Abstract
Six chlorpyrifos-degrading bacteria were isolated from an Australian soil and compared by biochemical and molecular methods. The isolates were indistinguishable, and one (strain B-14) was selected for further analysis. This strain showed greatest similarity to members of the order Enterobacteriales and was closest to members of the Enterobacter asburiae group. The ability of the strain to mineralize chlorpyrifos was investigated under different culture conditions, and the strain utilized chlorpyrifos as the sole source of carbon and phosphorus. Studies with ring or uniformly labeled [14C]chlorpyrifos in liquid culture demonstrated that the isolate hydrolyzed chlorpyrifos to diethylthiophospshate (DETP) and 3, 5, 6-trichloro-2-pyridinol, and utilized DETP for growth and energy. The isolate was found to possess mono- and diphosphatase activities along with a phosphotriesterase activity. Addition of other sources of carbon (glucose and succinate) resulted in slowing down of the initial rate of degradation of chlorpyrifos. The isolate degraded the DETP-containing organophosphates parathion, diazinon, coumaphos, and isazofos when provided as the sole source of carbon and phosphorus, but not fenamiphos, fonofos, ethoprop, and cadusafos, which have different side chains. Studies of the molecular basis of degradation suggested that the degrading ability could be polygenic and chromosome based. Further studies revealed that the strain possessed a novel phosphotriesterase enzyme system, as the gene coding for this enzyme had a different sequence from the widely studied organophosphate-degrading gene (opd). The addition of strain B-14 (106 cells g−1) to soil with a low indigenous population of chlorpyrifos-degrading bacteria treated with 35 mg of chlorpyrifos kg−1 resulted in a higher degradation rate than was observed in noninoculated soils. These results highlight the potential of this bacterium to be used in the cleanup of contaminated pesticide waste in the environment.
Microbial degradation of organophosphate pesticides is of particular interest because of the high mammalian toxicity of such compounds and their widespread and extensive use. For some organophosphates such as parathion, it has been relatively easy to isolate degrading bacteria: two different strains, Flavobacterium sp. strain ATCC 27551 and Pseudomonas diminuta strain Gm, have been isolated from soils in the Philippines and United States, respectively (32, 33). In addition, studies by Rani and Lalitha-Kumari (29) found that a strain of Pseudomonas putida could hydrolyze methyl parathion and use p-nitrophenol as a sole source of carbon. Although chlorpyrifos [O, O-diethyl O-(3,5,6-trichlor-2-pyridyl) phosphorothioate] has been widely used for agricultural and household pest control since 1965, it has been problematic isolating a degrading strain for this organophosphate. Several attempts to isolate a chlorpyrifos-degrading microbial system by repeated treatments or enrichment of soils and other media with chlorpyrifos have not been successful (20, 27). The resistance of chlorpyrifos to enhanced degradation in soil was attributed for this failure. Chlorpyrifos has been reported to be degraded cometabolically in liquid media by Flavobacterium sp. and also by an Escherichia coli clone with an opd gene (20, 30, 39). However these microbes did not utilize chlorpyrifos as a source of energy. Mallick et al. (20) reported degradation of chlorpyrifos in mineral salt medium by an Arthrobacter species that was initially isolated from methyl parathion-enriched soil. Recently we have reported enhanced degradation of chlorpyrifos, effects of soil pH on degradation, and the enrichment of a degrading strain in an Australian soil in which this insecticide has been used for several years, using bacterial profiling of the 16S rRNA gene by denaturing gradient gel electrophoresis (36). This culture was shown to mineralize chlorpyrifos in minimal medium in which the pesticide was the only source of carbon. However, the prevalence of degrading strains in soil and the pathway of degradation of chlorpyrifos by this bacterium in liquid culture are unknown.
The molecular basis of degradation of certain organophosphates has been studied extensively (13, 21, 32). A widely distributed organophosphate-degrading gene (opd) was isolated from temporally, geographically, and biologically different species (13, 32, 33). In most of the studies, opd genes were found to be plasmid based and had similar DNA sequences. However, Horne et al. (13) isolated an opd gene from Agrobacterium radiobacter, which was located on the chromosome but had a similar sequence to the opd gene from other bacteria. Recently, there have been reports of organophosphate degradation genes with similar function but different sequences from the opd gene: for example, a methyl parathion-degrading Plesiomonas species had a DNA sequence quite different from those of the known opd genes (40). Similarly, coumaphos-degrading Nocardioides simplex NRRL B-24074 (23) has been reported to have novel organophosphate-degrading enzyme and gene systems.
Chlorpyrifos has been one of the pesticides most used worldwide since 1965. Contamination of soil by the pesticide can result from bulk handling in the farmyard, and rinsing of containers and accidental release may occasionally lead to the contamination of surface and groundwater. Reports from the Environmental Protection Agency suggest that a wide range of water and terrestrial ecosystems may be contaminated with chlorpyrifos (1, 9). Use of pesticide-degrading microbial systems for removal of pollutants from the contaminated systems requires an understanding of ecological, physiological, and biochemical requirements of degrading organisms.
The objectives of the present experiments were to isolate degrading strains from the soil, characterize chlorpyrifos-degrading isolates, investigate the pathway of degradation, and study the ecological factors that govern chlorpyrifos degradation in soil.
MATERIALS AND METHODS
Chemicals and pesticide analysis.
Analytical (technical)-grade chlorpyrifos and 3, 5, 6-trichloro-2-pyridinol (TCP), fonofos, ethoprophos, coumaphos, diazinon, parathion (99% purity; British Greyhound, Ltd., Birkenhead, United Kingdom), fenamiphos, isazofos (Promochem, Teddington, United Kingdom), and cadusafos (FMC, Philadelphia, Pa.) were used throughout this study. Ring-labeled [2,6-ring-14C]chlorpyrifos (946 MBq mmol−1) was supplied by Dow Agroscience, Indianapolis, Ind., and uniformly labeled chlorpyrifos (388 MBq mmol−1) was supplied by International Isotopes, Munich, Germany. Detailed methods of pesticide residue measurement and radioactivity measurement have been described previously (35, 36).
Isolation of a chlorpyrifos-degrading bacterium by enrichment culture.
Three different media, a mineral salt medium (MSM), a mineral salt medium supplemented with nitrogen (MSMN), and a soil extract medium (SEM), were used in both enrichment culture of pretreated soils and liquid culture of isolated bacteria. The constituents of media and preparation methods have been described in detail previously (7, 14, 15). Nutrient agar (Difco), nutrient broth (Difco), and Luria broth were prepared according to the manufacturer's instructions.
Samples from an Australian soil (pH 8.4, with 27% sand, 42% silt, 31% clay, and 3.75% organic matter) in triplicate (100 g), which were reported earlier for enhanced degradation (36), were treated with chlorpyrifos to achieve a concentration of 25 mg kg−1, three times at intervals of 9 days to maximize the rapid-degrading capability of the soil. Immediately after 50% loss of chlorpyrifos following the third treatment, 0.5 g of soil from each replicate was used to inoculate separate Duran bottles containing 50 ml of MSM, MSMN, or SEM with a 25-mg-liter−1 concentration of chlorpyrifos. Samples including triplicate controls (medium plus pesticide, with and without chloroform sterile soil inoculations) were incubated in the dark on an orbital shaker at 20°C and 150 rpm. The concentration of chlorpyrifos was measured regularly by removing 0.5-ml subsamples of medium and transferring them to 2-ml high-performance liquid chromatography (HPLC) vials with 1.5 ml of acetonitrile and was analyzed by HPLC. Immediately after the loss of 50% of the initial concentration of chlorpyrifos, 0.5 ml from each replicate was transferred to the respective fresh media (20 ml). This dilution and transfer process was repeated three times. Following the third transfer, immediately after loss of 50% of the chlorpyrifos, a 10-fold-dilution series was prepared and 0.1 ml of each dilution was spread in triplicate on corresponding plates of MSM, MSMN, and SEM (containing 1.2% agar and 25 mg of chlorpyrifos liter−1) and incubated at 20°C for 2 days. Approximately 200 colonies from each medium combination were randomly transferred and streaked on respective fresh solid media. After overnight growth, all isolates were tested for their degrading capability by inoculation in liquid medium (10 ml in a universal bottle). The incubation conditions were the same as described previously. Chlorpyrifos degradation was monitored for 8 weeks.
Characterization of isolated chlorpyrifos-degrading bacteria.
Different isolates were tested for their oxidase and catalase activities; Gram stain, shape, and motility under the microscope were also observed. Strains were characterized by analysis of their 16S rRNA gene. Cells were grown overnight on MSMN or SEM agar plates. The cells were washed off with 2 ml of sterile MilliQ water and then pelleted by centrifugation at 6,000 × g for 5 min, and the DNA was extracted as described previously (36). The 16S rRNA genes of the bacterial isolates were amplified with a set of universal primers (Life Technologies, Paisley, United Kingdom) that allows amplification of most bacterial rRNA genes. The primers 8f (5′ CACGGATCCAGACTTTGATYMTGGCTCAG 3′, forward) and 1512r (5′ GTGAAGCTTACGGYTAGCTTGTTACGACTT 3′, reverse) were used. After amplification, 5 μl of each reaction mixture was run on 0.7% (wt/vol) agarose gel to confirm the size and purity of PCR products. The DNA was then purified with a QIAquick PCR purification kit following the protocol provided by the supplier (Qiagen, Ltd., Crawley, West Sussex, United Kingdom). Samples (10 μl) of purified PCR product from each isolate were digested in 20-μl reaction mixtures with the restriction enzyme RsaI (2 U). Digestion was performed following the instructions provided by the supplier (Life Technologies), and the DNA was analyzed on 2% (wt/vol) agarose gel.
Protein profiles of isolates were compared to type strains. Isolates were grown in Luria broth at 30°C overnight. The cells were pelleted by centrifugation at 5,000 × g for 5 min. Pellets were washed and resuspended in 100 μl of phosphate buffer (0.01 M, pH 7.4). Two samples from each isolate were stored at −70°C. One sample (50 μl) from each isolate was mixed with 50 μl of NuPAGE sodium dodecyl sulfate (SDS) loading buffer (Novex Experimental Technology). Samples were boiled for 5 min and then centrifuged at 13,000 × g for 2 min. SDS-polyacrylamide gel electrophoresis (PAGE) was performed with precast 3 to 8% polyacrylamide gels in Tris-acetate buffer. Samples were run at 30 V in 1× NUPAGE Tris-acetate SDS running buffer for the first 10 min and then at 150 V until the tracer dye had reached the bottom of the gel. Gels were stained with 0.25% (wt/vol) Coomassie brilliant blue (Sigma, Bookham, Surrey, United Kingdom) in 50% (vol/vol) methanol plus 10% (vol/vol) acetic acid for approximately 1 h and destained with methanol-water-acetic acid (50:40:10 [vol/vol]).
Characterization of isolate B-14.
The 16S rRNA PCR was sequenced with the primers 8f and 1512r and internal primers derived from these sequencings to obtain double-stranded coverage of the product (36). The DNA sequences were edited and assembled with the DNA* package and analyzed for similarity to other 16S rRNA sequences, using the ribosomal database online analysis service and Fasta 6.1 for the EMBL database. Selected sequences with the greatest sequence similarity to the B-14 sequence were extracted from the database and aligned. Sequences were compared by using the packages Dnadistance and Dnapars (with Seqboot), and trees were generated by Drawtree (all from within the PHYLIP and RDP suite of packages). Various trees were compared and produced essentially the same results. Therefore, one analysis was presented, which used the Jukes-Cantor algorithm and the treeing method unweighted pair group method for arithmetic means (UPGMA).
Fatty acid methyl ester analysis was carried out at the Central Science Laboratory, York, United Kingdom. The bacterial isolate was grown overnight on nutrient agar medium at 28°C. The cultures were then transferred to Trypticase soy broth for 24 h at 28°C, and fatty acid methyl esters were analyzed by gas chromatography (GC). The Hewlett-Packard column HP6890 series GC system (Ultra2 capillary column, 25 m, 0.2 mm, 0.33 μm) was used for chromatography. The operating temperature was set at 170°C, and the GC column was attached to a flame ionization detector. The results were compared to a commercial library (TSBA40) and against the CSL library (NCPPB3).
Inoculum preparation for degradation studies.
Unless otherwise stated, the inocula for all of the experiments were prepared by growing bacteria in 50 ml of MSMN overnight at 20°C on a rotary shaker at 150 rpm. Cultures were pelleted by centrifugation at 6,000 × g for 10 min. Cells were washed three times with 25 ml of sterile 0.0125 M phosphate buffer (pH 7.2) and quantified by the dilution plate count technique. For all experiments, 106 cells ml−1 were used and samples were incubated at 20°C at 150 rpm unless otherwise stated.
Degradation of chlorpyrifos by isolate B-14.
The degrading ability of the isolated bacterium was assayed with radiolabeled pesticide. Triplicate samples (10 ml) of MSMN were added to 50 μl of [14C]chlorpyrifos labeled either uniformly (388 MBq mmol−1) to achieve a concentration of 222-Bq-ml−1 MSMN or with 50 μl of [14C]chlorpyrifos on the pyridinyl ring (946 MBq mmol) to achieve a concentration of 418-Bq-ml−1 MSMN. Additionally, 1 ml of analytical chlorpyrifos solution was added to achieve a concentration of 25 mg liter−1. 14CO2 was collected in a vial containing 1 M NaOH. Radioactivity in samples, trapped CO2, and pesticide residues was measured at regular intervals. In order to investigate if the chlorpyrifos-degrading isolate could use chlorpyrifos as the sole source of carbon and phosphorus, MSMN was modified as described by Karpouzas et al. (15). In this modified medium, chlorpyrifos at 25 mg liter−1 was the only source of carbon and phosphorus. Effects of nutrient composition, incubation temperature, pesticide concentration, inoculum density, and medium pH were studied.
To examine the effect of nutrient composition on the chlorpyrifos-degrading ability of the isolated consortia, the MSMN and chlorpyrifos media were supplemented with extra carbon sources. Active degrading cultures (0.25 ml) were transferred to 5 ml of the following media: MSMN containing chlorpyrifos (25 mg liter−1), MSMN containing chlorpyrifos and glucose, and MSMN containing chlorpyrifos and succinate. Glucose and succinate were added as 0.1-ml filter-sterile solutions in distilled water to give a final concentration of 1 g liter−1. Also nutrient broth and Luria broth media were supplemented with chlorpyrifos (35 mg liter−1) and then inoculated with the isolate B-14 to study the effect of other nutrient-rich media on chlorpyrifos degradation. Triplicate sets of each composition without inoculation were kept as controls.
Enzymatic activities of isolate B-14.
Phosphodiesterase and alkaline phosphoatase (phosphomonoesterase) activity of the isolate was examined by measuring the rate of hydrolysis of bis (p-nitrophenyl) phosphate and p-nitrophenyl phosphate. In this experiment, 1 ml of 25 mM of either compound was inoculated with about one loop of the overnight-grown cultures. Samples were incubated for 30 min at 37°C. One control without inoculation and another control inoculated with E. coli were also incubated for both compounds. The reaction was stopped with 4 ml of 0.5 M NaOH and 1 ml of 0.5 M CaCl2. Formation of p-nitrophenol was measured by optical absorbance of the samples at 420 nm. To determine the presence of a chlorpyrifos-degrading gene or plasmid, standard methods were used as described previously (36, 38).
Substrate range.
Cross-feeding studies with other organophosphorus insecticides were also carried out. The liquid medium was supplemented with diazinon (O,O-diethyl O-2-isopropyl-6-methylpyrimidin-4-yl-phosphorothioate), parathion (O,O-diethyl O-4-nitrophenyl phosphorothioate), or coumaphos (O-3-chloro-4-methyl-2-oxo-2H-chromen-7-yl O,O-diethyl phosphorothioate) at 25 mg liter−1. The pesticide residues were measured by HPLC. Cross-feeding with organophosphorus nematicides, cadusofos (S, S-di-sec O-ethyl-phosphorodithioate), ethoprop (O-ethyl S,S-dipropyl phosphorodithioate), fenamiphos (ethyl 4-methylthio-m-tolyl isopropylphosphoramidate), fonofos [O-ethyl S-phenyl (RS)-ethyl phosphonodithioate], and isazofos (O-5-chloro-1-isopropyl-1H-1, 2, 4-triazol-3-yl O,O-diethyl phosphorothioate) was also carried out. Growth of isolates on methanol, ethanol, propanol, isopropanol, and acetone was tested by using MSMN medium with one of these compounds as the sole source of carbon.
Inoculation and degradation in soil.
Soil from the Cottage field at HRI, Wellesbourne, United Kingdom, was used for this study. This soil had characteristics of a sandy loam with 71% sand, 16% silt and 8% clay, 4.97% organic matter, and a pH of 6.7. Soil samples (1 kg) were sterilized by fumigation with chloroform for 10 days at 30°C. Similar soil samples were stored at 4°C in sealed polyethylene bags. Residual chloroform was removed from the fumigated soils by repeat evacuation in a vacuum desiccator. Subsamples (100 g) of the fumigated and nonfumigated soil were treated under aseptic conditions with chlorpyrifos (35 mg kg−1). One set of fumigated and nonfumigated soils in triplicate was inoculated with chlorpyrifos-degrading bacteria (106 cells g−1), and another set without inoculation was kept as controls. The inoculum was thoroughly mixed into soils under sterile conditions. All soil samples were retreated with the appropriate pesticides 6 and 10 days after the first application to indirectly monitor the survival and proliferation of the inoculated bacteria. Soil samples were incubated at 20°C and 40% of water-holding capacity in the dark. Soil samples were also treated with uniformly or ring-labeled [14C]chlorpyrifos as described previously (36) to determine the part of the molecule used as a source of carbon by the inoculated microorganisms.
Nucleotide sequence accession number.
The 16S rRNA gene sequence deposited in EMBL has been given the accession no. AJ639856.
RESULTS
Isolation of chlorpyrifos-degrading bacteria by enrichment culture.
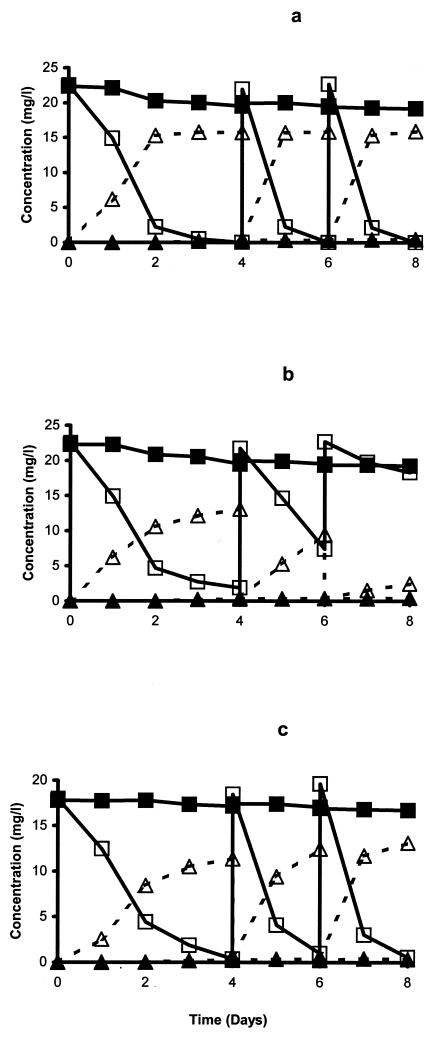
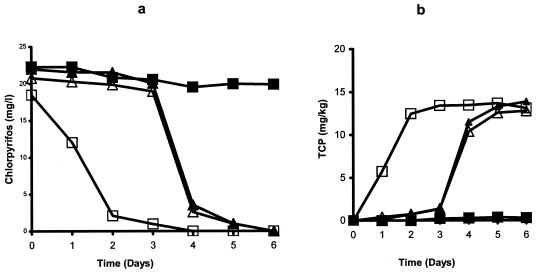
The degradation of chlorpyrifos was rapid in the Australian soil from the first application, with a 50% degradation time of less than 4 days. Most of the applied pesticide was released in the form of CO2. No significant accumulation of TCP was observed during soil enrichment. The degradation of chlorpyrifos during enrichment culture in three different liquid media is shown in Fig. 1. In SEM and MSMN, the degradation rate increased with the second and third treatments, where more than 50% of the applied pesticide was degraded within 1 day (Fig. 1a and c). The degradation rate of chlorpyrifos in MSM was similar to that in the other two media following the first application, but degradation of the second application was slow, and virtually no degradation occurred in the third treatment (Fig. 1b). In contrast to soil enrichment, TCP accumulated in all liquid media. No degradation of chlorpyrifos was observed in uninoculated media. Six isolates (four from MSMN and two from SEM) were obtained. In a previous publication, the four MSMN isolates were briefly described (36) but not characterized. No degrading isolates were obtained from MSM. Repeated attempts at isolating bacteria by TCP enrichment in liquid cultures failed, and therefore no isolates capable of degrading TCP were obtained.
FIG. 1.
Degradation of chlorpyrifos in liquid media amended with 1% (wt/vol) of enhanced soil from Australia. At 4 and 6 days, 0.5-ml aliquots were transferred to fresh SEM (a), MSM (b), and MSMN (c). ▪, chlorpyrifos control; □, chlorpyrifos inoculated; ▴, TCP control; ▵, TCP inoculated. The standard error was within 5% of the mean.
Characterization of an isolated chlorpyrifos-degrading bacterium.
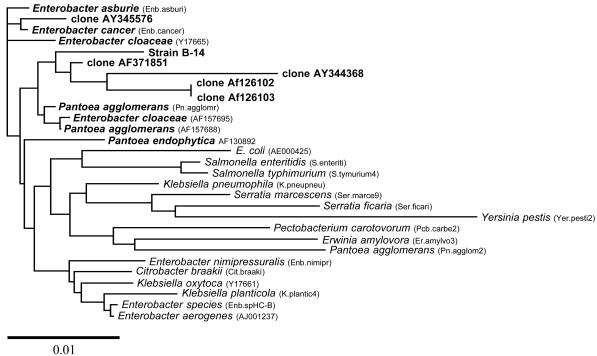
All six isolates were motile, gram negative, and rod shaped. They were catalase and oxidase positive. When the 16S rRNA gene was amplified and digested with RsaI, all isolates produced identical restriction fragment length polymorphism (RFLP) profiles. When the proteins of the six isolates were compared by SDS-PAGE, all isolates produced identical profiles. These results indicate that all the isolates belonged to the same bacterial group and were most likely reisolations of the same bacteria present in the Australian soil. Therefore, further analysis was done with only one isolate, B-14. The 16S rRNA sequence showed similarity to 16S rRNA sequences from members of the order Enterobacteriales. The results indicated that greatest similarity was seen to members of the Enterobacter asburiae group within the RDP database. A dendrogram illustrating the results of the 16S rRNA analysis using PHYLIP is presented in Fig. 2. These results again indicate that the isolate belongs within the E. asburiae group. Overall the B-14 sequence had greatest similarity to Enterobacter-like isolates within this group that are poorly characterized and isolated from environmental samples or represented in environmental clone libraries. Further characterization of this group may lead to a better affiliation in the future. The fatty acid profile of the isolate was compared with the commercial library TSBA40 and the Central Science Laboratory library NCPPB3. The fatty acid profile was found to be entirely typical of a member of the Enterobacteriaceae. The library matches showed close similarity to species within Enterobacter, Klebsiella, and Erwinia. The closest match (82%) was with strains that were classified as Enterobacter agglomerans. However, the environmental isolates within the E. asburiae group are not represented on the database, and it is not possible to determine the level of similarity to this group by this method. When all of these results are taken into account, the only unusual result is the oxidase test. Strains related to Enterobacter are generally oxidase negative; the positive oxidase reaction detected may be attributed to the metabolic versatility of the isolate. Therefore the isolate was designated Enterobacter strain B-14.
FIG. 2.
Dendrogram illustrating the similarity of the chlorpyrifos-degrading bacterium (B-14) 16S rRNA gene to that of members of the enteric bacteria and relatives that had highest sequence similarity (RDP analysis and FASTA). This tree was generated by using the Jukes-Cantor algorithm and the UPGMA linking method. A distance bar is illustrated.
Degrading ability of Enterobacter strain B-14.
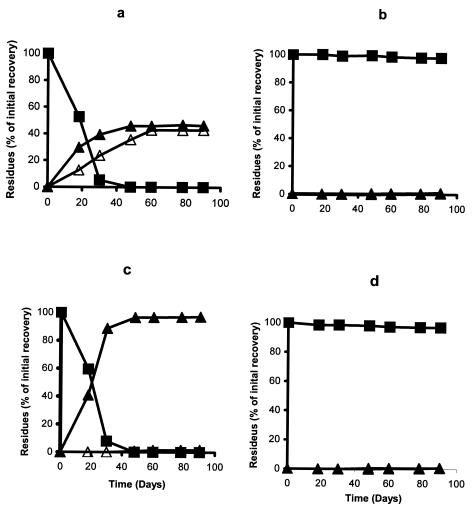
The isolate B-14 was able to degrade chlorpyrifos into diethyl thiophosphate (DETP) and TCP and utilized DETP as the sole source of carbon. When the isolate was grown in MSMN with uniformly labeled [14C]chlorpyrifos, more than 40% of applied pesticide was released in the form of 14CO2 within 48 h. The remaining radioactivity was associated with TCP (45%) (Fig. 3a and b) and bacterial biomass (3%). However, when the medium was supplemented with pyridine ring-labeled chlorpyrifos, no 14CO2 was obtained during the entire incubation, though chlorpyrifos was degraded (Fig. 3c and d). The degradation patterns of chlorpyrifos in MSMN and MSMN-P are shown in Fig. 4a and b. The degradation of chlorpyrifos was similar in both media and was not affected by the absence of a phosphorus supplement. However, the degradation pattern for the isolate was greatly influenced in the presence of other sources of carbon. There was almost no degradation of chlorpyrifos during the first 3 days in the presence of glucose or succinate (Fig. 5a and b). However, after 3 days, chlorpyrifos was degraded rapidly in these two amended media. In MSMN without any other source of carbon, all chlorpyrifos was degraded in 2 days. Repeated subculturing, especially in the nutrient-rich medium, resulted in permanent loss of bacterial capability to use chlorpyrifos as a source of energy, but in some cases, the strain retained the ability to hydrolyze chlorpyrifos. However, in other cases it lost both degradation and utilization capabilities.
FIG. 3.
Degradation of uniformly labeled [14C]chlorpyrifos by Enterobacter strain B-14 in inoculated (a) and noninoculated (b) MSMN and ring-labeled [14C]chlorpyrifos in inoculated (c) and noninoculated (d) MSMN. ▪, chlorpyrifos; ▴, TCP; ▵, 14CO2. The standard error was within 5% of the mean.
FIG. 4.
Degradation of chlorpyrifos by Enterobacter strain B-14 in MSMN in the presence and absence of an added phosphorus source (a) and accumulation of TCP (b). ▪, control; ▴, MSMN without P; ▵, MSMN with P. The standard error was within 5% of the mean.
FIG. 5.
Degradation of chlorpyrifos by Enterobacter strain B-14 in MSMN in the presence of added carbon sources (a) and accumulation of TCP in the medium (b). ▪, control; □, MSMN inoculated; ▴, MSM plus glucose; ▵, MSM plus succinate. The standard error was within 5% of the mean.
The most rapid degradation was observed at 35°C, and the slowest was observed at the two extreme temperatures (5 and 45°C). The degradation rates were similar at 15 and 25°C. The concentration of chlorpyrifos had little effect on degradation rate. All concentrations of pesticide were degraded rapidly in the liquid culture. No pesticide residue was observed after 3 days of incubation. At the high inoculum density (>104 cells ml−1), chlorpyrifos was degraded completely within 48 h. However, degradation was slower at lower density, but the lag phase was followed by rapid degradation. No degradation of chlorpyrifos was observed when the isolate was inoculated at a very low density (100 and 10 cells ml−1). Degradation of chlorpyrifos increased with an increase in medium pH, although the differences were small, except at pH 5, where degradation was very slow. Degradation of chlorpyrifos was negligible in all controls irrespective of medium pH (data not shown).
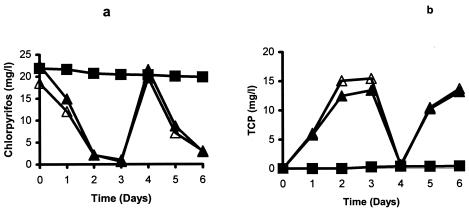
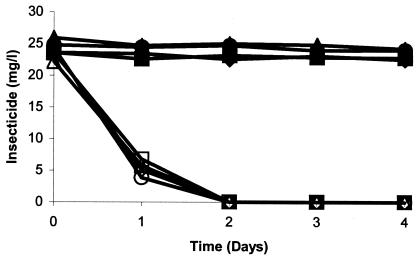
Enzymatic assays of the isolate gave positive results for both phosphodiesterase and phosphomonoesterase activity. PCR of B-14 genomic DNA with three different sets of opd primers did not give any amplification. Similarly hybridization of the opd gene probe did not give a positive signal. All organophosphorus insecticides tested in the cross-feeding experiment were degraded by the isolate (Fig. 6). All compounds were hydrolyzed at a phosphotriester bond, as evident by the accumulation of hydrolysis products. However, none of the organophosphorus nematicides except isazofos was degraded (data not shown). Isolates were not able to use any tested organic solvents as a source of carbon except ethanol.
FIG. 6.
Degradation of different organophosphate insecticides by Enterobacter strain B-14 in inoculated and noninoculated MSMN. ♦, chlorpyrifos control; ⋄, chlorpyrifos inoculated; •, coumaphos control; ○, coumaphos inoculated; ▪, parathion control; □, parathion inoculated; ▴, diazinon control; ▵, diazinon inoculated.
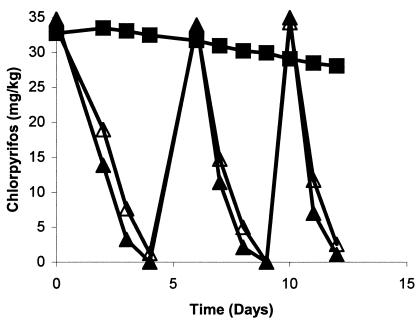
Inoculation of strains B-14 into soil and degradation of chlorpyrifos.
The addition of B-14 to cottage field soils resulted in a more rapid rate of chlorpyrifos degradation than that by indigenous microflora. The estimated half-life of chlorpyrifos was less than 2 days for the first application, which was further reduced to less than a day for the second and third treatments in both fumigated and nonfumigated inoculated soils. Degradation of chlorpyrifos in control nonfumigated soils (without inoculation) was minimal where less than 15% of the applied concentration was degraded in 12-day incubation studies (Fig. 7). Degradation of chlorpyrifos was insignificant in control fumigated soil (data not shown). Studies with radiolabeled pesticide suggested that the degradation mechanisms in inoculated soils were identical to that of B-14, for which chlorpyrifos was degraded in DETP and TCP and which utilized the former compound as a source of carbon. About 38% of applied pesticide was released from soil as 14CO2 in 5 days of incubation from soil samples that were treated with uniformly labeled chlorpyrifos. No radioactive CO2 was detected in soil samples that were treated with ring-labeled chlorpyrifos. TCP was observed to accumulate in a soil sample (data not shown).
FIG. 7.
Degradation of chlorpyrifos in fumigated and nonfumigated soils inoculated with chlorpyrifos-degrading strain B-14 at the rate of 106 cells g−1. ▪, control; ▴, fumigated soil; ▵, nonfumigated soil. The standard error was within 5% of the mean.
DISCUSSION
In this study, six chlorpyrifos-degrading isolates were obtained from an Australian soil. All isolates were shown to be the same species, as indicated by RFLP of the 16S rRNA gene and SDS-PAGE protein profiles. The isolate selected for further study, strain B-14, showed greatest similarity to members of the order Enterobacteriales and in particular the E. asburiae group. Different species of Enterobacter have been reported to degrade phosphonate (19), glyphosate (8), pentaerythritol tetranitrate (4), and trinitrotoluene (11). However, this is the first report of organophosphate degradation by an Enterobacter species. In our previous work, in which the same Australian soil used here was mixed with United Kingdom soil to enhance degradation of chlorpyrifos, a band with a similar mobility on a denaturing gradient gel electrophoresis gel and a sequence to that found for isolate B-14 dominated the degrading soil. This indicates that Enterobacter strain B-14 grows in and dominates soil samples and is not just a rare degrading isolate from the soil sample that has the ability to grow on the laboratory media that we used. This bacterium is unusual as it has been shown to hydrolyze chlorpyrifos and utilizes part of the compound (DEPT) as its sole source of carbon. The evidence from the studies with radiolabeled chlorpyrifos supports the proposed pathway. When ring-labeled [14C]chlorpyrifos was added to the media, no 14CO2 was recovered during incubation, although chlorpyrifos was degraded rapidly. However, when the MSMN medium was supplemented with uniformly 14C-labeled chlorpyrifos, about 40% of applied radioactivity was trapped as 14CO2 in 3 days. This amount of radioactivity is very near that associated with the side chain (44.4%) of chlorpyrifos.
Previous reports concerning isolation of organophosphorus-degrading microorganisms suggest that the bacteria mainly degrade these compounds cometabolically (13, 21, 30, 40). Some species of bacteria have been isolated that can utilize organophosphates as a source of carbon or phosphorus (31) from the hydrolysis products (25). The bacterium isolated in the present study had very strong phosphotriesterase (OPH) activity and hydrolyzed 35-mg-liter−1 concentrations of chlorpyrifos within 24 h when inoculated with 106 cells ml−1. However, in the presence of other carbon sources, it stopped degrading chlorpyrifos. When these carbon sources were depleted, it then degraded chlorpyrifos as a source of carbon, signifying the environmental adaptation of this bacterium. In the natural environment, the competition for carbon sources is immense and the utilization of chlorpyrifos as an energy source by this bacterium provides it with a substantial competitive advantage over other microorganisms. However, subculturing in nutrient-rich media several times led to the permanent loss of chlorpyrifos-degrading capability. Parekh et al. (26) found similar results for metamitron degradation by a Rhodococcus sp. However, the present results do not agree with those from previous studies with carbofuran and ethoprophos (3, 15), in which the presence of other carbon sources had no effect on the degrading ability of the bacteria.
Another significant observation was the utilization of organophosphorus insecticides as a source of phosphorus by Enterobacter sp. This is also the first report in which an organophosphate compound was used for the supply of two elements, carbon and phosphorus, by a single species. Sethunathan and Yoshida (33) isolated a Flavobacterium species that could use parathion as a source of phosphorus but not carbon and diazinon as a carbon source. However, a Flavobacterium strain was not able to use other organophosphorus pesticides either as a source of phosphorus or carbon. Similarly a variety of isolates that could use phosphorothionate or phosphorodithionate compounds as the sole source of phosphorus were unable to degrade these compounds as a source of carbon (31). Shelton (34) isolated a consortium that could use diethylthiophosphoric acid as a carbon source but was unable to degrade it as a phosphorus or sulfur source. Kertesz et al. (18) suggested that the conditions under which the environmental isolates are enriched are crucial in selecting for strains not only with the desired degradative enzyme systems, but also with specific regulation mechanisms for the degradation pathways. However, the utilization of chlorpyrifos as a source of phosphorus may be explained by the presence of phosphodiesterase and phosphomonoesterase activity in the B-14 strain. OPH degrades the triester bond of organophosphorus compounds. Phosphodiesterase and monoesterase are required to make the phosphorus atom available for uptake as a source of phosphorus (inorganic form) and to release ethanol for utilization as a carbon source.
Chlorpyrifos degradation by the isolate B-14 was very rapid at temperatures ranging from 15 to 35°C. The most rapid degradation was observed at 35°C, and the slowest was observed at 5 and 45°C. These results were expected, since most members of the family Enterobacteriaceae grow well at 37°C. The concentration of pesticide had no apparent effect on degradation rate. The Enterobacter sp. was able to degrade chlorpyrifos at concentrations as high as 250 mg liter−1 in less than 2 days. Degradation of different pesticides at high concentration by isolated microorganisms has been reported earlier (15, 16, 37). However, the inhibitory effects of chlorpyrifos on indigenous nonadapted soil microflora at a concentration of less than 30 mg kg−1 reported by Racke and Coats (28) are in contrast to the tolerance shown by the chlorpyrifos-degrading isolate Enterobacter strain B-14. One of the important reasons cited for lack of isolation of chlorpyrifos-utilizing microorganisms is antimicrobial activity of TCP at high concentrations (27). The growth of the isolated Enterobacter species was not affected by TCP even at concentrations of more than 150 mg liter−1. Although degradation of TCP by a Pseudomonas sp. has been reported (10), the Enterobacter strain isolated in this study did not degrade TCP.
In the present experiment, more than 100 cells ml of medium−1 was required to initiate the acclimation process. The lower levels of inoculum resulted in longer lag periods, and an ideal inoculum density for degradation appeared to be around 106 cells ml−1. These cell densities degraded chlorpyrifos without a lag phase. The isolated Enterobacter species were capable of degrading chlorpyrifos in the pH range from 5.5 to 7.6. The isolate was able to degrade all tested organophosphorus insecticides but none of the nematicides, except isazofos. All the studied insecticides as well as isazofos have diethyl phosphorothionate side chains, which may explain the reason for their degradation. Other nematicides, although organophosphorus, had a different overall structure.
Although the initial step in chlorpyrifos degradation by the Enterobacter sp. was similar to other OPH activities, the genes encoding the enzymes were different. Most of the opd genes are plasmid based (12, 21). Two opd genes from P. diminuta and Flavobacterium sp. strain ATCC 27551 have been cloned and sequenced. The two genes shared 86.3% nucleotide sequence identity on the DNA level at the coding region. Southern blot analysis of a methyl parathion-degrading strain of a Pseudomonas sp. against the opd gene from P. diminuta showed positive hybridization with the hydrolase gene probe (5). Recently, Horne et al. (13) isolated a chromosome-based opd gene from A. radiobacter, which shows DNA sequence homology with the opd genes mentioned above. In the bacterium isolated in the present studies, no plasmid DNA was detected. This suggests that the chlorpyrifos-degrading gene may be chromosome based. Amplification of isolated Enterobacter sp. DNA with three different sets of primers designed from the conserved region of opd did not give any PCR products. Hybridization of isolate DNA with the opd probe also failed, suggesting that the DNA sequence of the gene responsible for chlorpyrifos degradation is not similar to that of the known opd gene. There have been some reports of different DNA sequences for opd genes (22, 40). Also, the opd gene product has recently been expressed on the cell surface and has the potential to remediate organophosphate nerve compounds (6, 39). Due to its broad specificity against a range of organophosphorus compounds, the isolated Enterobacter sp. strain possesses a great potential to provide a versatile gene or enzyme system that may be used for the remediation of highly toxic organophosphate nerve agents.
Successful removal of pesticides by the addition of bacteria (bioaugmentation) had been reported earlier for many compounds, including parathion (2), coumaphos (17, 24), ethoprop (16), and atrazine (37). Results from the present study confirm that the isolated chlorpyrifos-degrading bacterium could be used successfully for the removal of these pesticides from contaminated soil. In the present work, the bacterial systems successfully degraded chlorpyrifos in fumigated and nonfumigated soils, suggesting that these bacterial systems can compete with and survive with the local microflora.
Acknowledgments
This work was funded in part by the United Kingdom Biotechnology and Biological Sciences Research Council (BBSRC).
REFERENCES
- 1.Anonymous. 1998. Fact sheet: chlorpyrifos. Pestic. News 41:18-19. [Google Scholar]
- 2.Barles, R. W., G. C. Daughton, and D. P. H. Hsieh. 1979. Accelerated parathion degradation in soil inoculated with acclimated bacteria under field conditions. Arch. Environ. Contam. Toxicol. 8:647-660. [DOI] [PubMed] [Google Scholar]
- 3.Bekhi, R. M., E. E. Topp, and B. A. Blackwell. 1994. Ring hydroxylation of N-methylcarbamate insecticide by Rhodococcus TE1. J. Agric. Food Chem. 42:1375-1378. [Google Scholar]
- 4.Binks, P. R., C. E. French, S. Nicklin, and N. C. Bruce. 1996. Degradation of pentaerythritol tetranitrate reductase by Enterobacter cloacae PB2. Appl. Environ. Microbiol. 62:1214-1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chaudry, G. R., A. N. Ali, and W. B. Wheeler. 1988. Isolation of a methyl parathion-degrading Pseudomonas sp. that posessess DNA homologous to the opd gene from Flavobacterium sp. Appl. Environ. Microbiol. 54:288-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cho, C. M.-H., A. Mulchandani, and W. Chen. 2002. Bacterial cell surface display of organophosphorus hydrolase for selective screening of improved hydrolysis of organophosphate nerve agents. Appl. Environ. Microbiol. 68:2026-2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cullington, J. E., and A. Walker. 1999. Rapid biodegradation of diuron and other phenylurea herbicides by a soil bacterium. Soil Biol. Biochem. 31:677-686. [Google Scholar]
- 8.Dick, R. E., and J. P. Quinn. 1995. Glyphosate-degrading isolates from environmental samples: occurrence and pathways of degradation. Appl. Microbiol. Biotechnol. 43:545-550. [DOI] [PubMed] [Google Scholar]
- 9.Environmental Protection Agency. 1997. Review of chlorpyrifos poisoning data. Environmental Protection Agency, Washington, D.C.
- 10.Feng, Y., K. D. Racke, and J.-M. Bollag. 1997. Isolation and characterization of a chlorinated-pyridinol-degrading bacterium. Appl. Environ. Microbiol. 63:4096-4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.French, C. E., S. Nicklin, and N. C. Bruce. 1998. Aerobic degradation of 2,4,6-trinitrotoluene by Enterobacter cloacae PB2 and by pentaerythritol tetranitrate reductase. Appl. Environ. Microbiol. 64:2864-2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harper, L. L., C. S. McDaniel, C. E. Miller, and J. R. Wild. 1988. Dissimilar plasmids isolated from Pseudomonas diminuta MG and a Flavobacterium sp. (ATCC 27551) contain identical opd genes. Appl. Environ. Microbiol. 54:2586-2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horne, I., T. D. Sutherland, R. L. Harcourt, R. J. Russell, and J. G. Oakeshott. 2002. Identification of an opd (organophosphate degradation) gene in an Agrobacterium isolate. Appl. Environ. Microbiol. 68:3371-3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karpouzas, D. G., J. A. W. Morgan, and A. Walker. 2000. Isolation and characterisation of ethoprophos-degrading bacteria. FEMS Microbiol. Ecol. 33:209-218. [DOI] [PubMed] [Google Scholar]
- 15.Karpouzas, D. G., and A. Walker. 2000. Factors influencing the ability of Pseudomonas putida strains epI and II to degrade the organophosphate ethoprophos. J. Appl. Microbiol. 89:40-48. [DOI] [PubMed] [Google Scholar]
- 16.Karpouzas, D. G., and A. Walker. 2000. Factors influencing the ability of Pseudomonas putida epI to degrade ethoprophos in soil. Soil Biol. Biochem. 32:1753-1762. [DOI] [PubMed] [Google Scholar]
- 17.Kearney, P. C., J. S. Karns, M. T. Muldoon, and J. M. Ruth. 1986. Coumaphos disposal by combined microbial and UV-ozonation reactions. J. Agric. Food Chem. 34:702-706. [Google Scholar]
- 18.Kertesz, M. A., A. M. Cook, and T. Leisinger. 1994. Microbial metabolism of sulfur and phosphorus-containing xenobiotics. FEMS Microbiol. Rev. 15:195-215. [DOI] [PubMed] [Google Scholar]
- 19.Lee, K.-S., W. W. Metcalf, and B. L. Wanner. 1992. Evidence for two phosphonate degradative pathways in Enterobacter aerogenes. J. Bacteriol. 174:2501-2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mallick, B. K., A. Banerji, N. A. Shakil, and N. N. Sethunathan. 1999. Bacterial degradation of chlorpyrifos in pure culture and in soil. Bull. Environ. Contam. Toxicol. 62:48-55. [DOI] [PubMed] [Google Scholar]
- 21.Mulbry, W. W., J. S. Karns, P. C. Kearney, J. O. Nelson, C. S. McDaniel, and J. R. Wild. 1986. Identification of a plasmid-borne parathion hydrolase gene from Flavobacterium sp. by Southern hybridization with opd from Pseudomonas diminuta. Appl. Environ. Microbiol. 51:926-930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mulbry, W. W. 1998. Selective deletion involving the organophosphorus hydrolase gene adp from Nocardia strain B-1. Microbiol. Res. 153:213-217. [DOI] [PubMed] [Google Scholar]
- 23.Mulbry, W. W. 2000. Characterisation of a novel organophosphorus hydrolase from Nocardioides simplex NRRL B-24074. Microbiol. Res. 154:285-288. [DOI] [PubMed] [Google Scholar]
- 24.Mulbry, W. W., P. L. Del Valle, and J. S. Karns. 1996. Biodegradation of the organophosphate insecticide coumaphos in highly contaminated soils and in liquid wastes. Pestic. Sci. 48:149-155. [Google Scholar]
- 25.Munnecke, D. M., and D. P. M. Hsieh. 1976. Pathways of microbial metabolism of parathion. Appl. Environ. Microbiol. 31:63-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parekh, N. R., D. L. Suett, S. J. Roberts, and S. J. Welch. 1994. Rapid degradation of the triazinone herbicide metamitron by a Rhodococcus sp. isolated from treated soil. J. Appl. Bacteriol. 77:467-475. [DOI] [PubMed] [Google Scholar]
- 27.Racke, K. D., D. A. Laskowski, and M. R. Schultz. 1990. Resistance of chlorpyrifos to enhanced biodegradation in soil. J. Agric. Food Chem. 38:1430-1436. [Google Scholar]
- 28.Racke, K. D., and J. R. Coats. 1990. Pesticides in soil microbial ecosystems. Am. Chem. Soc. Symp. Ser. 426:1-12. [Google Scholar]
- 29.Rani, N. L., and D. Lalitha-Kumari. 1994. Degradation of methyl parathion by Pseudomonas putida. Can. J. Microbiol. 4:1000-1004. [DOI] [PubMed] [Google Scholar]
- 30.Richnis, R., I. Kaneva, A. Mulchandani, and W. Chen. 1997. Biodegradation of organophosphorus pesticides using surface-expressed organophosphorus hydrolase. Nat. Biotechnol. 15:984-987. [DOI] [PubMed] [Google Scholar]
- 31.Rosenberg, A., and M. Alexander. 1979. Microbial cleavage of various organophosphorus insecticides. Appl. Environ. Microbiol. 37:886-891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Serdar, C. M., D. T. Gibson, D. M. Munnecke, and J. H. Lancaster. 1982. Plasmid involvement in parathion hydrolysis by Pseudomonas diminuta. Appl. Environ. Microbiol. 44:246-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sethunathan, N. N., and T. Yoshida. 1973. A Flavobacterium that degrades diazinon and parathion. Can. J. Microbiol. 19:873-875. [DOI] [PubMed] [Google Scholar]
- 34.Shelton, D. R. 1988. Mineralization of diethylthiophosphoric acid by an enriched consortium from cattle dip. Appl. Environ. Microbiol. 54:2572-2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Singh, B. K., A. Walker, and D. J. Wright. 2002. Degradation of chlorpyrifos, fenamiphos and chlorothalonil alone and in combination and their effects on soil microbial activity. Environ. Toxicol. Chem. 21:2600-2605. [PubMed] [Google Scholar]
- 36.Singh, B. K., A. Walker, J. A. W. Morgan, and D. J. Wright. 2003. Effect of soil pH on the biodegradation of chlorpyrifos and isolation of a chlorpyrifos-degrading bacterium. Appl. Environ. Microbiol. 69:5198-5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Struthers, J. K., K. Jayachandran, and T. B. Moorman. 1998. Biodegradation of atrazine by Agrobacterium radiobacter J14a and use of this strain in bioremediation of contaminated soil. Appl. Environ. Microbiol. 64:3368-3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Turnbull, G. A., M. Ousley, A. Walker, E. Shaw, and J. A. W. Morgan. 2001. Degradation of substituted phenylurea herbicide by Arthrobacter globiformis strain D47 and characterization of a plasmid-associated hydrolase gene, puhA. Appl. Environ. Microbiol. 67:2270-2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang, A. A., A. Mulchandani, and W. Chen. 2002. Specific adhesion to cellulose and hydrolysis of organophosphate nerve agents by a genetically engineered Escherichia coli strain with a surface-expressed cellulose-binding domain and organophosphorus hydrolase. Appl. Environ. Microbiol. 68:1684-1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhongli, C., L. Shunpeng, and F. Guoping. 2001. Isolation of methyl parathion-degrading strain M6 and cloning of the methyl parathion hydrolase gene. Appl. Environ. Microbiol. 67:4922-4925. [DOI] [PMC free article] [PubMed] [Google Scholar]