Abstract
In recent years, several new periodontal taxa have been associated with the etiology of periodontitis. A recent systematic review provides further support for the pathogenic role of 17 species/phylotypes. Thus, the aim of this study was to assess the prevalence and levels of these species in subjects with generalized chronic periodontitis (GChP; n = 30), generalized aggressive periodontitis (GAgP; n = 30), and periodontal health (PH; n = 30). All subjects underwent clinical and microbiological assessment. Nine subgingival plaque samples were collected from each subject and analyzed for their content of 20 bacterial species/phylotypes through the RNA-oligonucleotide quantification technique. Subjects from the GChP and GAgP groups presented the highest mean values for all clinical parameters in comparison with the PH group (P < 0.05). Subjects with GChP and GAgP showed significantly higher mean levels of Bacteroidetes sp. human oral taxon (HOT) 274, Fretibacterium sp. HOT 360, and TM7 sp. HOT 356 phylotypes, as well as higher mean levels of Filifactor alocis, Fretibacterium fastidiosum, Porphyromonas gingivalis, Tannerella forsythia, and Selenomonas sputigena species than PH subjects (P < 0.05). GAgP subjects presented higher mean levels of TM7 sp. HOT 356 and F. alocis than GChP subjects (P < 0.05). A significantly higher mean prevalence of Bacteroidales sp. HOT 274, Desulfobulbus sp. HOT 041, Fretibacterium sp. HOT 360, and Fretibacterium sp. HOT 362 was found in subjects with GChP and GAgP than in PH subjects. Mean levels of P. gingivalis (r = 0.68), T. forsythia (r = 0.62), F. alocis (r = 0.51, P = 0.001), and Fretibacterium sp. HOT 360 (r = 0.41) were correlated with pocket depth (P < 0.001). In conclusion, Bacteroidales sp. HOT 274, Desulfobulbus sp. HOT 041, Fretibacterium sp. HOT 360, Fretibacterium sp. HOT 362, and TM7 sp. HOT 356 phylotypes, in addition to F. alocis, F. fastidiosum, and S. sputigena, seem to be associated with periodontitis, and their role in periodontal pathogenesis should be further investigated.
Keywords: dental plaque, biofilms, RNA probes, chronic periodontitis, aggressive periodontitis, microbiota
Introduction
There is well-established evidence of the role of oral microbiota in the etiology of periodontal diseases and some specificity among certain bacterial species or groups of species and the various forms of periodontal disease (Socransky and Haffajee 2005). However, although the complexity and diversity of the periodontal microbiota have been confirmed by numerous studies (Paster et al. 2001; Socransky and Haffajee 2005; Ledder et al. 2007; Gonçalves et al. 2012; You et al. 2013; Vengerfeldt et al. 2014; Park et al. 2015), up to now, only 3 bacterial species have been recognized as true periodontal pathogens—namely, Aggregatibacter actinomycetemcomitans, Porphyromonas gingivalis, and Tannerella forsythia (American Academy of Peri-odontology 1996).
Yet, our understanding of the subgingival microbiota has expanded considerably since then. The composition, complexity, and diversity of the oral microbiome have been studied at much greater depth and breadth in recent years. These advances in knowledge were mainly a consequence of the technological progress in molecular methods, which permitted an open-ended analysis of the local microbiome and/or allowed the study of the uncultivated segment of the microbiota (Paster et al. 2001; Kumar et al. 2005; Matarazzo et al. 2011; Teles et al. 2011; Griffen et al. 2012; Abusleme et al. 2013; Park et al. 2015).
Collectively, the data provided by several studies conducted in the past 15 y suggest the existence of new potential periodontal pathogens. For instance, using pyrosequencing, Abusleme et al. (2013) reported that the shifts in community structure from health to periodontitis resembled ecologic succession, with emergence of newly dominant taxa in periodontitis without replacement of primary health-associated species. Their results showed that subgingival biofilm of periodontitis subjects had higher proportions of Spirochetes, Synergistetes, Firmicutes, and Chloroflexi, while the proportions of Actinobacteria, particularly Actinomyces, were higher in health. Griffen et al. (2012) demonstrated that Filifactor alocis, Fretibacterium sp. human oral taxon (HOT) 360, Desulfobulbus sp. HOT 041, and Selenomonas sputigena were among the most abundant microorganisms in deep periodontitis sites (throughout this manuscript, HOT designations for uncultivated/unrecognized taxa are provided in accord with the Human Oral Microbiome Database [www.homd.org]; Chen et al. 2010). Together, these results indicate that previously understudied/unknown taxa might participate in the initiation and progression of periodontitis.
In a recent systematic review, Pérez-Chaparro et al. (2014) compared the results of several association studies that used different culture-independent microbiological techniques to identify new candidate periodontal pathogens. The authors suggested a positive association of at least 17 novel species or phylotypes with periodontitis, including F. alocis, S. sputigena, Desulfobulbus sp. HOT 041, Fretibacterium sp. HOT 360, Bacteroidales sp. HOT 274, and TM7 [G-5] sp. HOT 356. Hence, the role of these species in the onset and progression of periodontal disease requires further investigation. The purpose of the present study was therefore to evaluate the levels of bacterial species that have been proposed as candidate new periodontal pathogens by Pérez-Chaparro et al. (2014) in subgingival biofilm samples from subjects with generalized chronic periodontitis (GChP), generalized aggressive periodontitis (GAgP), and periodontal health (PH).
Materials and Methods
Subject Population
Subjects were recruited between July 2012 and March 2014. Thirty PH subjects and 60 with GAgP (n = 30) and GChP (n = 30) were included in this investigation. They were all systemically healthy and were selected from the population referred to the Periodontal Clinic of Guarulhos University (Guarulhos, Brazil). Medical and dental histories were obtained, and a full-mouth periodontal examination was performed. The periodontal diagnosis was made, and subjects who fulfilled the inclusion/exclusion criteria were invited to participate in the study. The study protocol was explained to each subject, and a signed “Term of Free and Informed Consent” was obtained. The Guarulhos University Ethics Committee on Clinical Research approved the study protocol. This study was conducted in accordance with the STROBE (STrengthening the Reporting of OBservational studies in Epidemiology) guidelines for observational studies.
Clinical Examination
One trained and calibrated examiner performed the clinical examination of all subjects. Visible plaque (0/1), gingival bleeding (0/1), bleeding on probing (BOP; 0/1), suppuration (0/1), probing depth (PD; mm), and clinical attachment level (CAL; mm) were measured at 6 sites per tooth (mesiobuccal, buccal, distobuccal, distolingual, lingual, and mesiolingual) in all teeth, excluding third molars. PD and CAL measurements were recorded to the nearest millimeter with a North Carolina periodontal probe (Hu-Friedy, Chicago, IL, USA).
Inclusion Criteria
GAgP, GChP, and PH subjects were diagnosed according to the periodontal classification of the American Academy of Periodontology (Armitage 1999). Subjects were required to have at least 20 teeth and to meet the following criteria to be included in this study:
GAgP: <35 y of age, minimum of 6 permanent incisors and/or first molars with at least 1 site each with PD and CAL ≥5 mm, minimum of 6 teeth other than first molars and incisors with at least 1 site each with PD and CAL ≥5 mm, familial aggregation (at least 1 other member of the family presenting or with a history of periodontal disease; Faveri et al. 2009).
GChP: ≥35 y of age and a minimum of 6 teeth with at least 1 site each with PD and CAL ≥5 mm and at least 30% of the sites with PD and CAL ≥4 mm and BOP.
PH: ≥18 y of age, no sites with PD and CAL measurements >3 mm, and <10% of sites exhibiting BOP.
Exclusion Criteria
Exclusion criteria were pregnancy, lactation, smoking, subgingival periodontal therapy in the past 12 mo, any systemic condition that could affect the progression of periodontal disease (e.g., diabetes and immunologic disorders), long-term administration of anti-inflammatory medication, and antibiotic therapy in the previous 6 mo.
Microbiological Examination
Sample collection and extraction of total nucleic acids
After having recorded the clinical parameters, supragingival plaque was removed, and then 9 individual subgingival plaque samples were collected per subject. For the GAgP and GChP groups, samples were collected from 3 sites in each of following PD categories: shallow (PD ≤3 mm), moderate (PD 4 to 6 mm), and deep (PD ≥7 mm). Nine sites with PD ≤3 mm were collected from the PH group. The selected sites were randomized in different quadrants. The samples were taken with individual sterile Gracey curettes and immediately placed in separate microcentrifuge tubes containing 150 µL of RNAse-free TE (10mM Tris-HCl, 1mM EDTA, pH 7.6) and kept at −80 oC until extraction of total nucleic acids (TNAs). For TNA extraction, samples were mixed with lysozyme (35 KU/µL; Epicentre, Madison, WI, USA), incubated at 37 °C overnight, followed by incubation at 95 °C for 5 min. TNA was extracted with a Masterpure RNA & DNA Purification Kit (Epicentre, Madison, WI, USA), and after extraction, TNA samples were kept at −80 °C until analysis.
RNA-oligonucleotide quantification technique
The counts of the 20 bacterial species/phylotypes (Table 1) were determined in each sample, with the RNA-oligonucleotide quantification technique (Teles et al. 2011) at the Laboratory of Microbiology of Guarulhos University. The probe panel also included a universal (eubacterial) probe, based on a conserved region of the bacterial 16S rRNA gene. The universal probe is only a positive control in the test, and the levels were not considered. In brief, TNAs were extracted from the biofilm samples and fixed onto a nylon membrane. Oligonucleotides targeting the 16S ribosomal DNA gene were synthesized (Table 1) and labeled with digoxigenin. These probes were hybridized with the TNA samples. A universal probe was used as a positive control, and sequences complementary to the probes were used as standards for quantification at 0.004 and 0.04 picomolars (pM). Hybrids generated chemiluminescent signals, which were visualized with a CCD camera. Signals were converted to approximate counts by comparison with the standards on each membrane. The counts were computed by estimating that 0.04 pM of target sequences in the standard was equivalent to approximately 106 cells and that 0.004-pM target sequences approximated 105 cells. Absence of signal detection was recorded as zero.
Table 1.
Sequences for Oligonucleotide Probes and for the Standards for Quantification of the Species/Phylotypes Evaluated.
| Species/Phylotypes | Sequence (5′–3′) | Control (5′–3′) |
|---|---|---|
| Anaeroglobus geminatus HOT 121 | CGCUAAGAGGACCGUAUU | AAUACGGUCCUCUUAGCG |
| Bacteroidales sp. HOT 274 | ACGUGUCUCACUUUACUCC | GGAGUAAAGUGAGACACGU |
| Desulfobulbus sp. HOT 041 | UGUUAUUCGCUGCCUUGCA | UGCAAGGCAGCGAAUAACA |
| Enterococcus faecalis HOT 604 | UCCUCUUUCCAAUUGAGUGC | GCACUCAAUUGGAAAGAGGA |
| Eubacterium saphenum HOT 759 | CACUCAAGUCUGCCAGUU | AACUGGCAGACUUGAGUG |
| Filifactor alocis HOT 539 | GGCUCAUCUUUGUCCACU | AGUGGACAAAGAUGAGCC |
| Fretibacterium fastidiosum HOT 363 | GUGUUACCACUUCACGA | GUCGUGAAGUGGUAACAC |
| Fretibacterium sp. HOT 360 | GCAGCGUCGUCAAUGUUU | AAACAUUGACGACGCUGC |
| Fretibacterium sp. HOT 362 | ACACGAGUGCCUCCUGU | ACAGGAGGCACUCGUGU |
| Mogibacterium timidum HOT 042 | GCGUCAUUUCCAAGCUUC | GAAGCUUGGAAAUGACGC |
| Peptostreptococcus stomatis HOT 112 | GCGUCAUUUCCAAGCUUC | UCUGAGCAAAACCAAGCG |
| Porphyromonas endodontalis HOT 273 | UCUUUCCGUCUUUCCCCA | AUGGGGAAAGACGGAAAGA |
| Porphyromonas gingivalis HOT 619 | CACCAUCAGUCAUCUAC | GUAGAUGACUGAUGGUG |
| Prevotella denticola HOT 291 | ACACGAGUGCCUCCUGU | ACAGGAGGCACUCGUGU |
| Selenomonas sputigena HOT 151 | CCCUUAUGAAGCACUGAG | CUCAGUGCUUCAUAAGGG |
| Tannerella forsythia HOT 613 | UUGCGGGCAGGUUACAUA | UAUGUAACCUGCCCGCAA |
| TM7 sp. HOT 356 | CGAACAACAAGCUAUCGG | CCGAUAGCUUGUUGUUCG |
| Treponema lecithinolyticum HOT 653 | ACACCAAGCUUGCUCAUC | GAUGAGCAAGCUUGGUGU |
| Treponema medium HOT 667 | CCCUUAUGAAGCACUGAG | CUCAGUGCUUCAUAAGGG |
| Treponema vincentii HOT 029 | CCCUUAUGAAGCACUGAG | CUCAGUGCUUCAUAAGGG |
HOT, human oral taxon.
Oral taxon designations for uncultivated/unrecognized taxa are provided in accordance with the Human Oral Microbiome Database (www.homd.org), when available; GenBank accession numbers can also be found in the website.
Statistical Analysis
The mean age; the mean percentage of sites with visible plaque, gingival bleeding, BOP, and suppuration; and mean PD and CAL were computed for each subject and averaged across subjects in each group. Mean estimated levels (pM) of individual bacterial species were computed for each site, averaged within each subject and across subjects in each group. Prevalence of the test taxa was computed by determining the percentage of sites per subject colonized by ≥0.004 pM of each species (the lower level of the standard for quantification used). The mean percentage of “positive” sites was calculated for each subject and then averaged across subjects in each group. The significance of differences among groups for age and clinical and microbiological parameters was sought through the Kruskal-Wallis and Dunn tests with application of Bonferroni’s correction for multiple comparisons. The Friedman and Dunn tests were used to detect statistically significant differences within PD categories in the GAgP and GChP groups. The chi-square test was used to compare the differences in the frequency of sex. The Spearman correlation was used to assess possible associations between PD and mean levels of bacterial species.
Results
Clinical Findings
The demographic characteristics and clinical parameters of the studied population are presented in Table 2. No statistically significant differences were observed among groups for sex. The GAgP and GChP groups showed significantly higher mean PD and CAL and percentage of sites with BOP, gingival bleeding, and supuration (P < 0.05) in comparison with the PH group. The GAgP group presented a lower mean age than the PH and GChP groups (P < 0.05).
Table 2.
Demographic Characteristics and Mean (±SD) Full-mouth Clinical Parameters of the Subjects in Experimental Groups.
| Experimental Groups |
||||
|---|---|---|---|---|
| Variables | PH | GAgP | GChP | P Value |
| Subjects, n | 30 | 30 | 30 | |
| Age, y | 33.5 ± 11.0A | 26.3 ± 3.5B | 42.0 ± 5.7C | <0.001 |
| Sex, male:female | 12:18 | 14:16 | 13:17 | NS |
| Probing depth, mm | 1.8 ± 0.4A | 4.1 ± 0.9B | 4.0 ± 1.1B | <0.001 |
| Clinical attachment level, mm | 0.7 ± 0.3A | 3.8 ± 1.1B | 3.7 ± 1.0B | <0.001 |
| Sites with, % | ||||
| Plaque | 20.9 ± 7.7A | 78.1 ± 16.7B | 86.4 ± 18.9B | <0.001 |
| Gingival bleeding | 2.1 ± 1.1A | 35.6 ± 12.5B | 38.5 ± 20.5B | <0.001 |
| Bleeding on probing | 3.2 ± 1.8A | 45.5 ± 15.9B | 42.1 ± 29.9B | <0.001 |
| Suppuration | 0 ± 0A | 4.32 ± 3.49B | 3.09 ± 3.7B | <0.001 |
GAgP, generalized aggressive periodontitis; GChP, generalized chronic periodontitis; NS, not significant; PH, periodontal health.
The significance of differences among groups was assessed using the Kruskall-Wallis test. The significance of differences between pairs of comparisons was determined using Tukey’s multiple comparison test, and the significances are represented by different capital letters.
Microbiological Findings
The mean estimated levels of the 20 species evaluated in the subgingival biofilm samples from the PH, GAgP, and GChP groups are shown in Table 3. Subjects with GAgP and GChP showed significantly higher mean levels of Bacteroidales sp. HOT 274, F. alocis, Fretibacterium fastidiosum, Fretibacterium sp. HOT 360, P. gingivalis, S. sputigena, T. forsythia, and TM7 sp. HOT 356 (P < 0.002) than subjects in the PH group. However, there was no statistical difference between the GAgP and GChP groups for Bacteroidales sp. HOT 274, F. fastidiosum, P. gingivalis, and T. forsythia. Higher levels of F. alocis, S. sputigena, and TM7 sp. HOT 356 were found in the GAgP group than the GChP group (P < 0.05), while GChP subjects showed higher mean levels of Fretibacterium sp. HOT 360 than GAgP subjects.
Table 3.
Mean (±SD) Levels (10-2 pM) of 20 Bacterial Taxa in Subgingival Biofilm Samples Obtained from Subjects.
| Experimental Groups |
||||
|---|---|---|---|---|
| Species/Phylotypes | PH | GAgP | GChP | P Value |
| Anaeroglobus geminatus HOT 121 | 1.5 ± 2.0 | 2.5 ± 1.5 | 2.7 ± 2.5 | NS |
| Bacteroidales sp. HOT 274 | 0.2 ± 4.0A | 2.0 ± 4.1B | 3.1 ± 2.2B | 0.002 |
| Desulfobulbus sp. HOT 041 | 1.0 ± 1.0 | 0.5 ± 1.3 | 0.9 ± 2.1 | NS |
| Enterococcus faecalis HOT 604 | 3.1 ± 2.1 | 4.5 ± 3.7 | 3.1 ± 1.1 | NS |
| Eubacterium saphenum HOT 759 | 1.7 ± 2.9 | 2.4 ± 1.4 | 1.5 ± 1.3 | NS |
| Filifactor alocis HOT 539 | 0.3 ± 0.1A | 3.5 ± 3.3B | 2.2 ± 2.5C | <0.001 |
| Fretibacterium fastidiosum HOT 363 | 0.2 ± 0.8A | 2.1 ± 1.5B | 1.8 ± 1.9B | <0.001 |
| Fretibacterium sp. HOT 360 | 0.3 ± 0.7A | 1.1 ± 2.8B | 2.5 ± 1.5C | 0.001 |
| Fretibacterium sp. HOT 362 | 0.7 ± 1.1 | 1.3 ± 2.1 | 1.1 ± 0.9 | NS |
| Mogibacterium timidum HOT 042 | 0.2 ± 0.1 | 0.6 ± 0.5 | 0.9 ± 0.8 | NS |
| Peptostreptococcus stomatis HOT 112 | 2.1 ± 1.5 | 1.6 ± 1.1 | 2.3 ± 1.7 | NS |
| Porphyromonas endodontalis HOT 273 | 0.4 ± 0.3 | 2.1 ± 1.9 | 3.1 ± 0.9 | NS |
| Porphyromonas gingivalis HOT 619 | 0.2 ± 0.5A | 3.1 ± 2.8B | 4.5 ± 1.8B | <0.001 |
| Prevotella denticola HOT 291 | 3.1 ± 1.6 | 3.6 ± 2.1 | 3.9 ± 1.9 | NS |
| Selenomonas sputigena HOT 151 | 1.1 ± 0.8A | 3.2 ± 1.9B | 2.1 ± 1.1C | <0.001 |
| Tannerella forsythia HOT 613 | 0.8 ± 0.5A | 4.4 ± 3.1B | 4.9 ± 3.1B | <0.001 |
| TM7 sp. HOT 356 | 0 ± 0A | 1.0 ± 0.8B | 0.4 ± 0.5C | <0.001 |
| Treponema lecithinolyticum HOT 653 | 0.2 ± 0.3 | 0.8 ± 1.4 | 1.1 ± 0.9 | NS |
| Treponema medium HOT 667 | 0.1 ± 0.3 | 0.2 ± 0.5 | 0.4 ± 0.4 | NS |
| Treponema vincentii HOT 029 | 0.3 ± 0.2 | 0.8 ± 0.3 | 1.0 ± 0.7 | NS |
GAgP, generalized aggressive periodontitis; GChP, generalized chronic periodontitis; NS, not significant; PH, periodontal health.
The significance of differences among groups was assessed using the Kruskall-Wallis test. The significance of differences between pairs of comparisons was determined using Tukey’s multiple comparison test, and the significances are represented by different capital letters.
The percentage of sites colonized by the 20 species/phylotypes evaluated in subgingival biofilm samples taken from GAgP, GChP, and PH subjects are presented in Table 4. Subjects with GAgP and GChP showed higher mean percentage of sites colonized by Bacteroidales sp. HOT 274, Desulfobulbus sp. HOT 041, F. alocis, Fretibacterium sp. HOT 360, Fretibacterium sp. HOT 362, P. gingivalis, S. sputigena, T. forsythia, and TM7 sp. HOT 356 (P < 0.05) than PH subjects. F. alocis and Fretibacterium sp. HOT 360 were more prevalent in GAgP than in GChP (P < 0.0001). No differences in the prevalence of subjects colonized by all the species analyzed were observed among groups, except P. gingivalis and T. forsythia, which were more prevalent in GAgP and GChP subjects, and F. alocis, which was more prevalent in GAgP subjects (data not shown, P < 0.05).
Table 4.
Mean (±SD) Percentage of Sites Colonized by 20 Bacterial Taxa in Subgingival Biofilm Samples Obtained from Subjects.
| Experimental Groups |
||||
|---|---|---|---|---|
| Species/Phylotypes | PH | GAgP | GChP | P Value |
| Anaeroglobus geminatus HOT 121 | 5 ± 23 | 17 ± 11 | 16 ± 25 | NS |
| Bacteroidales sp. HOT 274 | 15 ± 33A | 54 ± 25B | 65 ± 32B | <0.0001 |
| Desulfobulbus sp. HOT 041 | 5 ± 15A | 43 ± 44B | 35 ± 33B | <0.0001 |
| Enterococcus faecalis HOT 604 | 25 ± 35 | 33 ± 23 | 41 ± 17 | NS |
| Eubacterium saphenum HOT 759 | 18 ± 39 | 32 ± 47 | 42 ± 45 | NS |
| Filifactor alocis HOT 539 | 12 ± 23A | 49 ± 23B | 32 ± 25C | <0.0001 |
| Fretibacterium fastidiosum HOT 363 | 12 ± 25 | 23 ± 37 | 30 ± 21 | NS |
| Fretibacterium sp. HOT 360 | 04 ± 16A | 25 ± 33B | 17 ± 45C | <0.0001 |
| Fretibacterium sp. HOT 362 | 09 ± 19A | 29 ± 16B | 22 ± 41B | <0.0001 |
| Mogibacterium timidum HOT 042 | 2 ± 19 | 15 ± 41 | 12 ± 22 | NS |
| Peptostreptococcus stomatis HOT 112 | 4 ± 11 | 12 ± 19 | 19 ± 41 | NS |
| Porphyromonas endodontalis HOT 273 | 18 ± 23 | 32 ± 18 | 29 ± 22 | NS |
| Porphyromonas gingivalis HOT 619 | 15 ± 21A | 75 ± 25B | 80 ± 15B | <0.0001 |
| Prevotella denticola HOT 291 | 23 ± 15 | 31 ± 24 | 24 ± 21 | NS |
| Selenomonas sputigena HOT 151 | 15 ± 18A | 68 ± 34B | 51 ± 32B | <0.0001 |
| Tannerella forsythia HOT 613 | 8 ± 18A | 65 ± 36B | 71 ± 20B | <0.0001 |
| TM7 sp. HOT 356 | 0 ± 0A | 16 ± 22B | 23 ± 39B | <0.0001 |
| Treponema lecithinolyticum HOT 653 | 6 ± 23 | 12 ± 33 | 15 ± 29 | NS |
| Treponema medium HOT 667 | 2 ± 8 | 5 ± 11 | 4 ± 4 | NS |
| Treponema vincentii HOT 029 | 8 ± 11 | 15 ± 25 | 21 ± 32 | NS |
GAgP, generalized aggressive periodontitis; GChP, generalized chronic periodontitis; NS, not significant; PH, periodontal health.
The significance of differences among groups was assessed using the Kruskall-Wallis test. The significance of differences between pairs of comparisons was determined using Tukey’s multiple comparison test, and the significances are represented by different capital letters.
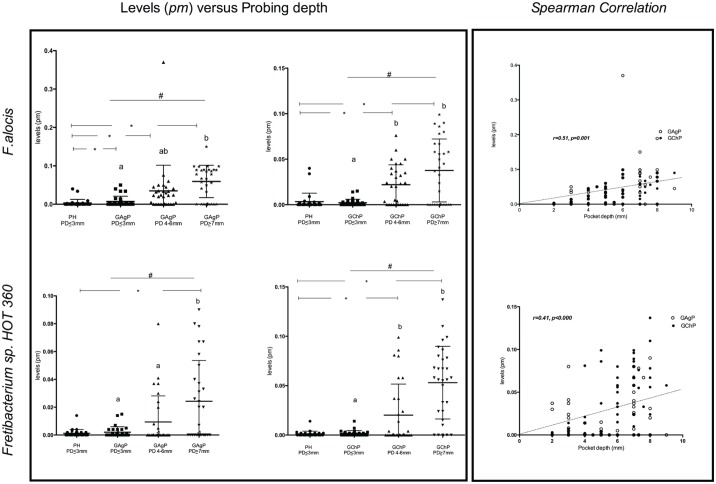
The mean estimated levels of F. alocis and Fretibacterium sp. HOT 360 detected in shallow, intermediate, and deep periodontal pockets (≤3, 4 to 6, and ≥7 mm, respectively) of GAgP and GChP subjects and in the shallow pocket (≤3 mm) of the PH subjects are shown in the Figure. In subjects with GAgP and GChP, F. alocis and Fretibacterium sp. HOT 360 were detected in significantly higher mean counts in deep than in shallow pockets and in comparison with the shallow sites of PH subjects. Furthermore, shallow sites of GAgP subjects harbored higher mean levels of F. alocis when compared with the shallow sites of PH subjects. In addition, Spearman correlations revealed positive correlations between PD and mean levels of P. gingivalis (r = 0.68, P = 0.0001), T. forsythia (r = 0.62, P < 0.001), F. alocis (r = 0.51, P = 0.001), and Fretibacterium sp. HOT 360 (r = 0.41, P < 0.001; Fig.).
Figure.
Mean estimated levels (pM) of Filifactor alocis and Fretibacterium sp. Human oral taxon (HOT) 360 in the PH subjects and in sites with PD ≤3 mm, 4 to 6 mm, and ≥7 mm in the GAgP and GChP subjects. Scatterplots show the median, minimum, and maximum values. The significance of differences between shallow sites of PH subjects and sites from different PD categories of GAgP and GChP subjects was assessed through the Mann-Whitney U test (*P < 0.05). The significance of differences within different PD categories of GAgP and GChP subjects was assessed through Friedman (#P < 0.05) and Dunn tests, and the significances are represented by different capital letters. GAgP, generalized aggressive periodontitis; GChP, generalized chronic periodontitis; PD, probing depth; PH, periodontal health.
Discussion
The subgingival pocket is a complex environment that harbors a highly diverse microbiota. Studies employing targeted or open-ended molecular techniques have identified new potential periodontal pathogens (Paster et al. 2001; Kumar et al. 2005; Ledder et al. 2007; Faveri et al. 2008; Griffen et al. 2012; Abusleme et al. 2013). Findings from those studies were summarized in a systematic review by Pérez-Chaparro et al. (2014), where 17 bacterial species/phylotypes presented a moderate weight of a role in the etiology of periodontitis, based on several association studies. Thus, the purpose of the present study was to evaluate the levels of a subset of those new candidate pathogens in subgingival biofilm samples from patients with GChP, GAgP, and PH.
The results of the present study support the association of 5 uncultivated/unrecognized phylotypes with periodontal disease, based on the differences in levels and prevalence across the groups examined. Bacteroidales sp. HOT 274 (P < 0.002), TM7 sp. HOT 356 (P < 0.0001), Desulfobulbus sp. HOT 041, Fretibacterium sp. HOT 360, and Fretibacterium sp. HOT 362 were found in higher levels of GChP and GAgP subjects than PH (Table 3). Regarding their prevalence (percentage of sites colonized), all those phylotypes were more prevalent in the GChP and GAgP group than in the PH subjects (P < 0.0001; Table 4). Based on the same parameters, our findings also support the association of 3 cultivated species with periodontitis—namely, S. sputigena, F. alocis, and F. fastidiosum.
These associations are further supported by the comparison of the data pertaining to the 8 taxa mentioned above with the results obtained for P. gingivalis and T. forsythia: 2 members of the red complex (Socransky et al. 1998) and well-recognized periodontal pathogens. We included those species in our probe panel to provide a frame of reference for making inferences in the results obtained from the new candidate periodontal pathogens. We observed that the taxa mentioned above behaved similarly to P. gingivalis and T. forsythia in that their levels and prevalence were substantially higher in GChP and GAgP than PH. Still, the abundance and prevalence of P. gingivalis and T. forsythia in GChP and GAgP were considerably higher that in the 5 candidate phylotypes. It should also be highlighted that our results for P. gingivalis and T. forsythia were in line with those of previous investigations using different microbiological techniques, such as cultivation (Moore et al. 1985; Dzink et al. 1998), polymerase chain reaction (Darby et al. 2000; Cortelli et al. 2005), checkerboard DNA-DNA hybridization (Faveri et al. 2009; Teles et al. 2010), and pyrosequencing (Griffen et al. 2012; Abusleme et al. 2013).
It is noteworthy that the 8 new candidate taxa proposed herein have also been proposed as being relevant to periodontitis in 2 recent studies using pyrosequencing, a sensitive open-ended next-generation sequencing platform. Abusleme et al. (2013) demonstrated that all of them were among the taxa classified as the “core subgingival microbiome” in periodontitis, along with P. gingivalis and T. forsythia. Griffen et al. (2012) showed that F. alocis was the most abundant species in deep periodontitis sites, followed by P. gingivalis. Furthermore, the relative abundance of F. alocis, Fretibacterium sp. HOT 360, Desulfobulbus sp. HOT 041, S. sputigena, as well as T. forsythia and P. gingivalis was significantly higher in deep sites from periodontitis patients when compared with shallow sites from the same patients and sites from PH individuals. In fact, findings from that study revealed a “dose response” effect in the relative abundance of those taxa across the 3 categories of sites: the highest abundance was observed in deep sites, followed by shallow sites in periodontitis patients, then sites from healthy individuals.
Our results indicate that levels of Fretibacterium sp. HOT 360 were positively correlated with increased PD in both GChP and GAgP groups (Fig.). This finding could be interpreted as evidence that they are the consequence of the changes of environment brought by disease development, rather than its cause. However, Griffen et al. (2012) demonstrated that shallow sites from diseased patients present a pathogen-enriched microbial profile in comparison with clinically similar sites. Hence, the fact that they were already colonized by pathogens (P. gingivalis and T. forsythia) and new candidate pathogens (Bacteroidales sp. HOT 274, Desulfobulbus sp. HOT 041, Fretibacterium sp. HOT 360, F. alocis, and S. sputigena) before the clinical presentation of periodontitis is a good indicator that they are not a consequence of disease but likely key players in its initiation.
Among the candidate new pathogens, TM7 is a recently described subgroup of gram-positive uncultivated bacteria originally found in natural environmental habitats. An association of TM7 bacterial division with the inflammatory pathogenesis of periodontitis has been previously shown (Paster et al. 2001; Kumar et al. 2005; Griffen et al. 2012). In addition, Al-Hebshi et al. (2015) found a strong association between TM7 and Parvimonas micra, suggesting that this phylotype may be a member of the orange complex of the subgingival biofilm. In the present study, TM7 sp. HOT 356 was not detected in the PH group but was found in higher levels in GAgP than GChP subjects (Table 3), which is in accord with the previously reported association between TM7 and chronic periodontitis (Camelo-Castillo et al. 2015). In addition, those authors reported a significant positive correlation between this phylum and Eubacterium, Porphyromonas, and Tannerella genera, as well as a positive correlation with PD, CAL, and BOP.
Other postulates to be fulfilled to classify a species as a pathogen is its virulence properties and ability to elicit host immune response. In that regard, periodontitis patients have shown high antibody titers against S. sputigena (Socransky et al. 1982): a gram-negative, multiflagellate, motile, anaerobic rod (Kolenbrander et al. 1989). Additionally, F. alocis—a non-spore-forming gram-positive obligate slow-growing anaerobic rod (Cato et al. 1985; Jalava and Eerola 1999; Schlafer et al. 2010)—has been shown to induce secretion of proinflammatory cytokines, triggering apoptosis of gingival epithelial cells (Moffatt et al. 2011). Other interactions with the host have stimulated in F. alocis the upregulation of several proteins (e.g., proteases, proteins involved in secretion systems, and proteins with cell wall anchor motifs) that are considered to have virulence properties in other bacteria (Aruni et al. 2011). Moreover, in coculture with P. gingivalis, F. alocis showed an increased invasive capacity of HeLa cells (Aruni et al. 2011). In addition, higher levels of F. alocis and Eubacterium nodatum were found in sites that showed disease progression (≥2 mm of clinical attachment loss) than in sites that remained nonprogressing for a period of up to 1 y (Yost et al. 2015). Therefore, these observations suggest that F. alocis and S. sputigena may have an important role in the pathogenic process of periodontal disease.
Unfortunately, there is no information available regarding the virulence properties of the other 5 new candidate periodontal pathogens. That is due to the fact that these are as-of-yet uncultured/unrecognized organisms, from which not much is known beyond its 16S rRNA gene sequence, which represents a small fraction of their genome. Therefore, not much can be inferred from their functional potential, metabolic properties, or direct effects on the host. However, studies such as the present investigation provide valuable insights into the selection of which uncultured taxa merit further pursuit and cultivation efforts.
The data from the present study support the continuing investigation of the potential role of those new candidate periodontal pathogens in the etiology of periodontal disease. In addition, the absence of a possible association with disease for some phylotypes present in our study could be due to the study design as well as the size of the experimental groups. Therefore, the present data encourage additional studies of the interactions between those species/phylotypes and the host and how the host responds to this microbiota. Collectively, those studies will contribute to our understanding of the role of these microorganisms in the etiology of periodontitis and help in the development of better treatment strategies.
Conclusion
In conclusion, uncultivated/unrecognized Bacteroidales sp. HOT 274, Desulfobulbus sp. HOT 041, Fretibacterium sp. HOT 360, Fretibacterium sp. HOT 362, and TM7 sp. HOT 356 phylotypes, in addition to F. alocis, F. fastidiosum, and S. sputigena, seem to be associated with periodontitis, and their role in its pathogenesis should be further investigated.
Author Contributions
R.R.D.S. Oliveira, D. Fermiano, contributed to data acquisition, critically revised the manuscript; M. Feres, F.R.F. Teles, M. Faveri, contributed to conception, design, and data interpretation, drafted and critically revised the manuscript; L.C. Figueiredo, G.M.S. Soares, contributed to data interpretation, critically revised the manuscript. All authors gave final approval and agree to be accountable for all aspects of the work.
Footnotes
This study was supported by research grants 2013/10139-6 and 2012/23503-5 from the Fundação de Amparo a Pesquisa do Estado de São Paulo (Brazil) and in part by the National Institutes of Health / National Institute of Dental and Craniofacial Research (grants R01 DE024767 and DE021127-01) to F.R.T.
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
References
- Abusleme L, Dupuy AK, Dutza N, Silva N, Burleson JA, Strasbaugh LD, Gamonal J, Diaz PI. 2013. The subgingival microbiome in health and periodontitis and its relationship with community biomass and inflammation. ISME J. 7(5):1016–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Hebshi NN, Al-Alimi A, Taiyeb-Ali T, Jaafar N. 2015. Quantitative analysis of classical and new putative periodontal pathogens in subgingival biofilm: a case-control study. J Periodontal Res. 50(3):320–329. [DOI] [PubMed] [Google Scholar]
- American Academy of Periodontology. 1996. Consensus report. Periodontal diseases: pathogenesis and microbial factors. Ann Periodontol. 1(1):926–932. [DOI] [PubMed] [Google Scholar]
- Armitage GC. 1999. Development of a classification system for periodontal diseases and conditions. Ann Periodontol. 4(1):1–6. [DOI] [PubMed] [Google Scholar]
- Aruni AW, Roy F, Fletcher HM. 2011. Filifactor alocis has virulence attributes that can enhance its persistence under oxidative stress conditions and mediate invasion of epithelial cells by Porphyromonas gingivalis. Infect Immun. 79(10):3872–3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camelo-Castillo AJ, Mira A, Pico A, Nibali L, Henderson B, Donos N, Tomás I. 2015. Subgingival microbiota in health compared to periodontitis and the influence of smoking. Front Microbiol 6:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cato EP, Moore LV, Moore WE. 1985. Fusobacterium alocis sp. nov. and Fusobacterium sulci sp. nov. from the human gingival sulcus. Int J Syst Bacteriol. 35(4):475–477. [Google Scholar]
- Chen T, Yu WH, Izard J, Baranova OV, Lakshmanan A, Dewhirst FE. 2010. The Human Oral Microbiome Database: a web accessible resource for investigating oral microbe taxonomic and genomic information. Database (Oxford). 2010:baq013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortelli JR, Cortelli SC, Jordan S, Haraszthy VI, Zambon JJ. 2005. Prevalence of periodontal pathogens in Brazilians with aggressive or chronic periodontitis. J Clin Periodontol. 32(8):860–866. [DOI] [PubMed] [Google Scholar]
- Darby IB, Hodge PJ, Riggio MP, Kinane DF. 2000. Microbial comparison of smoker and non-smoker adult and early-onset periodontitis patients by polymerase chain reaction. J Clin Periodontol. 27(6):417–424. [DOI] [PubMed] [Google Scholar]
- Dzink JL, Socransky SS, Haffajee AD. 1988. The predominant cultivable microbiota of active and inactive lesions of destructive periodontal diseases. J Clin Periodontol. 15(5):316–323. [DOI] [PubMed] [Google Scholar]
- Faveri M, Figueiredo LC, Duarte PM, Mestnik MJ, Mayer MP, Feres M. 2009. Microbiological profile of untreated subjects with localized aggressive periodontitis. J Clin Periodontol. 36(9):739–749. [DOI] [PubMed] [Google Scholar]
- Faveri M, Mayer MP, Feres M, de Figueiredo LC, Dewhirst FE, Paster BJ. 2008. Microbiological diversity of generalized aggressive periodontitis by 16S rRNA clonal analysis. Oral Microbiol Immunol 23(2):112–118. [DOI] [PubMed] [Google Scholar]
- Gonçalves LF, Fermiano D, Feres M, Figueiredo LC, Teles FR, Mayer MP, Faveri M. 2012. Levels of Selenomonas species in generalized aggressive periodontitis. J Periodontal Res. 47(6):711–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffen AL, Beall CJ, Campbell JH, Firestone ND, Kumar PS, Yang ZK, Podar M, Leys EJ. 2012. Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. ISME J. 6(6):1176–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalava J, Eerola E. 1999. Phylogenetic analysis of Fusobacterium alocis and Fusobacterium sulci based on 16S rRNA gene sequences: proposal of Filifactor alocis (Cato, Moore and Moore) comb. nov. and Eubacterium sulci (Cato, Moore and Moore) comb. nov. Int J Syst Bacteriol. 49(Pt 4):1375–1379. [DOI] [PubMed] [Google Scholar]
- Kolenbrander PE, Andersen RN, Moore LV. 1989. Coaggregation of Fusobacterium nucleatum, Selenomonas flueggei, Selenomonas infelix, Selenomonas noxia, and Selenomonas sputigena with strains from 11 genera of oral bacteria. Infect Immun. 57(10):3194–3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar PS, Griffen AL, Moeschberger ML, Leys EJ. 2005. Identification of candidate periodontal pathogens and beneficial species by quantitative 16S clonal analysis. J Clin Microbiol. 43(8):3944–3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledder RG, Gilbert P, Huws SA, Aarons L, Ashley MP, Hull PS, McBain AJ. 2007. Molecular analysis of the subgingival microbiota in health and disease. Appl Environ Microbiol. 73(2):516–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matarazzo F, Ribeiro AC, Feres M, Faveri M, Mayer MP. 2011. Diversity and quantitative analysis of Archaea in aggressive periodontitis and periodontally healthy subjects. J Clin Periodontol. 38(7):621–627. [DOI] [PubMed] [Google Scholar]
- Moffatt CE, Whitmore SE, Griffen AL, Leys EJ, Lamont RJ. 2011. Filifactor alocis interactions with gingival epithelial cells. Mol Oral Microbiol. 26(6):365–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore WE, Holdeman LV, Cato EP, Smibert RM, Burmeister JA, Palcanis KG, Ranney RR. 1985. Comparative bacteriology of juvenile periodontitis. Infect Immun. 48(2):507–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park OJ, Yi H, Jeon JH, Kang SS, Koo KT, Kum KY, Chun J, Yun CH, Han SH. 2015. Pyrosequencing analysis of subgingival microbiota in distinct periodontal conditions. J Dent Res. 94(7):921–927. [DOI] [PubMed] [Google Scholar]
- Paster BJ, Boches SK, Galvin JL, Ericson RE, Lau CN, Levanos V, Sahasrabudhe A, Dewhirst FE. 2001. Bacterial diversity in human subgingival plaque. J Bacteriol. 183(12):3770–3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Chaparro PJ, Gonçalves C, Figueiredo LC, Faveri M, Lobão E, Tamashiro N, Duarte P, Feres M. 2014. Newly identified pathogens associated with periodontitis: a systematic review. J Dent Res. 93(9):846–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlafer S, Riep B, Griffen AL, Petrich A, Hübner J, Berning M, Friedmann A, Göbel UB, Moter A. 2010. Filifactor alocis-involvement in periodontal biofilms. BMC Microbiol. 10:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Socransky SS, Haffajee AD. 2005. Periodontal microbial ecology. Periodontol 2000. 38:135–187. [DOI] [PubMed] [Google Scholar]
- Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL., Jr. 1998. Microbial complexes in subgingival plaque. J Clin Periodontol. 25(2):134–144. [DOI] [PubMed] [Google Scholar]
- Socransky SS, Tanner AC, Goodson JM, Haffajee AD, Walker CB, Ebersole JL, Sornberger GC. 1982. An approach to the definition of periodontal disease syndromes by cluster analysis. J Clin Periodontol. 9(6):460–471. [DOI] [PubMed] [Google Scholar]
- Teles FR, Teles RP, Siegelin Y, Paster B, Haffajee AD, Socransky SS. 2011. RNA-oligonucleotide quantification technique (ROQT) for the enumeration of uncultivated bacterial species in subgingival biofilms. Mol Oral Microbiol. 26(2):127–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teles RP, Gursky LC, Faveri M, Rosa EA, Teles FR, Feres M, Socransky SS, Haffajee AD. 2010. Relationships between subgingival microbiota and GCF biomarkers in generalized aggressive periodontitis. J Clin Periodontol. 37(4):313–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vengerfeldt V, Špilka K, Saag M, Preem JK, Oopkaup K, Truu J, Mändar R. 2014. Highly diverse microbiota in dental root canals in cases of apical periodontitis (data of illumina sequencing). J Endod. 40(11):1778–1783. [DOI] [PubMed] [Google Scholar]
- Yost S, Duran-Pinedo AE, Teles R, Krishnan K, Frias-Lopez J. 2015. Functional signatures of oral dysbiosis during periodontitis progression revealed by microbial metatranscriptome analysis. Genome Med. 7(1):27 Erratum in: Genome Med. 2015;7(1): 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You M, Mo S, Watt RM, Leung WK. 2013. Prevalence and diversity of Synergistetes taxa in periodontal health and disease. J Periodontal Res. 48(2):159–168. [DOI] [PubMed] [Google Scholar]