Abstract
Cornelia de Lange Syndrome (CdLS) is a multisystem birth defects disorder that affects every tissue and organ system in the body. Understanding the factors that contribute to the origins, prevalence, and severity of these developmental defects provides the most direct approach for developing screens and potential treatments for individuals with CdLS. Since the majority of cases of CdLS are caused by haploinsufficiency for NIPBL (Nipped-B-like, which encodes a cohesin-associated protein), we have developed mouse and zebrafish models of CdLS by using molecular genetic tools to create Nipbl-deficient mice and zebrafish (Nipbl+/− mice, zebrafish nipbl morphants). Studies of these vertebrate animal models have yielded novel insights into the developmental etiology and genes/gene pathways that contribute to CdLS-associated birth defects, particularly defects of the gut, heart, craniofacial structures, nervous system, and limbs. Studies of these mouse and zebrafish CdLS models have helped clarify how deficiency for NIPBL, a protein that associates with cohesin and other transcriptional regulators in the nucleus, affects processes important to the emergence of the structural and physiological birth defects observed in CdLS: NIPBL exerts chromosome position-specific effects on gene expression; it influences long-range interactions between different regulatory elements of genes; and it regulates combinatorial and synergistic actions of genes in developing tissues. Our current understanding is that CdLS should be considered as not only a cohesinopathy, but also a “transcriptomopathy,” that is, a disease whose underlying etiology is the global dysregulation of gene expression throughout the organism.
Keywords: Nipped-B-like (NIPBL) gene, Protocadherin genes, Hox genes, Shh genes, chromatin conformation
INTRODUCTION
Cornelia de Lange syndrome (CdLS) is a birth defects disorder that affects every system in the body. Although long understood by clinicians to be a distinctive syndrome [Opitz, 1985], it was not until the discovery that the majority of individuals with CdLS carry mutations in NIPBL [Krantz et al., 2004; Tonkin et al., 2004], which encodes a protein that binds to the cohesin complex of chromosomal structural proteins, that CdLS was recognized as the prototypic cohesinopathy [Dorsett and Krantz, 2009]. Subsequent to the identification of NIPBL as the major causative gene for CdLS, researchers have gone on to identify a number of cohesin-and/or cohesin regulatory protein-encoding genes which, when mutated, give rise to the classical spectrum of features and deficits associated with CdLS: these genes include NIPBL, SMC1A, SMC3, RAD21, and HDAC8 [Deardorff and Krantz, 2005]. For most of these genes, animal models have been generated, including models from Drosophila, Xenopus, zebrafish, and mouse; these model systems have been widely used as tools for understanding the etiology of CdLS related syndromes [Horsfield et al., 2012; Singh and Gerton, 2015]. In the present paper, we review studies from our group on two vertebrate animal models of CdLS: nipbl-morphant zebrafish and Nipbl-deficient mice. These two model systems have been particularly useful for understanding how Nipbl deficiency leads to global changes in gene expression, and how global gene expression changes act additively and synergistically to generate the structural birth defects that are the hallmarks of CdLS.
DEVELOPMENT OF MOUSE AND ZEBRAFISH ANIMAL MODELS OF CDLS
The discovery that most individuals with CdLS carry mutations in, and are likely haploinsufficient for, NIPBL [Krantz et al., 2004; Tonkin et al., 2004], led to the rapid realization that understanding the etiology of CdLS would require a deeper understanding of how the NIPBL protein, which associates with the cohesin complex of chromosomal proteins, functions in the regulation of chromatin structure and gene expression [Dorsett, 2009]. Since a Drosophila model of Nipbl deficiency already existed [Rollins et al., 1999], and increased insight into the interplay between gene expression and the origins of structural birth defects in CdLS was most likely to arise from studies of vertebrate models of the syndrome, our group decided to generate mouse and zebrafish models of CdLS. Mouse and zebrafish models of Nipbl deficiency recapitulate many of the important structural and functional deficits observed in individuals with CdLS, and have proven invaluable for gaining information about the molecular etiology of the syndrome.
Mouse and zebrafish models of Nipbl deficiency recapitulate many of the important structural and functional deficits observed in individuals with CdLS, and have proven invaluable for gaining information about the molecular etiology of the syndrome.
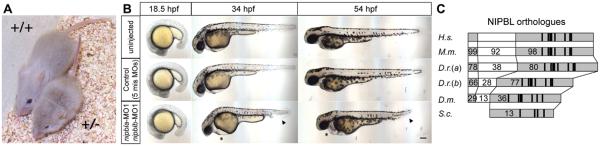
In mouse, we generated a line carrying one null allele of Nipbl. Nipbl+/− mice, created by introduction of a gene trap into Intron 1 of the Nipbl gene, show a high degree of perinatal lethality (~18% of mutant animals survive to weaning), are much smaller than their wildtype littermates, and have numerous structural, physiological, and gene expression defects that mimic the defects seen in CdLS (see below and Fig. 1A). The high incidence of perinatal lethality and small size of Nipbl+/− mice are heritable phenotypes and have been reproduced over many generations [Kawauchi et al., 2009]. Nipbl-deficient zebrafish embryos (Fig. 1B) were created by using translation-blocking morpholinos (MOs) to knock down expression of the two zebrafish nipbl genes (nipbla and nipblb), which our group first cloned (Fig. 1C; since the two zebrafish nipbl genes have functions that do not overlap completely, both are knocked down for most analyses [Muto et al., 2011]). Zebrafish nipbl morphants die at larval stages and exhibit stunted growth: embryos begin to exhibit obvious phenotypes within 24 hr postfertilization (hpf), with growth retardation—especially of the head and tail—and deficits in heart and circulatory system becoming progressively more pronounced over time (Fig. 1B).
Figure 1.
Nipbl-deficient mutant animals show growth abnormalities. (A) Nipbl+/− mouse is markedly smaller than wildtype (+/+) sibling at 4 weeks of age. (B) Nipbl a+b morpholino (MO) co-injected zebrafish larvae (nipbl a/b morphants) resemble uninjected and mixed MO controls at 18.5 hr post-fertilization (hpf), but show marked circulatory defects, pericardial edema (asterisk) and tail defects (arrowheads) by 34 hpf. Scale bar ¼ 100 mm. (C) Structural comparison of NIPBL orthologues among different species. H.s., Homo sapiens, 2,804 amino acids (aa).; M.m., Mus musculus, 2,798 aa; D.r. (a), Danio rerio, form a, 2,876 aa; D.r. (b), form b, 2,381 aa; D.m., Drosophila melanogaster, 2,077 aa; S.c., Saccharomyces cerevisiae, 1,493 aa. Numbers indicate percent amino acid sequence identity compared to human NIPBL. Adapted from [Kawauchi et al., 2009; Muto et al., 2011].
NIPBL-DEFICIENT MICE AND ZEBRAFISH SHOW PRONOUNCED DEFECTS IN HEART AND GUT DEVELOPMENT RELATED TO CHANGES IN ENDODERMAL GENE EXPRESSION
Heart and gut malformations are a prominent part of the pathology of CdLS. Structural heart defects are well-documented, and occur in about 30–40% of individuals with CdLS [Selicorni et al., 2009; Chatfield et al., 2012]. In our examinations of Nipbl+/− mice, we also noted structural defects in the heart. In particular, defects in atrial septum formation (ASD) were noted in the hearts of about half of embryos taken between E15.5 and 18.5 (Fig. 2A and B). Although no ASDs or other obvious cardiac malformations are observed in Nipbl+/− mice that survive to adulthood, these results strongly suggest that perinatal mortality in Nipbl-deficient mice may be caused by cardiac defects [Kawauchi et al., 2009]. Because of the prominence and severity of cardiac defects in Nipbl+/− mice, their underlying cellular and genetic etiology is being studied in detail [Santos, Kawauchi, Calof; unpublished results].
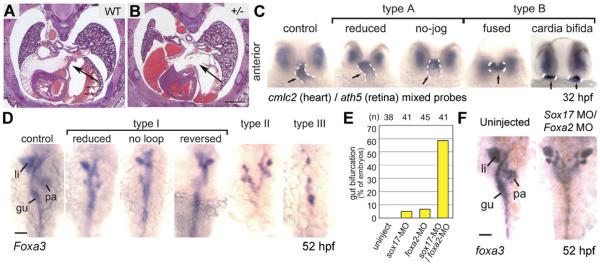
Figure 2.
Heart and gut abnormalities in Nipbl-deficient animals. (A and B) Cryosections, stained with hematoxylin/eosin, of wildtype and Nipbl+/− littermate embryos at gestational day 17.5 (E17.5). A well-formed atrial septum is apparent in the wildtype heart, but absent in a significant fraction of mutants. Scale bar ¼ 1 mm. (C) Zebrafish embryos injected with nipbla/b MOs show a range of defects in heart tube formation at 32 hpf, ranging from mild (type A) to severe (type B). Dashed white lines indicate heart tube, labeled by in situ hybridization (ISH) for a cmlc2 probe (ath5 ISH shows that eye is relatively unaffected). (D) Gut and visceral organ morphology assessed by ISH for foxa3 in nipbla/b morphants at 52 hpf. Defects ranged from mild looping defects (type I) to more severe defects such as bifurcation (type II) and even absence (type III) of gut parenchyma. gu, gut tube; li, liver; pa, pancreas. (E) Frequency of gut bifurcation in sox17 and foxa2 single and double morphants at 52hpf. (F) Example of severe gut bifurcation in sox17/foxa2 double morphant embryo. Scale bar in D and F ¼ 50 mm. Adapted from [Kawauchi et al., 2009; Muto et al., 2011].
Defects in early development of internal organs are readily visualized in zebrafish embryos, so we took advantage of nipbla/b morphants to study the early development of heart and other visceral organs when Nipbl is deficient in this model. As shown in Figure 2C, the developing heart tube of zebrafish embryos is readily visualized by in situ hybridization (ISH) for cmlc2. Pronounced defects in early heart development, ranging from reduced heart tube looping (“type A defects”) to cardia bifida (“type B” defects), are observed in the majority of nipbla/b morphants. Early development of organs such as eyes is not affected (Fig. 2C, ath5 ISH), suggesting that heart development is particularly sensitive to decreases in Nipbl levels. As development proceeds, impaired heart function is observed in nipbla/b morphants, accompanied by circulation defects, pericardial edema, and premature death [Muto et al., 2011].
Gastrointestinal (GI) reflux is a hallmark of CdLS. Many individuals with CdLS die as a result of adverse events affecting the GI system, with gut obstructions and volvulus being reported frequently [Luzzani et al., 2003; Schrier et al., 2011]. We therefore examined nipbla/b morphant embryos for malformations of the developing gut and associated organs (pancreas, liver) using ISH for foxa3, which is expressed by early gut and associated endodermal derivatives. The defects observed in nipbla/b morphants, which range from decreases in gut size to severe patterning defects as extreme as duplications and even absence of associated organs, are shown in Figure 2D [Muto et al., 2011].
To gain insight into the genetic origins of these defects, we performed transcriptome analysis using mRNA obtained from gastrulation-stage (6-hr postfertilization [hpf]) uninjected and nipbla/b morphant embryos. Many small, but significant, gene expression changes were observed when Nipbl levels were reduced, consistent with our findings in mice (see below) and the findings of our colleagues in human CdLS cell lines [Liu et al., 2009]. In particular, genes that function as regulators of endoderm development, such as sox17 and foxa2, were found to be significantly downregulated in their expression. These genes play important roles in heart and visceral organ development [Ober et al., 2003; Fukuda and Kikuchi, 2005; Pfister et al., 2011], and since our experiments indicated that they are directly affected by decreases in Nipbl levels, we performed further studies to determine if knockdown of one or both would result in gut defects similar to those observed in nipbla/b morphants. As shown in Figure 2E and F, reduction of sox17 or foxa2 levels alone causes gut defects in a significant fraction (~5–7%) of morphants. Importantly, when levels of both sox17 and foxa2 are reduced simultaneously, the fraction of morphants with gut defects increases drastically, with almost 60% of morphants displaying severe defects such as gut bifurcation (Fig. 2E and F). Thus, reductions in expression of sox17 and foxa2 act synergistically to reproduce the types of defects in gut development seen in nipbla/b morphants [Muto et al., 2011]. These data strongly support the hypothesis that defects in GI structure/function, observed in individuals with CdLS, arise from the collective effects of multiple gene expression changes resulting from reductions in NIPBL levels.
CRANIOFACIAL, NEUROANATOMICAL, AND BRAIN GENE EXPRESSION ABNORMALITIES IN NIPBL+/− MICE
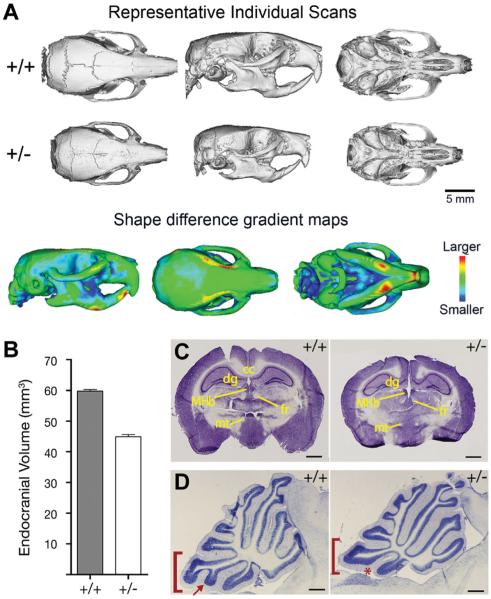
Individuals with CdLS display distinctive craniofacial features that play an important role in clinical diagnosis [Jackson et al., 1993]. Micro-CTanalysis of adult Nipbl+/− mice demonstrated that they exhibit microbrachycephaly and an upward deflection of the snout (“upturned nose”), two of the most distinctive features of CdLS (Fig. 3A and [Kawauchi et al., 2009]). The smaller skull size of Nipbl+/− mice is reflected in their smaller endocranial volume and smaller overall brain size relative to control littermates (Fig. 3B and C). Overall the brains of adult Nipbl+/− mice are grossly normal (aside from their reduced size), although cerebellar hypoplasia is observed (Fig. 3D), consistent with clinical reports [Hayashi et al., 1996; Ozkinay et al., 1998]. The neuroanatomical abnormalities observed in Nipbl+/− mice are reflected in aberrant behaviors and in impairment of neurological functions such as hearing (which is frequently impaired in CdLS [Sakai et al., 2002; Kawauchi et al., 2009]).
Figure 3.
Craniofacial and neuroanatomical abnormalities in Nipbl+/− mice. (A) Top two panels show representative reconstructions of individual scans based on micro-CT analysis of wildtype (top panel) and Nipbl+/− (middle panel) adult mice. Dorsal, relateral, and ventral views are shown. Bottom panel shows shape distance heat maps of wildtype overlaid with mutant skulls, from the entire dataset. The degree of shape change in specific bones is shown by different colors, as indicated in the heat map at right. (B) Endocranial volume (measured from CT scans) is reduced by 25% in in Nipbl+/− mice compared to wildtype. (C) Nissl-stained coronal sections of wildtype (+/+) and Nipbl-deficient (+/−) adult brains. MHb, medial habenular nucleus; fr, fasiculus retroflexus; mt, mammillothalamic tract; dg, dentate gyrus; cc, corpus callosum. Scale bar ¼1 mm. (D) Cerebellar vermal hypoplasia in Nipbl+/− mice. Folium IX (bracket) and its ventral subfolium (arrow, asterisk) are affected. Scale bar ¼ 500 mm. Adapted from [Kawauchi et al., 2009].
To gain insight into the gene expression changes that underlie the behavioral and neurological abnormalities we observe in Nipbl+/− mice, we performed transcriptome analysis on gestational day 13.5 (E13.5) brain RNA from large cohorts of mutant and wildtype embryos. Expression of 978 genes was found to be changed significantly in E13.5 Nipbl+/− brain compared to normal; interestingly, all gene expression changes (which included both increases and decreases in expression) were small, with the single greatest change being only 2.5-fold. Significant expression changes were observed for numerous genes with obvious roles in neuronal function, including genes encoding ion channels, neuropeptides, neurotransmitter receptors, axon guidance molecules, and so on. However, expression changes for genes encoding proteins associated with chromatin structure and function, metabolism, lipoprotein binding, and many other functions and were also observed [Kawauchi et al., 2009]. These data are consistent with studies by us and others, which indicate that Nipbl deficiency results in global dysregulation of gene expression in many—if not all—tissues [Kawauchi et al., 2009; Liu et al., 2009; Muto et al., 2011; Muto et al., 2014; Yuan et al., 2015].
EFFECTS OF NIPBL DEFICIENCY ON DEVELOPMENT AND GENE EXPRESSION IN LIMBS
Alterations in limb and digit development, ranging from mild oligodactyly to frank truncations of the arms, are a hallmark of CdLS [Jackson et al., 1993]. Interestingly, limb truncations are not seen in Nipbl+/− mice, in which only aberrations in ossification, formation of the olecranon process, and mild digit defects are observed (see Lopez-Burks et al. [2016]; and Kawauchi et al. [2009]). As a result, we have concentrated our studies of how Nipbl deficiency affects limb development on the zebrafish model system, where we hypothesized that Nipbl deficiency may be more effective at triggering limb defects than in the mouse.
We performed an extensive analysis of alterations in gene expression and cellular/structural changes in the development of pectoral fins (the structural homologues of mammalian forelimbs) in zebrafish morphants, which yielded a number of significant findings. As predicted, zebrafish embryos injected with nipbla/b MOs showed impaired pectoral fin development, which was dramatic at 76 hpf, when the limbs of morphants were 40% shorter than those of controls. Importantly, partial rescue of limb defects was observed when morphants were co-injected with nipbla mRNA, indicating that effects of MOs were specific to Nipbl depletion (Fig. 4A and B).
Figure 4.
Limb defects and gene expression changes in Nipbl-deficient zebrafish and mice. (A) Defects in pectoral fin development are observed in nipbla/b morphants; defects are partially rescued by injection of exogenous nipbla mRNA. Dorsal view of zebrafish larvae at 76 hpf. (B) Plots of fin lengths in morphants and controls at 76 hpf. (C) Expression of shha (ISH) in pectoral limb buds (arrows) is reduced in nipbla/b morphants at three indicated stages of development. (D) Whole-mount ISH for Shh in E10.5 mouse embryos; expression is reduced in Nipbl+/− hindlimb buds compared to wildtype (WT). Arrowheads indicate hindlimbs; arrows indicate forelimbs. Scale bar ¼ 0.5 mm. (E) Quantification of Shh ISH shown in D; *P < 0.05, **P < 0.01. (F) Zebrafish larvae injected with low doses of med12 or nipbla/b MOs display small reductions in fin size compared to controls; defects are enhanced when morphants are treated with combined nipbla/b and med12 MOs. (G) Plots of fin lengths in morphants and controls in the four conditions. (H) Changes in hox gene expression in morphants treated with nipbla/b, med12, and nipbla/b + med12 MOs at 36 hpf. Adapted from [Muto et al., 2014]
Genes known to be of general importance in vertebrate limb development were examined in nipbla/b morphants, and many were found to be misregulated in their expression. Some of the most important of these were fgf genes (expression of fgf4, fgf8a, and fgf16 were altered in the fin bud apical ectodermal ridge/limb mesenchyme of morphants) and shha, whose expression in the fin bud zone of polarizing activity (ZPA) was reduced in morphants (Fig. 4C). Importantly, Shh expression was also reduced in the ZPA of E10.5 limb buds in Nipbl+/− mice (Fig. 4D and E), indicating that regulatory genes for limb development are similarly affected by Nipbl deficiency in both zebrafish and mice. Transcriptome analysis of E10.5 Nipbl+/− and wildtype littermate limb buds demonstrated that approximately 1,000 genes are significantly altered in their expression when Nipbl is deficient, and that many changes in gene expression observed in mouse limb bud mirrored those observed in nipbla/b morphant zebrafish fin buds [Muto et al., 2014].
Appropriate spatial and temporal expression of Hox genes is important for limb development in all vertebrates [Zakany and Duboule, 2007]. Many hox genes show altered expression in the fin buds of nipbla/b morphant embryos, and expression of different hox genes is either up-or down-regulated in morphants [Muto et al., 2014]. Regulation of hox gene expression by Nipbl is dependent on the relative location of a given gene within its cluster (5′ versus 3′), and the implications of such positional effects for understanding the mechanism(s) by which Nipbl levels effect global changes in gene expression is discussed below. Our analysis of gene expression changes in mouse limb buds also indicated that genes encoding some subunits of Mediator, a protein complex affecting chromatin conformation and global regulation of transcription [Yin and Wang, 2014], are altered in their expression when Nipbl levels are reduced (Table I in Muto et al. [2014]). Because it had been shown by Kagey et al. [2010] that Mediator, cohesin, and Nipbl all interact to regulate gene expression, and since other investigators showed that loss-of-function mutations in med12 disrupt fin development in zebrafish [Rau et al., 2006], we generated med12 morphants and studied pectoral fin development in them. Zebrafish med12 morphants display disrupted fin development and misregulated hox gene expression similar to that observed in nipbla/b morphants. Importantly, when zebrafish embryos are injected with low doses of both med12 and nipbla/b MOs (each of which gives only partial reductions in fin size), a strong synergistic effect is observed (Fig. 4F and G). Moreover, simultaneous reduction of nipbls and med12 have a synergistic effect on alterations in hox gene expression (Fig. 4H). Altogether, these data are consistent with Nipbl and Mediator acting in a common pathway to regulate gene expression and development in the vertebrate limb.
NIPBL, CHROMATIN CONFORMATION, AND GLOBAL REGULATION OF GENE EXPRESSION
Despite the fact that we know that small changes in the expression of many genes contribute to the spectrum of birth defects in CdLS, we do not yet understand mechanistically how this occurs. Past and ongoing studies of Nipbl-deficient animal models do, however, provide a way forward. Transcriptome analyses performed on multiple Nipbl+/− tissues yield common findings, among them the observations that tens to hundreds of transcripts are significantly altered in a given tissue, with effects on gene expression being modest in size (the vast majority of gene expression changes are less than two-fold). Distinctive profiles of gene expression changes for specific tissues are also evident, and have provided insights into genetic pathways that underlie defects in different tissues. One instance is the dysregulation of endodermal gene expression that is hypothesized to underlie, at least in part, the development of heart and visceral organ defects in CdLS (Fig. 2 and Muto et al. [2011]). Similarly, the depletion of fat observed in Nipbl+/−mice is likely attributable to dysregulation of genes regulating the adipocyte differentiation pathway: expression of genes in the pathway that regulates adipogenesis is altered in Nipbl+/− mouse embryonic fibroblasts (MEFs), which are themselves defective in their ability to differentiate into adipocytes [Kawauchi et al., 2009]. Our findings are consistent with the hypothesis that the severe developmental defects observed in CdLS and in Nipbl-deficient animal models are the consequence of the collective action of many small changes in gene expression in every tissue throughout the organism.
Our findings are consistent with the hypothesis that the severe developmental defects observed in CdLS and in Nipbl-deficient animal models are the consequence of the collective action of many small changes in gene expression in every tissue throughout the organism.
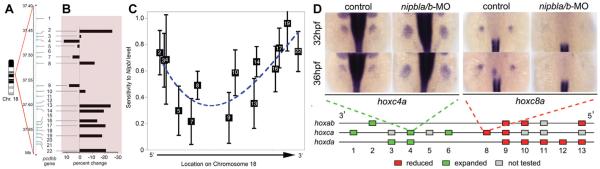
Are there types of genes, and/or gene loci, that are especially sensitive to Nipbl deficiency? One clue comes from the analysis of transcriptional effects that are shared across multiple tissues. In the Nipbl+/− mouse, for example, significant changes in the expression of genes in the protocadherin-beta (Pcdhb) cluster, located on chromosome 18, have been observed repeatedly in embryonic brain, MEFs, limbs, and even skin ([Kawauchi et al., 2009; Muto et al., 2011] and unpublished observations). As shown in Figure 5A and B, Pcdhb genes are both up- and down-regulated in their expression when Nipbl is deficient, with genes at the 3′ and 5′ ends of the cluster showing the greatest percent change in expression. An in-depth analysis, in which changes in expression of individual Pcdhb genes were evaluated as a function of Nipbl levels in the same samples, demonstrated that expression of Pcdhb genes closest to the 5−and 3− ends of the cluster are most sensitive to Nipbl levels, while expression of genes in the middle of the cluster are relatively insensitive to Nipbl levels (Fig. 5C). Therefore, within the Pcdhb gene cluster, position-specific effects of Nipbl on gene expression can clearly be demonstrated [Kawauchi et al., 2009]. Similarly, studies of hox gene expression during zebrafish limb development demonstrate a relationship between the positions of specific hox genes in a given chromosomal cluster and their expression changes when Nipbl levels are depleted (Fig. 5D and Muto et al. [2014]). Altogether, our results from studies in both mouse and zebrafish models support the hypothesis that chromosomal location is an important factor in the sensitivity of gene expression to NIPBL levels.
Figure 5.
Position-specific effects of Nipbl on gene expression. (A) Schematic of protocadherin beta (Pcdhb) locus (consisting of 22 genes) on chromosome 18 in mouse. (B) Quantitative RT-PCR results showing changes in gene expression in 14 of the 22 Pcdhb genes in E17.5 Nipbl+/− brain compared to wildtype controls. (C) Representative graph showing sensitivity of Pcdhb gene expression levels to Nipbl levels in the same samples. Sensitivities and error bars were plotted on an abscissa corresponding to the location of the transcriptional start sites of each of the Pcdhb genes. The dashed line is a smooth polynomial fit to the data. Data indicate that sensitivity of Pcdhb gene expression to Nipbl levels is highest at the 50 and 30 ends of the cluster, lowest in the middle. (D) ISH demonstrating mis-expression of hox genes in nipbla/b morphant zebrafish embryos (over-expression: hoxc4a; under-expression: hoxc8a). Schematic diagram represents effects of reduced nipbl levels on hox gene expression, with 50 hox genes showing reductions in expression (red) and 30 hox genes showing expanded expression (green). Adapted from [Kawauchi et al., 2009; Muto et al., 2011].
Altogether, our results from studies in both mouse and zebrafish models support the hypothesis that chromosomal location is an important factor in the sensitivity of gene expression to NIPBL levels.
How do these topological changes in NIPBL sensitivity come about? One possibility, for which there is increasing evidence, is that Nipbl—together with cohesin and proteins of the Mediator complex—physically brings together gene enhancers and their promoters, resulting in global changes in chromatin architecture that bring about widespread changes in gene expression [Kagey et al., 2010; Whyte et al., 2013; Yin and Wang, 2014]. Indeed, both Nipbl and Mediator are important in limb development; and in the zebrafish model of CdLS, deficiencies of nipbls and med12 together act synergistically to regulate gene expression and the severity of limb defects (Fig. 4F–H and Muto et al., [2014]). Importantly, depletion of both Nipbl and Med 12 have been shown in 3D-fluorescence ISH experiments to affect long-range chromosomal interactions at the hoxda locus in zebrafish limb buds [Muto et al., 2014]. The importance of Mediator, cohesin, and other general transcriptional regulators as collaborators of Nipbl in regulating chromatin conformation and gene expression is an ongoing subject of investigation in our attempt to understand the etiology of birth defects in CdLS.
Acknowledgments
Grant sponsor: National Institutes of Health; Grant numbers: P01-HD052860; P50-GM076516; Grant sponsor: Cornelia de Lange Syndrome Foundation.
Contributor Information
SHIMAKO KAWAUCHI, Associate Project Scientist who has been a primary researcher in developing and analyzing mouse models of CdLS since she was a Postdoctoral Scholar in the Calof lab. Dr. Kawauchi's research interests are in the regulation of gene expression and pattern formation during mammalian development
ROSAYSELA SANTOS, Postdoctoral Scholar who has been a lead researcher in developing and analyzing mouse models of CdLS since she was a graduate student in the Calof lab. Dr. Santos's research interests lie in using animal models to understand the origins of birth defects associated with developmental and genetic disorders in man
AKIHIKO MUTO, Assistant Professor of Biological Sciences at Hiroshima University in Japan. He was formerly a Project Scientist at the University of California, Irvine, where he began his research on zebrafish models of CdLS in collaboration with Thomas Schilling, Arthur Lander, and Anne Calof. Dr. Muto's research interests are in understanding the molecular mechanisms by which chromatin structure affects gene expression, and the implications of this process for disease pathogenesis
MARTHA E. LOPEZ-BURKS, Staff Research Associate who specializes in molecular biology. During the past decade, her research has focused on molecular analysis of Nipbl-deficient mice. Currently, she leads the molecular and phenotypic analysis of limb and skeletal development in the CdLS mouse model
THOMAS F. SCHILLING, Professor and Chair of the Department of Developmental & Cell Biology at the University of California, Irvine. Dr. Schilling has a long-standing interest in craniofacial development, with work in his lab largely focused on developmental genetics in zebrafish as a model for human structural birth defects
ARTHUR D. LANDER, Donald Bren Professor of Developmental & Cell Biology, Director of the Center for Complex Biological Systems, and Co-Director the Center of Excellence in Research on Cornelia de Lange Syndrome at the University of California, Irvine. Dr. Lander's research uses experimental, mathematical, and computational approaches to elucidate the engineering principles that underlie human development, with a focus on fundamental problems in patterning and growth control, and the origins of complex traits
ANNE L. CALOF, Professor of Anatomy & Neurobiology at the University of California, Irvine. She serves as Co-Director of UCI's Center of Excellence in Research on Cornelia de Lange Syndrome and as a Theme Leader in the Center for Complex Biological Systems. Dr. Calof's research is focused on understanding how cell differentiation, tissue morphogenesis, and tissue size are regulated, both during normal development and when pathological conditions result in birth defects.
REFERENCES
- Chatfield KC, Schrier SA, Li J, Clark D, Kaur M, Kline AD, Deardorff MA, Jackson LS, Goldmuntz E, Krantz ID. Congenital heart disease in Cornelia de Lange syndrome: Phenotype and genotype analysis. Am J Med Genet A. 2012;158A:2499–2505. doi: 10.1002/ajmg.a.35582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deardorff MA NS, Krantz ID. Cornelia de Lange Syndrome. In: Pagon RA, Adam MP, Ardinger HH, editors. GeneReviews® [Internet] University of Washington, Seattle; Seattle (WA): Sep 16, 2005. [Updated 2016 Jan 28] 1993–2016. Available from: http://www.ncbi.nlm.nih.gov/books/NBK1104/ [PubMed] [Google Scholar]
- Dorsett D. Cohesin, gene expression and development: Lessons from Drosophila. Chromosome Res. 2009;17:185–200. doi: 10.1007/s10577-009-9022-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett D, Krantz ID. On the molecular etiology of Cornelia de Lange syndrome. Ann N Y Acad Sci. 2009;1151:22–37. doi: 10.1111/j.1749-6632.2008.03450.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda K, Kikuchi Y. Endoderm development in vertebrates: Fate mapping, induction and regional specification. Dev Growth Differ. 2005;47:343–355. doi: 10.1111/j.1440-169X.2005.00815.x. [DOI] [PubMed] [Google Scholar]
- Hayashi M, Sakamoto K, Kurata K, Nagata J, Satoh J, Morimatsu Y. Septo-optic dysplasia with cerebellar hypoplasia in Cornelia de Lange syndrome. Acta Neuropathol (Berl) 1996;92:625–630. doi: 10.1007/s004010050571. [DOI] [PubMed] [Google Scholar]
- Horsfield JA, Print CG, Monnich M. Diverse developmental disorders from the one ring: Distinct molecular pathways underlie the cohesinopathies. Front Genet. 2012;3:171. doi: 10.3389/fgene.2012.00171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson L, Kline AD, Barr MA, Koch S. De Lange syndrome: A clinical review of 310 individuals. Am J Med Genet. 1993;47:940–946. doi: 10.1002/ajmg.1320470703. [DOI] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, Taatjes DJ, Dekker J, Young RA. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawauchi S, Calof AL, Santos R, Lopez-Burks ME, Young CM, Hoang MP, Chua A, Lao T, Lechner MS, Daniel JA, Nussenzweig A, Kitzes L, Yokomori K, Hallgrimsson B, Lander AD. Multiple organ system defects and transcriptional dysregulation in the Nipbl(+/−) mouse, a model of Cornelia de Lange syndrome. PLoS Genet. 2009;5:e1000650. doi: 10.1371/journal.pgen.1000650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krantz ID, McCallum J, DeScipio C, Kaur M, Gillis LA, Yaeger D, Jukofsky L, Wasserman N, Bottani A, Morris CA, Nowaczyk MJ, Toriello H, Bamshad MJ, Carey JC, Rappa-port E, Kawauchi S, Lander AD, Calof AL, Li HH, Devoto M, Jackson LG. Cornelia de Lange syndrome is caused by mutations in NIPBL, the human homolog of Drosophila melanogaster Nipped-B. Nat Genet. 2004;36:631–635. doi: 10.1038/ng1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Zhang Z, Bando M, Itoh T, Deardorff MA, Clark D, Kaur M, Tandy S, Kondoh T, Rappaport E, Spinner NB, Vega H, Jackson LG, Shirahige K, Krantz ID. Transcriptional dysregulation in NIPBL and cohesin mutant human cells. PLoS Biol. 2009;7:e1000119. doi: 10.1371/journal.pbio.1000119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Burks ME, Santos R, Kawauchi S, Calof AL, Lander AD. Genetic enhancement of limb defects in a mouse model of Cornelia de Lange syndrome. Am J Med Genet Part C Semin Med Genet. 2016 doi: 10.1002/ajmg.c.31491. (in this issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luzzani S, Macchini F, Valad'e A, Milani D, Selicorni A. Gastroesophageal reflux and Cornelia de Lange syndrome: Typical and atypical symptoms. Am J Med Genet. 2003;119A:283–287. doi: 10.1002/ajmg.a.20191. [DOI] [PubMed] [Google Scholar]
- Muto A, Calof AL, Lander AD, Schilling TF. Multifactorial origins of heart and gut defects in nipbl-deficient zebrafish, a model of Cornelia de Lange syndrome. PLoS Biol. 2011;9:e1001181. doi: 10.1371/journal.pbio.1001181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto A, Ikeda S, Lopez-Burks ME, Kikuchi Y, Calof AL, Lander AD, Schilling TF. Nipbl and mediator cooperatively regulate gene expression to control limb development. PLoS Genet. 2014;10:e1004671. doi: 10.1371/journal.pgen.1004671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ober EA, Field HA, Stainier DY. From endoderm formation to liver and pancreas development in zebrafish. Mech Dev. 2003;120:5–18. doi: 10.1016/s0925-4773(02)00327-1. [DOI] [PubMed] [Google Scholar]
- Opitz JM. The Brachmann-de Lange syndrome. Am J Med Genet. 1985;22:89–102. doi: 10.1002/ajmg.1320220110. [DOI] [PubMed] [Google Scholar]
- Ozkinay F, Cogulu O, Gunduz C, Levent E, Ozkinay C. A case of Brachman de Lange syndrome with cerebellar vermis hypoplasia. Clin Dysmorphol. 1998;7:303–305. doi: 10.1097/00019605-199810000-00013. [DOI] [PubMed] [Google Scholar]
- Pfister S, Jones VJ, Power M, Truisi GL, Khoo PL, Steiner KA, Kanai-Azuma M, Kanai Y, Tam PP, Loebel DA. Sox17-dependent gene expression and early heart and gut development in Sox17-deficient mouse embryos. Int J Dev Biol. 2011;55:45–58. doi: 10.1387/ijdb.103158sp. [DOI] [PubMed] [Google Scholar]
- Rau MJ, Fischer S, Neumann CJ. Zebrafish Trap230/Med12 is required as a coactivator for Sox9-dependent neural crest, cartilage and ear development. Dev Biol. 2006;296:83–93. doi: 10.1016/j.ydbio.2006.04.437. [DOI] [PubMed] [Google Scholar]
- Rollins RA, Morcillo P, Dorsett D. Nipped-B, a Drosophila homologue of chromosomal adherins, participates in activation by remote enhancers in the cut and Ultrabithorax genes. Genetics. 1999;152:577–593. doi: 10.1093/genetics/152.2.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai Y, Watanabe T, Kaga K. Auditory brainstem responses and usefulness of hearing aids in hearing impaired children with Cornelia de Lange syndrome. Int J Pediatr Otorhinolaryngol. 2002;66:63–69. doi: 10.1016/s0165-5876(02)00214-8. [DOI] [PubMed] [Google Scholar]
- Schrier SA, Sherer I, Deardorff MA, Clark D, Audette L, Gillis L, Kline AD, Ernst L, Loomes K, Krantz ID, Jackson LG. Causes of death and autopsy findings in a large study cohort of individuals with Cornelia de Lange syndrome and review of the literature. Am J Med Genet A. 2011;155A:3007–3024. doi: 10.1002/ajmg.a.34329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selicorni A, Colli AM, Passarini A, Milani D, Cereda A, Cerutti M, Maitz S, Alloni V, Salvini L, Galli MA, Ghiglia S, Salice P, Danzi GB. Analysis of congenital heart defects in 87 consecutive patients with Brachmann-de Lange syndrome. Am J Med Genet A. 2009;149A:1268–1272. doi: 10.1002/ajmg.a.32838. [DOI] [PubMed] [Google Scholar]
- Singh VP, Gerton JL. Cohesin and human disease: Lessons from mouse models. Curr Opin Cell Biol. 2015;37:9–17. doi: 10.1016/j.ceb.2015.08.003. [DOI] [PubMed] [Google Scholar]
- Tonkin ET, Wang TJ, Lisgo S, Bamshad MJ, Strachan T. NIPBL, encoding a homolog of fungal Scc2-type sister chromatid cohesion proteins and fly Nipped-B, is mutated in Cornelia de Lange syndrome. Nat Genet. 2004;36:636–641. doi: 10.1038/ng1363. [DOI] [PubMed] [Google Scholar]
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin JW, Wang G. The Mediator complex: A master coordinator of transcription and cell lineage development. Development. 2014;141:977–987. doi: 10.1242/dev.098392. [DOI] [PubMed] [Google Scholar]
- Yuan B, Pehlivan D, Karaca E, Patel N, Charng WL, Gambin T, Gonzaga-Jauregui C, Sutton VR, Yesil G, Bozdogan ST, Tos T, Koparir A, Koparir E, Beck CR, Gu S, Aslan H, Yuregir OO, Al Rubeaan K, Alnaqeb D, Alshammari MJ, Bayram Y, Atik MM, Aydin H, Geckinli BB, Seven M, Ulucan H, Fenercioglu E, Ozen M, Jhangiani S, Muzny DM, Boerwinkle E, Tuysuz B, Alkuraya FS, Gibbs RA, Lupski JR. Global tran scriptional disturbances underlie Cornelia de Lange syndrome and related phenotypes. J Clin Invest. 2015;125:636–651. doi: 10.1172/JCI77435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakany J, Duboule D. The role of Hoxgenes during vertebrate limb development. Curr Opin Genet Dev. 2007;17:359–366. doi: 10.1016/j.gde.2007.05.011. [DOI] [PubMed] [Google Scholar]