Abstract
Nonsense-mediated mRNA decay (NMD) is a eukaryotic mRNA quality control and regulatory process that plays direct roles in human health and disease. In this Minireview, we discuss how understanding the molecular events that trigger NMD can facilitate strategic targeting of genes via CRISPR/Cas9 technologies and also inform disease diagnostics and treatments.
Setting the Stage: mRNA Biogenesis and NMD
Within the nucleus, pre-mRNAs are synthesized and immediately undergo cis-modifications and binding by various proteins prior to and during the formation of mRNA. At the 5′ end, a 7-methylguanosine residue is added to protect the transcript from 5′-to-3′ exoribonucleases and provide a binding platform for the cap-binding protein (CBP) complex (CBC), composed of CBP80–CBP20. Mammalian pre-mRNAs often contain intervening regions, or introns, that are excised by complexes of RNAs and proteins called spliceosomes. The act of pre-mRNA splicing contributes to the accurate production of mRNAs and thus proteins, in part by depositing macromolecular landmarks of proteins, named exon-junction complexes (EJCs), upstream of spliced exon-exon junctions. At the 3′ end, a series of non-templated adenosine residues generally form a platform for poly(A)-binding proteins that protect the transcript from 3′-to-5′ exoribonucleases. With an assortment of bound proteins, the mRNA is now termed an mRNP complex that—providing that pre-mRNA processing has been successful—is ready to be exported from the nucleus into the cytoplasm to engage with functional ribosomes and other factors that concomitantly direct protein synthesis and mRNA quality inspection via NMD, a process by which a premature stop codon that would lead to a truncated protein product triggers mRNA degradation.
NMD is tightly intertwined with mRNA biogenesis even at its earliest stages, since NMD is promoted by both the CBC and EJCs (Lykke-Andersen and Jensen, 2015). EJCs, which usually consist of four core proteins—eukaryotic translation initiation factor 4A3 (eIF4A3), cancer susceptibility candidate 3 (CASC3), RNA-binding motif protein 8A (RBM8A or Y14), and mago-nashi homolog (MAGOH)—are deposited ~20–24 nucleotides (nt) upstream of ~80% of all exon-exon junctions (Le Hir et al., 2016). This splicing-dependent “mark” serves to orient the NMD machinery once the mRNP is exported to the cytoplasm for translation-dependent inspection by NMD factors.
Generally, faulty transcripts are triaged for destruction by NMD immediately (i.e., < 1 min) after entry into the cytoplasm, largely before the CBC is replaced by eIF4E, which is the cap-binding protein of the bulk of cellular mRNA (Trcek et al., 2013). NMD uses an initial CBC-promoted “pioneer round” of translation to identify the termination codon within the reading frame. Concomitantly, pioneer ribosomes remove EJCs, which record mRNA splicing history, from within but not downstream of the reading frame, thereby establishing whether the termination codon falls ≥ 50–55 nt upstream of an exon-exon junction—a general indication that the termination codon is premature, since normal termination codons are largely located in the last exon.
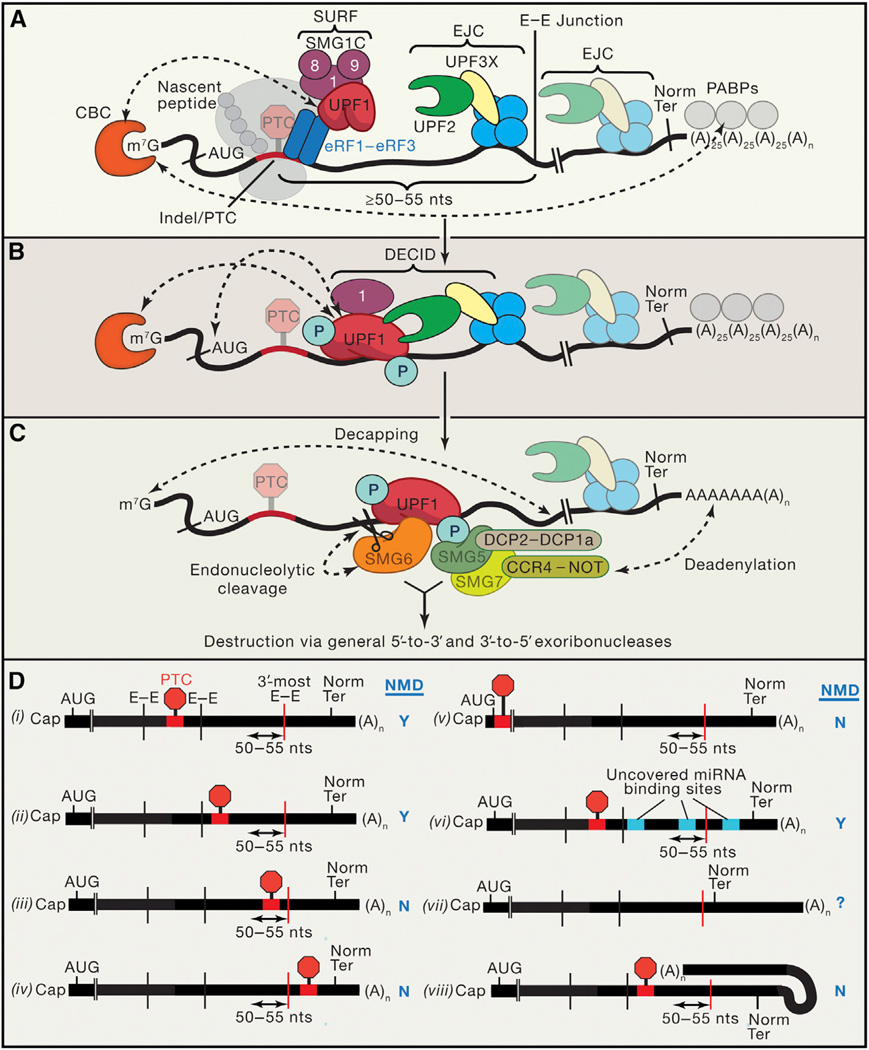
The molecular gymnastics of proteins that enable EJC-mediated NMD are partially understood. Like the situation for a normal termination codon, in the case of a premature termination codon (PTC) with a downstream EJC that was not removed by pioneer ribosomes, eukaryotic release factor 1 (eRF1) and eRF3 associate with the terminating ribosome. In the case of a PTC, however, additional factors form a transient so-called SURF complex with eRF1-eRF3 (Yamashita, 2013) (Figure 1A). These include suppressor with morphogenic effect on genitalia 1 (SMG1), which together with SMG8 and SMG9 forms the complex SMG1C, and up-frameshift 1 (UPF1). UPF1 is the central NMD factor: it is an ATP-dependent RNA helicase containing an abundance of serine and threonine residues located within its N and C termini that function as phosphate-acceptor sites. SMG1 is a phosphatidylinositol 3-kinase-related protein kinase that phosphorylates many of these residues, and this activity is initially held in check by the SMG8-SMG9 complex (Yamashita, 2013). If a terminating ribosome-SURF complex forms ≥ 50–55 nt upstream of an exon-exon junction (Nagy and Maquat, 1998) that has a splicing-generated EJC, then NMD ensues. This is a consequence of an interaction between minimally SMG1 and UPF1 of SURF and the EJC, whose core constituents may be further outfitted with the UPF3X (also called UPF3b) and UPF2 proteins (Shum et al., 2016). When UPF2, anchored to the EJC by UPF3X, interacts with the cysteine- and histidine-rich domain of UPF1, it triggers a conformational change that activates UPF1 helicase activity (Chamieh et al., 2008). This promotes mRNA unwinding and protein removal by UPF1, which are essential for substrate decay. The resulting complex is the “decay-inducing complex” (DECID) (Figure 1B), and it is at this stage that SMG1 phosphorylates the UPF1 termini. Tissue-specific expression of the NMD antagonist UPF3 disrupts this complex and confers fine-tuning NMD activity (Shum et al., 2016).
Figure 1. Understanding NMD Can Improve Generating CRISPR/Cas9-Mediated Gene Knockouts.
(A) Cytoplasmic mRNA bound by the cap (m7G)-binding protein complex (CBC) and poly(A)-binding proteins (PABPs) undergoes a pioneer round of translation coupled to inspection by the NMD machinery (Maquat et al., 2010). When the translating ribosome undergoes an altered termination event at the premature termination codon (PTC) (for example, as a result of indel formation), the SMG1 complex (SMG1C) joins factors that also typify normal termination events, namely eukaryotic release factor 1 (eRF1) and eRF3, to form the SURF complex. SMG1C is composed of the UPF1 kinase SMG1 and its repressors SMG8 and SMG9. EJCs typically display UPF2 anchored by UPF3X. Dashed lines signify that CBC promotes SURF formation and that CBC and PABPs may “circularize” the mRNA via bridging proteins. Notably, PABPs are known antagonists of NMD. AUG, initiation codon; Norm Ter, normal termination codon; nts, nucleotides.
(B) As a rule, if an EJC-associated exon-exon junction (E–E) resides ≥50–55 nt downstream of the termination event, then NMD initiates. UPF2 interacts with UPF1, causing a large conformational change that activates UPF1 helicase activity. Within the resulting decay-inducing complex (DECID), SMG1 phosphorylates UPF1. CBC also promotes SMG1-UPF1 binding to the EJC (dashed arrow), and phosphorylated UPF1 inhibits further translation initiation at the AUG codon (dashed arrow).
(C) Phosphorylated UPF1 recruits the SMG6 endonuclease, which cleaves between the PTC and EJC, and SMG5-SMG7 recruits the multisubunit CCR4-NOT complex, causing deadenylation, and/or DCP2-DCP1a, causing decapping. In all cases, destruction of mRNA fragments ensues via general exoribonucleases.
(D) Examples of indel/PTC placement within a typical transcript and its result on NMD as well as additional scenarios. Y, yes NMD occurs; N, no NMD. See text for details.
UPF1 phosphorylation has two consequences: first, phosphorylated UPF1 represses further translation initiation through an interaction with eIF3 that is critical for mRNA decay; second, phosphorylated UPF1 residues form platforms and expose new regions of UPF1 to which RNA-degradative enzymes are either directly or indirectly recruited, resulting in targeted, rather than indiscriminate, mRNA destruction (Figure 1C). Recruitment of the endonuclease SMG6 leads to NMD substrate cleavage in the vicinity of the PTC. This generates unstable decay intermediates that lack either the 5′ cap or 3′ poly(A) and, thus, are susceptible to cellular 5′-to-3′ or 3′-to-5′ exonucleases, respectively. Recruitment of the SMG5-SMG7 complex, which is devoid of intrinsic nuclease activity but has the capacity to bring in the CCR4-NOT deadenylase complex via SMG7 and the DCP2/DCP1a mRNA-decapping proteins via SMG5, also results in target degradation. SMG5 and SMG6 additionally recruit protein phosphatase 2A to dephosphorylate UPF1, recycling it for further rounds of NMD (Lykke-Andersen and Jensen, 2015).
NMD in Action: CRISPR/Cas9-Generated Knockouts
A promising means for creating loss-of-function alleles involves using the highly publicized CRISPR/Cas9 approach (Shalem et al., 2015). We provide only a thumbnail sketch of how this tool is used to knock out protein-encoding genes so as to set the context for how NMD contributes to this process.
Owing to its portability, the microbial type II clustered regularly interspaced short palindromic repeat (CRISPR) adaptive-immune system is now widely used to modify genomic DNA. Requirements for gene manipulation are minimal: the Streptococcus pyogenes Cas9 nuclease (SpCas9) need only be co-expressed with a single-guide RNA (sgRNA) composed of a fusion of CRISPR RNA (crRNA) and short trans-activating RNA (tracrRNA). Unlike other gene-editing nucleases that rely on amino acids to bring the nuclease to its correct location, SpCas9 uses the sgRNA to locate complementary regions in the genome. The only limitation is the requirement for a protospacer-adjacent motif (PAM), which for SpCas9 is the trinucleotide 5′-NGG-3′. Thus, by obeying the PAM requirement and modifying the 20-nt targeting sequence of the sgRNA, nuclease activity can be directed virtually anywhere within the genome. Many variations of this are emerging, such as using nicking enzymes to separately target Watson and Crick DNA strands to improve fidelity, orthogonal enzymes derived from other bacterial species with different PAM requirements, and CRISPR arrays for multiplex genome editing. Many CRISPR/Cas9 approaches utilize the cell’s own nonhomologous end-joining (NHEJ) pathway of DNA repair to generate the disruption (Shalem et al., 2015). Upon sensing a Cas9-induced double-stranded DNA break, the NHEJ pathway undertakes non-templated repair, which ultimately results in DNA insertions or deletions (indels).
Indel formation through the action of Cas9 nucleases can create loss-of-gene function in several ways. The indel may alter a protein-coding region, generating an unstable and/or nonfunctional protein as a result of, e.g., a frameshift mutation. Alternatively or concomitantly, useful indels often introduce premature termination codons (PTCs) that signal NMD to destroy the resulting mRNA before it has a chance to produce much protein. In any case, the key to success is to strategically choose the gene region to be targeted. In light of this, we suggest that designing high-quality sgRNAs requires a basic understanding of NMD, and we use NMD mediated by an EJC—the most-studied and robust type of NMD—to illustrate why.
EJC-mediated NMD follows the “50–55 nt rule” (Nagy and Maquat, 1998). According to the rule, if a PTC resides ≥50–55 nt upstream of an exon-exon junction, then the leading edge of the terminating ribosome fails to remove (i.e., is sufficiently far away from) the corresponding EJC, allowing for EJC-facilitated activation of UPF1. That this rule is predictive of whether a termination codon yields an NMD-sensitive or NMD-resistant transcript has important implications for the generation of CRISPR/Cas9-mediated knockout reagents and also for human health. With respect to CRISPR/Cas9 experiments, sgRNAs should be designed so that DNA cleavages fall sufficiently upstream of an exon-exon junction and, thus, the associated EJC. The rule is concretely illustrated in several scenarios that the experimentalist may encounter, diagrammed in Figure 1D, where an mRNA product is generated as a result of at least one splicing event. In the case of the mRNA exemplified, placement of an indel (for simplicity, the PTC is diagrammed within the indel) downstream of the first or second exon-exon junction satisfies this rule, provided that there is at least one exon-exon boundary residing at least 50–55 nt downstream of the PTC (Figures 1D(i) and 1D(ii)). Although the efficiency of NMD may be enhanced in particular cases by splicing events upstream of the PTC, the one or more EJCs produced from splicing upstream of the PTC are stripped off of the mRNA by the translating ribosome. It is at the PTC that the ribosome halts its progression, and if this occurs at least 50–55 nt upstream from the 3′-most exon-exon junction, the associated EJC(s) remain and NMD commences. If, however, the PTC resides closer than 50–55 nt upstream of the 3′-most exon-exon junction (Figure 1D(iii)) or downstream of this junction (Figure 1D(iv)), then NMD fails to occur since ribosomes will have removed all EJCs from the mRNA.
Given that the actual mutagens are the indels created as a result of random DNA repair, it may not be possible to predict the exact placement of a resulting PTC. However, since most mRNAs result from multiple splicing events and since approximately two-thirds of indels that reside within the reading frame shift the frame to yield a termination codon, placing sgRNAs upstream of several predicted exon-exon junctions will often satisfy the 50–55 nt rule. Nevertheless, precise placement of a PTC may be advantageous to ensure adherence to the rule. This may be accomplished using cell types capable of homology-directed DNA repair (HDR) and an exogenous repair template in addition to the requisite CRISPR/Cas9 components (Shalem et al., 2015). One caveat derives from the observation that start codon (AUG)-proximal PTCs trigger NMD inefficiently, possibly due to circularization of the mRNA and/or translation re-initiation in the proper reading frame (Figure 1D(v)). However, inefficiency diminishes rapidly with distance of the PTC from the initiation codon, e.g., >25 nt for β-globin mRNA (Pereira et al., 2015). A final consideration is that introduction of a novel PTC into a transcript effectively converts what was once a coding region to a new 3′ UTR. Without the ribosome translating through this region, new factors may bind and alter transcript regulation. MicroRNAs, for example, may bind to cryptic sites previously occluded by translating ribosomes (Figure 1D(vi)), potentially adding to NMD-mediated downregulation (Zhao et al., 2014).
Different branches of NMD exist, as illustrated by varying requirements among different NMD targets for one or more trans-acting factors, including UPF2 or UPF3X. Also, mRNA features other than a PTC-distal EJC can promote NMD. As exemplified by “failsafe” NMD, also called 3′ UTR EJC-independent NMD, an exceptionally long (≥420 nt) 3′ UTR that is devoid of an EJC can trigger NMD, possibly because poly(A)-binding protein C1 (a known NMD antagonist) lies distant from the termination event (Figure 1D(vii)). Long 3′ UTRs may also harbor largely uncharacterized sequences situated immediately downstream of a termination codon that inhibit NMD (Toma et al., 2015) or, in some cases, CU-rich sequences that bind polypyrimidine tract-binding protein 1 and antagonize NMD (Ge et al., 2016). The 3′ UTR EJC-independent mode of NMD is less efficient at eliciting NMD than the 3′ UTR EJC-dependent mode; however, if the transcript in question derives from a splicing event, it may be possible to use CRISPR-Cas9 to convert it to an EJC-mediated NMD target by adhering to the 50–55 nt rule.
In general, natural NMD targets cannot be defined by their number of 3′ UTR nucleotides likely because of their ability to fold, which would shorten 3′ UTR length in three-dimensional space (Figure 1D(viii)). Because of the ambiguities surrounding why some natural (i.e., non-mutated) transcripts with long 3′ UTRs are NMD targets, it is less clear how to experimentally shunt an mRNA to NMD using CRISPR/Cas9 techniques. The most reliable method for predicting whether a transcript is an NMD target remains the 50–55 nt rule.
Personalized Medicine
The hope that CRISPR/Cas9 technology can be used to edit and correct genomes to deliver personalized medicine, which tailors healthcare to an individual’s disease-associated mutation(s), is contingent on rapid and accurate diagnosis. Results defining the rules governing EJC-promoted NMD already contribute to diagnosis. As the trend toward personalized medicine increases, an increasing number of patient genomes will be sequenced to pinpoint mutations and polymorphisms that drive disease and/or determine responses to therapeutics. Although genome sequencing results will allow physicians and scientists to define alterations in the DNA code, the resulting information may be insufficient to understand how the alterations affect gene function and thus contribute to disease.
Discoveries made through studies of NMD provide rapid, testable ideas about whether a particular mutation leads to loss or toxic gain of gene function, ultimately providing insight into the molecular etiology of diseases and information useful to devise the best possible treatments. The former is already borne out in the literature. A recent report documents a 4-month-old girl who presented with congenital diarrhea polyuria; sequencing revealed that she was homozygous for a splice-site mutation within the proprotein convertase 1 (PCSK1) gene that was predicted (through the 50–55 nt rule) to generate a PTC resulting in NMD (Härter et al., 2016). Experimental evidence then confirmed that disease was due to the loss of functional PCSK1 mRNA. Reports of diseases assessed in this way are increasing. This workflow—from defining symptoms in the clinic, to sequencing, to discovering the molecular underpinnings of a particular person’s disease—is greatly assisted by knowing the rules governing NMD, considering that ~one-third of inherited and acquired human diseases result from the erroneous generation of a PTC and that ~20% of the ~43,000 disease-associated single-base-pair substitutions are caused by nonsense mutations (Mort et al., 2008).
While evidence points to the evolution of NMD as a protective mechanism, destroying aberrant mRNAs with the potential to encode toxic truncated proteins, it is increasingly clear that NMD has additionally been co-opted by cells to function in regulating the levels of endogenous, non-mutated human mRNAs (Smith and Baker, 2015). Only some of these transcripts have features that are predicted to render them subject to NMD, but they are all upregulated upon experimental ablation of UPF1 or other NMD factors. This imparts on NMD the ability to control a large segment (as much as ~10%) of the human transcriptome, and these transcripts have an array of functions. As a result, any perturbation that purposefully modulates NMD activity will have physiological consequences. Hypoxia (a stress encountered in the tumor microenvironment) and other stresses down-modulate NMD activity in an incompletely understood manner that involves eIF2α phosphorylation (Gardner, 2010). Among the endogenous NMD-sensitive transcripts that are consequently upregulated are those encoding transcription factors (ATF-4, ATF-3, and CHOP) involved in restoring normal cellular homeostasis. Alternative splicing (AS) may be coupled to NMD (AS-NMD), whereby a PTC is introduced during a splicing event that purposefully downregulates the transcript. Between 10% and 30% of mammalian genes may be regulated in this way (Smith and Baker, 2015). Alternative polyadenylation that introduces a splicing event downstream of the normal termination codon offers additional opportunities to downregulate transcripts via NMD. Caspase proteases activated as a consequence of chemotherapeutic treatments attenuate NMD activity by cleaving UPF1, resulting in upregulation of mRNAs encoding pro-apoptotic factors that accelerate cell death (Jia et al., 2015; Popp and Maquat, 2015). Normal developmental programs also modulate NMD to achieve a desired transcriptomic output. During brain development, expression of the microRNA miR-128 increases dramatically. miR-128, in turn, downregulates UPF1 and CASC3 mRNAs, blunting NMD activity to upregulate a suite of mRNAs encoding pro-neurogenic factors (Bruno et al., 2011). Several other examples are documented and reviewed elsewhere (Lykke-Andersen and Jensen, 2015). Clearly the function of NMD goes beyond mere quality control, governing cellular homeostasis and adaptation in ways that have yet to be discovered.
Current readthrough therapeutics aimed at salvaging some functional protein from NMD-sensitive transcripts by forcing the ribosome to insert a non-cognate amino acid when it encounters a PTC suffer from the disadvantage that little if any transcript remains to be translated after NMD has occurred. Small molecules inhibiting the core NMD machinery may offer a useful additional treatment, and efforts to discover such molecules are ongoing (Martin et al., 2014). Given the roles that NMD plays beyond quality control, these molecules may also be useful in other situations. Moreover, knowledge regarding the NMD pathway is poised to assist in making major technological and clinical advances in ways that we can’t yet anticipate.
Acknowledgments
Work on NMD in the Maquat laboratory is supported by NIH R01 GM59614. M.W.P. was an HHMI Fellow of the Damon Runyon Cancer Research Foundation (DRG-2119-12). We thank R.A. Elbarbary for comments on the manuscript and apologize to those colleagues whose work could not be cited due to word and reference limitations.
REFERENCES
- Bruno IG, Karam R, Huang L, Bhardwaj A, Lou CH, Shum EY, Song HW, Corbett MA, Gifford WD, Gecz J, et al. Mol. Cell. 2011;42:500–510. doi: 10.1016/j.molcel.2011.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamieh H, Ballut L, Bonneau F, Le Hir H. Nat. Struct. Mol. Biol. 2008;15:85–93. doi: 10.1038/nsmb1330. [DOI] [PubMed] [Google Scholar]
- Gardner LB. Mol. Cancer Res. 2010;8:295–308. doi: 10.1158/1541-7786.MCR-09-0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Z, Quek BL, Beemon KL, Hogg JR. eLife. 2016;5:e11155. doi: 10.7554/eLife.11155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Härter B, Fuchs I, Müller T, Akbulut UE, Cakir M, Janecke AR. J. Pediatr. Gastroenterol. Nutr. 2016;62:577–580. doi: 10.1097/MPG.0000000000001018. [DOI] [PubMed] [Google Scholar]
- Jia J, Furlan A, Gonzalez-Hilarion S, Leroy C, Gruenert DC, Tulasne D, Lejeune F. Cell Death Differ. 2015;22:1754–1763. doi: 10.1038/cdd.2015.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H, Sauliere J, Wang Z. Nat. Rev. Mol. Cell Biol. 2016;17:41–54. doi: 10.1038/nrm.2015.7. [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen S, Jensen TH. Nat. Rev. Mol. Cell Biol. 2015;16:665–677. doi: 10.1038/nrm4063. [DOI] [PubMed] [Google Scholar]
- Maquat LE, Tarn WY, Isken O. Cell. 2010;142:368–374. doi: 10.1016/j.cell.2010.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin L, Grigoryan A, Wang D, Wang J, Breda L, Rivella S, Cardozo T, Gardner LB. Cancer Res. 2014;74:3104–3113. doi: 10.1158/0008-5472.CAN-13-2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mort M, Ivanov D, Cooper DN, Chuzhanova NA. Hum. Mutat. 2008;29:1037–1047. doi: 10.1002/humu.20763. [DOI] [PubMed] [Google Scholar]
- Nagy E, Maquat LE. Trends Biochem. Sci. 1998;23:198–199. doi: 10.1016/s0968-0004(98)01208-0. [DOI] [PubMed] [Google Scholar]
- Pereira FJ, Teixeira A, Kong J, Barbosa C, Silva AL, Marques-Ramos A, Liebhaber SA, Romão L. Nucleic Acids Res. 2015;43:6528–6544. doi: 10.1093/nar/gkv588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popp MW, Maquat LE. Nat. Commun. 2015;6:6632. doi: 10.1038/ncomms7632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Zhang F. Nat. Rev. Genet. 2015;16:299–311. doi: 10.1038/nrg3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shum EY, Jones SH, Shao A, Dumdie J, Krause MD, Chan WK, Lou CH, Espinoza JL, Song HW, Phan MH, et al. Cell. 2016;165:382–395. doi: 10.1016/j.cell.2016.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JE, Baker KE. BioEssays. 2015;37:612–623. doi: 10.1002/bies.201500007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toma KG, Rebbapragada I, Durand S, Lykke-Andersen J. RNA. 2015;21:887–897. doi: 10.1261/rna.048637.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trcek T, Sato H, Singer RH, Maquat LE. Genes Dev. 2013;27:541–551. doi: 10.1101/gad.209635.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita A. Genes Cells. 2013;18:161–175. doi: 10.1111/gtc.12033. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Lin J, Xu B, Hu S, Zhang X, Wu L. eLife. 2014;3:e03032. doi: 10.7554/eLife.03032. [DOI] [PMC free article] [PubMed] [Google Scholar]