigned to the correct source groups by the use of HFERP-generated DNA fingerprints and Pearson's correlation coefficient. The average rate of correct classification was 88.2 and 86.1% for analyses done with Pearson's and Jaccard's algorithms, respectively. The lower percentage of correctly classified human isolates may, in part, be due to the smaller size of fingerprints analyzed for this category. Taken together, these results indicated that (i) broader classifications of source groups should be used when appropriate or (ii) a targeted subset of the DNA fingerprint database should be used to more precisely determine sources of fecal pollutants in watersheds where specific source groups are known to be present. The pooling of source groups into a more limited number of categories has previously been shown to increase the average rate of correct classification following discriminant analysis of antibiotic resistance (10, 15, 45), ribotype analysis (3, 4), and rep-PCR DNA fingerprint analyses (4).
TABLE 4.
Percentage of E. coli isolates correctly classified into human and animal source groups by the HFERP DNA fingerprinting method
| Source group | No. of DNA fingerprints | % (No.) correctly classifieda
|
|
|---|---|---|---|
| Pearson | Jaccard | ||
| Animal | 1,321 | 93.7 (1,237) | 92.6 (1,223) |
| Human | 210 | 53.8 (113) | 45.2 (95) |
| Overall | 1,531 | 88.2 (1,350) | 86.1 (1,318) |
Done using Jackknife analysis with 1% optimization and maximum similarities with curve-based Pearson's product-moment correlation coefficient and band-based Jaccard similarity calculations.
In summary, our results suggest that HFERP-generated Box A1R DNA fingerprints of E. coli are useful to differentiate between different E. coli subtypes of human and animal origin and that this method reduces within-gel groupings of DNA fingerprints and ensures more proper alignment and normalization of fingerprint data. However, our results further indicate that other important issues must also be resolved to more fully understand the potential applications and limitations of this and other library-based microbial source-tracking methodologies. Among these are questions concerning the inclusion of identical DNA fingerprints from the same animal in the library and the number of fingerprints that must be included in an E. coli known-source library to adequately capture the diversity of E. coli genotypes that exist among potential host animals and, ultimately, whether E. coli exhibits a sufficient level of host specificity to allow unambiguous assignment of unknown environmental E. coli isolates to specific host animals.
Acknowledgments
This work was supported, in part, by funding from the Legislative Commission on Minnesota Resources through the Environment and Natural Resources Trust Fund and the MN Future Resource Fund, the Metropolitan Council Environmental Services, and The University of Minnesota Agricultural Experiment Station (to M.J.S.).
We thank Linda Kinkel and Anita Davelos for help with rarefaction analysis and Vivek Kapur, Todd Markowski, and Bruce Witthun for help with laser image analysis.
REFERENCES
- 1.Albert, J. M., J. Munkata-Marr, L. Tenorio, and R. L. Siegrist. 2003. Statistical evaluation of bacterial source tracking data obtained by rep-PCR DNA fingerprinting of Escherichia coli. Environ. Sci. Technol. 37:4554-4560. [DOI] [PubMed] [Google Scholar]
- 2.Burnes, B. S. 2003. Antibiotic resistance analysis of fecal coliforms to determine fecal pollution sources in a mixed-use watershed. Environ. Monit. Assess. 85:87-98. [DOI] [PubMed] [Google Scholar]
- 3.Carson, C. A., B. L. Shear, M. R. Ellersieck, and A. Asfaw. 2001. Identification of fecal Escherichia coli from humans and animals by ribotyping. Appl. Environ. Microbiol. 67:1503-1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carson, C. A., B. L. Shear, M. R. Ellersieck, and J. D. Schnell. 2003. Comparison of ribotyping and repetitive extragenic palindromic-PCR for identification of fecal Escherichia coli from humans and animals. Appl. Environ. Microbiol. 69:1836-1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Bruijn, F. J. 1992. Use of repetitive (repetitive extragenic palindromic and enterobacterial repetitive intergeneric consensus) sequences and the polymerase chain reaction to fingerprint the genomes of Rhizobium meliloti isolates and other soil bacteria. Appl. Environ. Microbiol. 58:2180-2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dombek, P. E., L. K. Johnson, S. T. Zimmerley, and M. J. Sadowsky. 2000. Use of repetitive DNA sequences and the PCR to differentiate Escherichia coli isolates from human and animal sources. Appl. Environ. Microbiol. 66:2572-2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gordon, D. M. 2001. Geographical structure and host specificity in bacteria and the implications for tracing the source of coliform contamination. Microbiology 147:1079-1085. [DOI] [PubMed] [Google Scholar]
- 8.Graves, A. K., A. T. Hagedorn, M. Mahal, A. M. Booth, and R. B. Reneau, Jr. 2002. Antibiotic resistance profiles to determine sources of fecal contamination in a rural Virginia watershed. J. Environ. Qual. 31:1300-1308. [DOI] [PubMed] [Google Scholar]
- 9.Guan, S., R. Xu, S. Chen, J. Odumeru, and C. Gyles. 2002. Development of a procedure for discriminating among Escherichia coli isolates from animal and human sources. Appl. Environ. Microbiol. 68:2690-2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hagedorn, C., S. L. Robinson, J. R. Filtz, S. M. Grubbs, T. A. Angier, and R. B. Reneau, Jr. 1999. Determining sources of fecal pollution in a rural Virginia watershed with antibiotic resistance patterns in fecal streptococci. Appl. Environ. Microbiol. 65:5522-5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Häne, B. G., K. Jäger, and H. Drexler. 1993. The Pearson product-moment correlation coefficient is better suited for identification of DNA fingerprinting profiles than band matching algorithms. Electrophoresis 14:967-972. [DOI] [PubMed] [Google Scholar]
- 12.Hartel, P. G., J. D. Summer, J. L. Hill, J. Collins, J. A. Entry, and W. I. Segars. 2002. Geographic variability of Escherichia coli ribotypes from animals in Idaho and Georgia. J. Environ. Qual. 31:1273-1278. [DOI] [PubMed] [Google Scholar]
- 13.Hartel, P. G., J. D. Summer, and W. I. Segars. 2003. Deer diet affects ribotype diversity of Escherichia coli for bacterial source tracking. Water Res. 37:3263-3268. [DOI] [PubMed] [Google Scholar]
- 14.Hartl, D. L., and D. E. Dykhuizen. 1984. The population genetics of Escherichia coli. Annu. Rev. Genet. 18:31-68. [DOI] [PubMed] [Google Scholar]
- 15.Harwood, V. J., J. Whitlock, and V. H. Withington. 2000. Classification of the antibiotic resistance patterns of indicator bacteria by discriminant analysis: use in predicting the source of fecal contamination in subtropical Florida waters. Appl. Environ. Microbiol. 66:3698-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jenkins, M. B., P. G. Hartel, T. J. Olexa, and J. A. Stuedemann. 2003. Putative temporal variability of Escherichia coli ribotypes from yearling steers. J. Environ. Qual. 32:305-309. [DOI] [PubMed] [Google Scholar]
- 17.Judd, A. K., M. Schneider, M. J. Sadowsky, and F. J. de Bruijn. 1993. Use of repetitive sequences and the polymerase chain reaction technique to classify genetically related Bradyrhizobium japonicum serocluster 123 strains. Appl. Environ. Microbiol. 59:1702-1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krebs, C. J. 1999. Ecological methodology. Benjamin/Cummings, Menlo Park, Calif.
- 19.Legendre, P., and L. Legendre. 1998. Numerical ecology, 2nd English ed. Elsevier Science, Amsterdam, The Netherlands.
- 20.Lipman, J. A., A. de Nijs, T. J. G. M. Lam, and W. Gaastra. 1995. Identification of Escherichia coli strain from cows with clinical mastitis by serotyping and DNA polymorphism patterns with REP and ERIC primers. Vet. Microbiol. 43:13-19. [DOI] [PubMed] [Google Scholar]
- 21.Louws, F. J., J. L. W. Rademaker, and F. J. de Bruijn. 1999. The three Ds of PCR-based genomic analysis of phytobacteria: diversity, detection, and disease diagnosis. Annu. Rev. Phytopathol. 37:81-125. [DOI] [PubMed] [Google Scholar]
- 22.Martin, B., O. Humbert, M. Camara, E. Guenzi, J. Walker, T. Mitchell, P. Andrew, M. Prudhomme, G. Alloing, R. Hakenbeck, D. A. Morrison, G. J. Boulnois, and J.-P. Claverys. 1992. A highly conserved repeated DNA element located in the chromosome of Streptococcus pneumoniae. Nucleic Acids Res. 20:3479-3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McLellan, S. L., A. D. Daniels, and A. K. Salmore. 2003. Genetic characterization of Escherichia coli populations from host sources of fecal pollution using DNA fingerprinting. Appl. Environ. Microbiol. 69:2587-2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nebra, Y., X. Bonjoch, and A. R. Blanch. 2003. Use of Bifidobacterium dentium as an indicator of the origin of fecal water pollution. Appl. Environ. Microbiol. 69:2651-2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parveen, S., N. C. Hodge, R. E. Stall, S. R. Farrah, and M. L. Tamplin. 2001. Phenotypic and genotypic characterization of human and nonhuman Escherichia coli. Water Res. 35:379-386. [DOI] [PubMed] [Google Scholar]
- 26.Parveen, S., R. L. Murphree, L. Edmiston, C. W. Kasper, K. M. Portier, and M. L. Tamplin. 1997. Association of multiple-antibiotic-resistance profiles with point and nonpoint sources of Escherichia coli in Apalachicola Bay. Appl. Environ. Microbiol. 63:2607-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rademaker, J. L. W., F. J. Louws, and F. J. de Bruijn. 1998. Characterization of the diversity of ecologically important microbes by rep-PCR genomic fingerprinting, suppl. 3, p. 1-26. In A. D. L. Akkermans, J. D. van Elsas, and F. J. de Bruijn (ed.), Molecular microbial ecology manual. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 28.Rademaker, J. L. W., F. J. Louws, U. Rossbach, P. Vinuesa, and F. J. de Bruijn. 1999. Computer-assisted pattern analysis of molecular fingerprints and database construction, suppl. 4, p. 1-33. In A. D. L. Akkermans, J. D. van Elsas, and F. J. de Bruijn (ed.), Molecular microbial ecology manual. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 29.Sadowsky, M. J., and H.-G. Hur. 1998. Use of endogenous repeated sequences to fingerprint bacterial genomic DNA, p. 399-413. In J. R. Lupski, G. Weinstock, and F. J. de Bruijn (ed.), Bacterial genomes: structure and analysis. Chapman and Hall, New York, N.Y.
- 30.Sadowsky, M. J., L. L. Kinkel, J. H. Bowers, and J. L. Schottel. 1996. Use of repetitive intergenic DNA sequences to classify pathogenic and disease-suppressive Streptomyces strains. Appl. Environ. Microbiol. 62:3489-3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scott, T. M., S. Parveen, K. M. Portier, J. B. Rose, M. L. Tamplin, S. R. Farrah, A. Koo, and J. Lukasik. 2003. Geographical variation in ribotype profiles of Escherichia coli isolates from humans, swine, poultry, beef, and dairy cattle in Florida. Appl. Environ. Microbiol. 69:1089-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scott, T. M., J. B. Rose, T. M. Jenkins, S. R. Farrah, and J. Lukasik. 2002. Microbial source tracking: current methodology and future directions. Appl. Environ. Microbiol. 68:5796-5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seurinck, S., W. Verstraete, and S. D. Siciliano. 2003. Use of 16S-23S rRNA intergenic spacer region PCR and repetitive extragenic palindromic PCR analyses of Escherichia coli isolates to identify nonpoint fecal sources. Appl. Environ. Microbiol. 69:4942-4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Simpson, J. M., J. W. Santo Domingo, and D. J. Reasoner. 2003. Microbial source tracking: state of the science. Environ. Sci. Technol. 36:5280-5288. [DOI] [PubMed] [Google Scholar]
- 35.Souza, V., M. Rocha, A. Valera, and L. Eguiarte. 1999. Genetic structure of natural populations of Escherichia coli in wild hosts on different continents. Appl. Environ. Microbiol. 65:3373-3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tyler, K. D., G. Wang, S. D. Tyler, and W. M. Johnson. 1997. Factors affecting reliability and reproducibility of amplification-based DNA fingerprinting of representative bacterial pathogens. J. Clin. Microbiol. 35:339-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.U.S. Environmental Protection Agency. 2000. National water quality inventory: 1998 report to Congress. EPA-841-R-00-001. Office of Water, U.S. Environmental Protection Agency, Washington, D.C.
- 38.U.S. Environmental Protection Agency. 2001. Protocol for developing pathogen TMDLs. EPA 841-R-00-002. Office of Water, U.S. Environmental Protection Agency, Washington, D.C.
- 39.Versalovic, J., V. Kapur, T. Koeuth, G. H. Mazurek, T. S. Whittam, J. M. Musser, and J. R. Lupski. 1995. DNA fingerprinting of pathogenic bacteria by fluorophore-enhanced repetitive sequence-based polymerase chain reaction. Arch. Pathol. Lab. Med. 119:23-29. [PubMed] [Google Scholar]
- 40.Versalovic, J., T. Koeuth, and J. R. Lupski. 1991. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 19:6823-6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Versalovic, J., M. Schneider, F. J. de Bruijn, and J. R. Lupski. 1994. Genomic fingerprinting of bacteria using repetitive sequence-based polymerase chain reaction. Methods Mol. Cell. Biol. 5:25-40. [Google Scholar]
- 42.Wheeler, A. L., P. G. Hartel, D. G. Godfrey, J. L. Hill, and W. I. Segars. 2002. Potential of Enterococcus faecalis as a human fecal indicator for microbial source tracking. J. Environ. Qual. 31:1286-1293. [DOI] [PubMed] [Google Scholar]
- 43.Whitlock, J. E., D. T. Jones, and V. J. Harwood. 2002. Identification of the sources of fecal coliforms in an urban watershed using antibiotic resistance analysis. Water Res. 36:4273-4282. [DOI] [PubMed] [Google Scholar]
- 44.Wiggins, B. A. 1996. Discriminant analysis of antibiotic resistance patterns in fecal streptococci, a method to differentiate human and animal sources of fecal pollution in natural waters. Appl. Environ. Microbiol. 62:3997-4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wiggins, B. A., R. W. Andrews, R. A. Conway, C. L. Corr, E. J. Dobratz, D. P. Dougherty, J. R. Eppard, S. R. Knupp, M. C. Limjoco, J. M. Mettenburg, J. M. Rinehardt, J. Sonsino, R. L. Torrijos, and M. E. Zimmerman. 1999. Use of antibiotic resistance analysis to identify nonpoint sources of fecal pollution. Appl. Environ. Microbiol. 65:3483-3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wiggins, B. A., P. W. Cash, W. S. Creamer, S. E. Dart, P. P. Garcia, T. M. Gerecke, J. Han, B. L. Henry, K. B. Hoover, E. L. Johnson, K. C. Jones, J. G. McCarthy, J. A. McDonough, S. A. Mercer, M. J. Noto, H. Park, M. S. Phillips, S. M. Purner, B. M. Smith, E. N. Stevens, and A. K. Varner. 2003. Use of antibiotic resistance analysis for representativeness testing of multiwatershed libraries. Appl. Environ. Microbiol. 69:3399-3405. [DOI] [PMC free article] [PubMed] [Google Scholar]