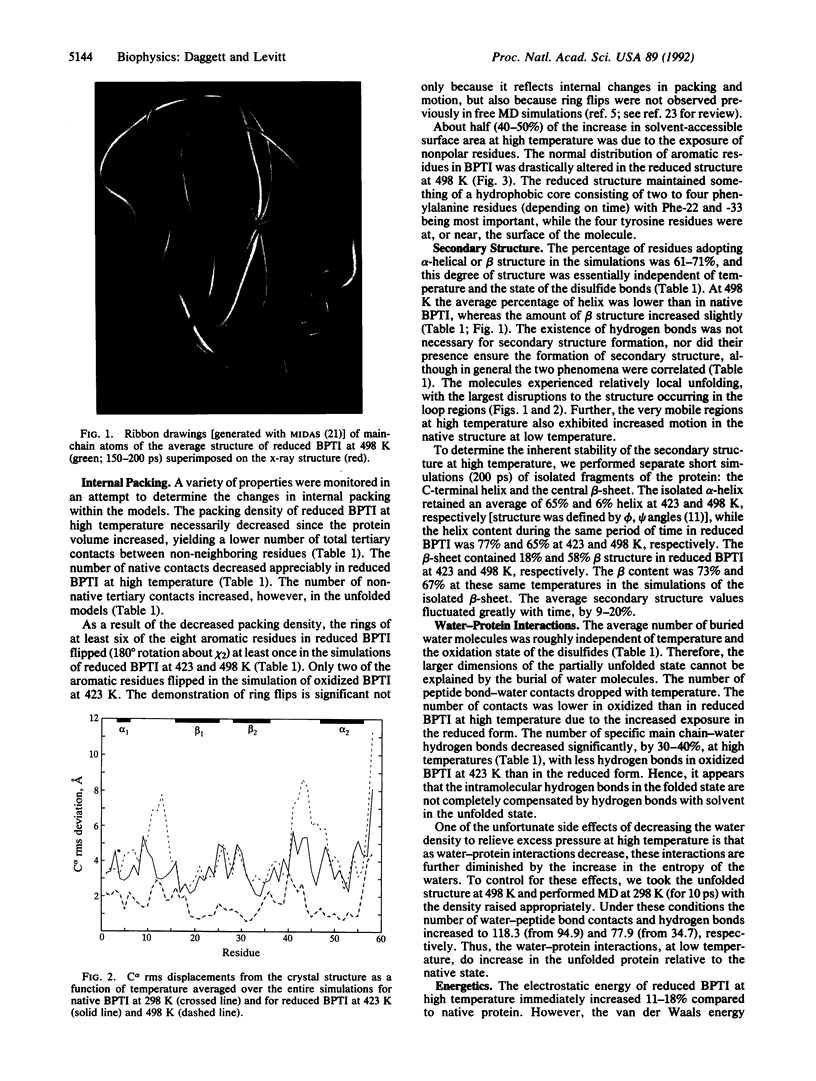
Abstract
It is generally accepted that a protein's primary sequence determines its three-dimensional structure. It has proved difficult, however, to obtain detailed structural information about the actual protein folding process and intermediate states. We present the results of molecular dynamics simulations of the unfolding of reduced bovine pancreatic trypsin inhibitor. The resulting partially "denatured" state was compact but expanded relative to the native state (11-25%); the expansion was not caused by an influx of water molecules. The structures were mobile, with overall secondary structure contents comparable to those of the native protein. The protein experienced relatively local unfolding, with the largest changes in the structure occurring in the loop regions. A hydrophobic core was maintained although packing of the side chains was compromised. The properties displayed in the simulation are consistent with unfolding to a molten globule state. Our simulations provide an in-depth view of this state and details of water-protein interactions that cannot yet be obtained experimentally.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amir D., Haas E. Reduced bovine pancreatic trypsin inhibitor has a compact structure. Biochemistry. 1988 Dec 13;27(25):8889–8893. doi: 10.1021/bi00425a003. [DOI] [PubMed] [Google Scholar]
- Baum J., Dobson C. M., Evans P. A., Hanley C. Characterization of a partly folded protein by NMR methods: studies on the molten globule state of guinea pig alpha-lactalbumin. Biochemistry. 1989 Jan 10;28(1):7–13. doi: 10.1021/bi00427a002. [DOI] [PubMed] [Google Scholar]
- Beasty A. M., Hurle M. R., Manz J. T., Stackhouse T., Onuffer J. J., Matthews C. R. Effects of the phenylalanine-22----leucine, glutamic acid-49----methionine, glycine-234----aspartic acid, and glycine-234----lysine mutations on the folding and stability of the alpha subunit of tryptophan synthase from Escherichia coli. Biochemistry. 1986 May 20;25(10):2965–2974. doi: 10.1021/bi00358a035. [DOI] [PubMed] [Google Scholar]
- Bychkova V. E., Pain R. H., Ptitsyn O. B. The 'molten globule' state is involved in the translocation of proteins across membranes? FEBS Lett. 1988 Oct 10;238(2):231–234. doi: 10.1016/0014-5793(88)80485-x. [DOI] [PubMed] [Google Scholar]
- Cooke R., Kuntz I. D. The properties of water in biological systems. Annu Rev Biophys Bioeng. 1974;3(0):95–126. doi: 10.1146/annurev.bb.03.060174.000523. [DOI] [PubMed] [Google Scholar]
- Creighton T. E. Experimental studies of protein folding and unfolding. Prog Biophys Mol Biol. 1978;33(3):231–297. doi: 10.1016/0079-6107(79)90030-0. [DOI] [PubMed] [Google Scholar]
- Daggett V., Kollman P. A., Kuntz I. D. A molecular dynamics simulation of polyalanine: an analysis of equilibrium motions and helix-coil transitions. Biopolymers. 1991 Aug;31(9):1115–1134. doi: 10.1002/bip.360310911. [DOI] [PubMed] [Google Scholar]
- Daggett V., Kollman P. A., Kuntz I. D. Molecular dynamics simulations of small peptides: dependence on dielectric model and pH. Biopolymers. 1991 Feb 15;31(3):285–304. doi: 10.1002/bip.360310304. [DOI] [PubMed] [Google Scholar]
- Daggett V., Levitt M. Molecular dynamics simulations of helix denaturation. J Mol Biol. 1992 Feb 20;223(4):1121–1138. doi: 10.1016/0022-2836(92)90264-k. [DOI] [PubMed] [Google Scholar]
- Dolgikh D. A., Gilmanshin R. I., Brazhnikov E. V., Bychkova V. E., Semisotnov G. V., Venyaminov SYu, Ptitsyn O. B. Alpha-Lactalbumin: compact state with fluctuating tertiary structure? FEBS Lett. 1981 Dec 28;136(2):311–315. doi: 10.1016/0014-5793(81)80642-4. [DOI] [PubMed] [Google Scholar]
- Goodman E. M., Kim P. S. Folding of a peptide corresponding to the alpha-helix in bovine pancreatic trypsin inhibitor. Biochemistry. 1989 May 16;28(10):4343–4347. doi: 10.1021/bi00436a033. [DOI] [PubMed] [Google Scholar]
- Goto Y., Calciano L. J., Fink A. L. Acid-induced folding of proteins. Proc Natl Acad Sci U S A. 1990 Jan;87(2):573–577. doi: 10.1073/pnas.87.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding M. M., Williams D. H., Woolfson D. N. Characterization of a partially denatured state of a protein by two-dimensional NMR: reduction of the hydrophobic interactions in ubiquitin. Biochemistry. 1991 Mar 26;30(12):3120–3128. doi: 10.1021/bi00226a020. [DOI] [PubMed] [Google Scholar]
- Hughson F. M., Wright P. E., Baldwin R. L. Structural characterization of a partly folded apomyoglobin intermediate. Science. 1990 Sep 28;249(4976):1544–1548. doi: 10.1126/science.2218495. [DOI] [PubMed] [Google Scholar]
- Karplus M., McCammon J. A. Dynamics of proteins: elements and function. Annu Rev Biochem. 1983;52:263–300. doi: 10.1146/annurev.bi.52.070183.001403. [DOI] [PubMed] [Google Scholar]
- Kuwajima K. The molten globule state as a clue for understanding the folding and cooperativity of globular-protein structure. Proteins. 1989;6(2):87–103. doi: 10.1002/prot.340060202. [DOI] [PubMed] [Google Scholar]
- Lee B., Richards F. M. The interpretation of protein structures: estimation of static accessibility. J Mol Biol. 1971 Feb 14;55(3):379–400. doi: 10.1016/0022-2836(71)90324-x. [DOI] [PubMed] [Google Scholar]
- Levitt M., Chothia C. Structural patterns in globular proteins. Nature. 1976 Jun 17;261(5561):552–558. doi: 10.1038/261552a0. [DOI] [PubMed] [Google Scholar]
- Levitt M., Meirovitch H. Integrating the equations of motion. J Mol Biol. 1983 Aug 15;168(3):617–620. doi: 10.1016/s0022-2836(83)80305-2. [DOI] [PubMed] [Google Scholar]
- Levitt M., Sharon R. Accurate simulation of protein dynamics in solution. Proc Natl Acad Sci U S A. 1988 Oct;85(20):7557–7561. doi: 10.1073/pnas.85.20.7557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews C. R., Crisanti M. M., Manz J. T., Gepner G. L. Effect of a single amino acid substitution on the folding of the alpha subunit of tryptophan synthase. Biochemistry. 1983 Mar 15;22(6):1445–1452. doi: 10.1021/bi00275a019. [DOI] [PubMed] [Google Scholar]
- Oas T. G., Kim P. S. A peptide model of a protein folding intermediate. Nature. 1988 Nov 3;336(6194):42–48. doi: 10.1038/336042a0. [DOI] [PubMed] [Google Scholar]
- Roder H., Elöve G. A., Englander S. W. Structural characterization of folding intermediates in cytochrome c by H-exchange labelling and proton NMR. Nature. 1988 Oct 20;335(6192):700–704. doi: 10.1038/335700a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirado-Rives J., Jorgensen W. L. Molecular dynamics simulations of the unfolding of an alpha-helical analogue of ribonuclease A S-peptide in water. Biochemistry. 1991 Apr 23;30(16):3864–3871. doi: 10.1021/bi00230a009. [DOI] [PubMed] [Google Scholar]
- Udgaonkar J. B., Baldwin R. L. NMR evidence for an early framework intermediate on the folding pathway of ribonuclease A. Nature. 1988 Oct 20;335(6192):694–699. doi: 10.1038/335694a0. [DOI] [PubMed] [Google Scholar]