Abstract
STAT (Signal Transducer and Activator of Transcription) transcription factors are constitutively activated in most hematopoietic cancers. We previously identified a target gene, LPP/miR-28 (LIM domain containing preferred translocation partner in lipoma), induced by constitutive activation of STAT5, but not by transient cytokine-activated STAT5. miR-28 exerts negative effects on thrombopoietin receptor signaling and platelet formation. Here, we demonstrate that, in transformed hematopoietic cells, STAT5 and p53 must be synergistically bound to chromatin for induction of LPP/miR-28 transcription. Genome-wide association studies show that both STAT5 and p53 are co-localized on the chromatin at 463 genomic positions in proximal promoters. Chromatin binding of p53 is dependent on persistent STAT5 activation at these proximal promoters. The transcriptional activity of selected promoters bound by STAT5 and p53 was significantly changed upon STAT5 or p53 inhibition. Abnormal expression of several STAT5-p53 target genes (LEP, ATP5J, GTF2A2, VEGFC, NPY1R and NPY5R) is frequently detected in platelets of myeloproliferative neoplasm (MPN) patients, but not in platelets from healthy controls. In conclusion, persistently active STAT5 can recruit normal p53, like in the case of MPN cells, but also p53 mutants, such as p53 M133K in human erythroleukemia cells, leading to pathologic gene expression that differs from canonical STAT5 or p53 transcriptional programs.
Introduction
JAK-STAT (Janus Kinase-Signal Transducer and Activator of Transcription) signaling is transiently activated by cytokines during hematopoiesis. Genetic lesions inducing constitutively activated JAK-STAT signaling have been described for several cancers, especially myeloproliferative neoplasms (MPNs) and leukemia,1 whereas constitutively active STATs have been detected in many solid and hematopoietic cancers.2 The JAK2 V617F gain-of-function mutation is a common acquired somatic mutation associated with MPNs, which leads to constitutive activation of STAT5 and of several cytokine receptor downstream pathways.3–6 Activation of STAT5 is essential for the establishment of the MPN phenotype as recently demonstrated by studies performed in STAT5A/B double knockout mice.7,8 This is reminiscent of the constitutive activation of STAT5 in hematopoietic cells transformed by expression of the spleen focusforming virus gp55 envelope protein, which leads to persistent activation of erythropoietin receptor signaling via JAK2 and STAT5, with STAT5B being the most sensitive to activation by low levels of constitutive signals.9 Chromatin immunoprecipitation (ChIP) and sequencing showed that the constitutive activation of STAT5 leads not only to STAT5 binding to common promoters, but also to different promoters containing low affinity binding sites.9 Interestingly, although STAT5A and STAT5B share 96% sequence similarity and have redundant targets and effects, it has recently been shown that STAT5B and STAT5A bind target gene promoters with different kinetics10 and they contribute differently to protection against stress in hematopoietic cells, with STAT5B being essential for survival, whereas STAT5A regulating the redox state.11 However, a more complete picture needs to be drawn and such differences might come also from expression differences, and more studies are necessary to obtain a deep mechanistic understanding of the common and distinct functions of STAT5A and STAT5B.
We have recently identified a microRNA, miR-28 which downmodulates translation of thrombopoietin receptor (TpoR, MPL) mRNA, along with mRNAs coding for other signaling proteins involved in megakaryocyte differentiation.12 The host gene for miR-28 is LPP (LIM domain lipoma preferred partner). As miR-28 and LPP are not induced in cell lines by acute cytokine stimulation, but are induced by JAK2 V617F, by active mutants forms of STAT5, by BCR-ABL or by active mutants of TpoR,12 we hypothesized that the LPP promoter is a prototype of those promoters regulated mainly by persistent STAT5 activity.
We report here that the constitutively activated STAT5 transcription factor associates with p53 to induce the transcription of LPP/miR-28 and genes other than classical STAT5 or p53 target genes. Furthermore, both wild-type and mutated p53 (such as the M133K mutant in human erythroleukemia (HEL) cells) cooperate with STAT5 in this manner. We find genes induced by this mechanism to be pathologically overexpressed in platelets from MPN patients.
Results
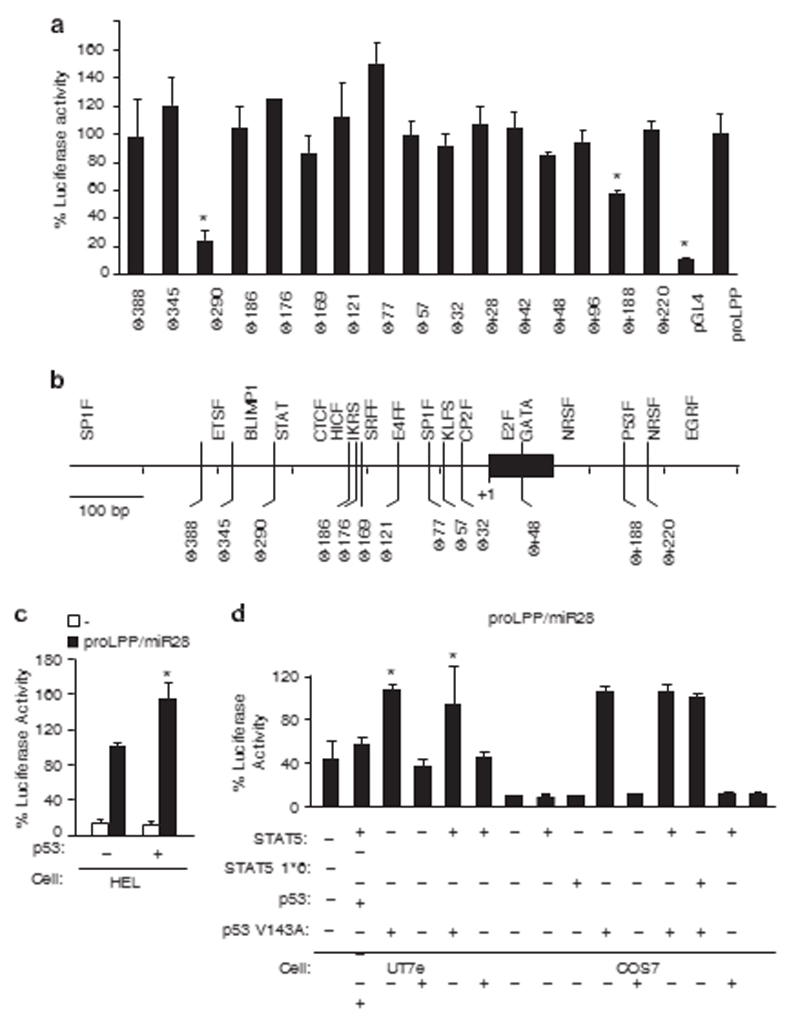
We first conducted an in-depth promoter analysis (Figure 1) of a gene coding for both a microRNA (miR-28) and a protein (LPP). In a previous study we observed that this gene is overexpressed in platelets of MPN patients carrying an activating mutation of JAK2 (Janus Kinase 2) (JAK2 V617F),12 responsible for the constitutive phosphorylation of STAT5.4 miR-28 negatively regulates translation of TpoR (c-MPL), and of several other signaling proteins, resulting in altered megakaryocyte differentiation.12 The LPP/ miR-28 promoter cloned in a luciferase vector was able to drive transcription in a JAK2 V617F positive cell line HEL cells. We previously reported that a dominant negative STAT5 mutant inhibits LPP/miR-28 transcription in HEL cells.12 Here we show that deletion of the STAT consensus site or of a p53 predicted binding site abrogated 80 or 40% of the luciferase activity, respectively. This suggests that STAT5 and, unexpectedly, p53, are both necessary for the transcription of the LPP/miR-28 promoter
Figure 1.
STAT- and p53-binding sites are required for transcriptional activity of the LPP/miR-28 promoter. (a) The cloned DNA sequence corresponding to the LPP/miR-28 promoter (chr3:187 871 155–187 872 054) was tested for its promoting activity compared with an empty luciferase vector (*Po0.05) by transfection in human HEL cells, that express LPP/miR-28 from its endogenous locus. The mutant promoters Δ− 290 and Δ+188 expressed significantly less luciferase, demonstrating that the transcriptional activity of the original promoter require a STAT (Δ− 290) and a p53 (Δ+188) binding site. (b) Predicted consensus binding sites for known transcription factors were deleted individually by directed mutagenesis and tested for luciferase activity compared with the original promoter. (c) A p53 expressing vector was cotransfected in HEL cells with a luciferase reporter under the control of the LPP/miR-28 promoter. The transcriptional activity was significantly increased by the expression of p53 WT (*Po0.05). (d) The transcription in UT-7e cells via the LPP/miR-28 promoter was increased by p53 WT (*Po0.05) and in the presence of both STAT5 and p53 WT (*Po0.05), but not by the inactive p53 mutant (V143A). The average of two experiments performed in triplicates is represented. Error bars represent s.e.m.
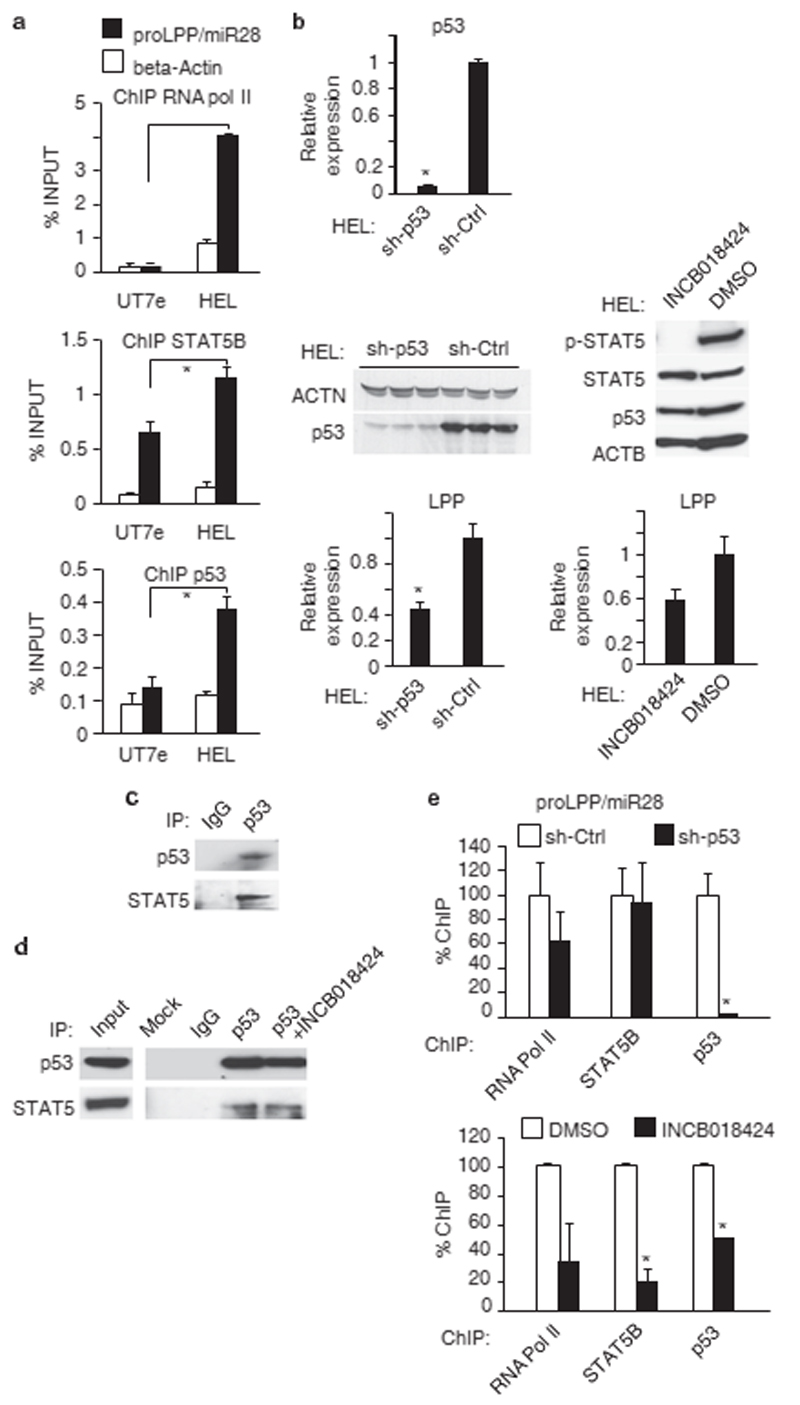
Because HEL cells (homozygous for JAK2 V617F) basally express high levels of LPP/miR-28, and the promoter induction required the p53 site, we hypothesized that both the wild-type and the M133K mutant p53 can cooperate with STAT5 in inducing LPP/ miR-28 expression. Next, we confirmed by ChIP that both STAT5B and p53 M133K were bound to the endogenous LPP/miR-28 promoter in HEL cells (Figure 2a). We focused mainly on STAT5B in this study, as constitutively activating mutations in STAT5B were found in large granulocytic leukemia patients,14 and due to significant expression levels in myeloid leukemia cell lines and the longer activation status after tyrosine phosphorylation compared with STAT5A.15 Thus, we chose STAT5B for genome-wide ChIP, after having tested for binding by other STATs, that is, STAT3, which did not bind significantly to the LPP/miR-28 promoter. STAT5A also bound to the LPP/miR-28 promoter but weakly, possibly because of a shorter half-life in the nucleus than STAT5B, and different kinetics of activation.10 In hematopoietic cells STAT5B is often activated at the lowest levels of constitutive signaling, is inducing survival and is effective at low levels associated with gene induction for survival.9,11 Using shRNAs targeting p53 and a JAK2 inhibitor (Ruxolitinib), we dissected the relative contributions of p53 and STAT5B to transcription of LPP/ miR-28. We used the INCB018424 (Ruxolitinib) as a clinically relevant JAK2 inhibitor, but for a broader spectrum of distinct JAK2 inhibitors and to exclude off-target effects, we also use TG101209, AZD1480 and JAK inhibitor 1, as we have shown already that they were preventing induction of LPP/miR-28 gene.12 Knockdown of p53 or inhibition of STAT5 phosphorylation by a JAK2 inhibitor were both able to reduce LPP expression by 50% (Figure 2b, bottom left and right panels). The JAK2 inhibitor prevented STAT5 tyrosine phosphorylation, but did not influence STAT5 or p53 protein levels (Figure 2b, middle panel, right and Nakatake et al.16). STAT5 co-immunoprecipitates with p53 in spleens from JAK2 V617F heterozygous knockin mice, where STAT5 is constitutively active17 (Figure 2c). When tested in HEL cells both in the absence or presence of JAK2 inhibitor, the coimmunoprecipitation between STAT5 and p53 was similar (Figure 2d), suggesting that association between p53 and STAT5 is independent of STAT5 activation. However, for a definitive conclusion, STAT5 mutants should be used in a STAT5-deficient background. Although ruxolitinib and other JAK2 inhibitors inhibited LPP transcription and STAT5 tyrosine phosphorylation, it is still possible that the association between unphosphorylated STAT5 (U-STAT5) and p53 might have a regulatory role relevant for induction of LPP. One scenario could involve U-STAT5 sustaining LPP expression after initial transient induction of LPP via tyrosine phosphorylated STAT5 and p53, a model that would reproduce examples from other systems describing effects of U-STATs.18 It would be of great interest to test the influence of p53 on noncanonical STAT signaling.
Figure 2.
STAT5B and p53 both are necessary for transcriptional activation of the LPP/miR-28 promoter. (a) Chromatin immunoprecipitations (ChIP) using antibodies against RNA pol II, STAT5B or p53 were performed on LPP/miR-28 non-expressing (UT7) or expressing (HEL) cell lines. Both STAT5B and p53 are bound on the LPP/miR-28 promoter in HEL cells whereas only STAT5B is bound in UT7 cells (*Po1e − 4; P = 3.7e − 3 and P = 1.4e − 3, respectively). The beta-actin amplification produced by primers in the beta-actin gene body was used as a negative control for STAT5B and p53 binding. (b) (Upper left) quantitative RT–PCR for p53 transcript in p53 knockdown HEL cell line compared with control HEL cell line (*P = 1.5e − 5). (Middle left) western blotting of p53 from HEL cell lines expressing an shRNA against p53 or a control shRNA. (Bottom left) quantitative RT–PCR for LPP transcript in HEL cell lines knockdown for p53 (*P = 4.4e − 3) or control. (Middle right) western blot analysis of p53 and STAT5 levels and tryrosine phosphorylated STAT5 in the presence and absence of JAK2 inhibitor in total lysates of HEL cells. (Bottom right) quantitative RT–PCR for LPP transcript in HEL cell lines treated with the JAK2 inhibitor INCB018424 (*P = 8.2e − 3). (c) Coimmunoprecipitation of endogenous STAT5 and p53 from enlarged spleens of JAK2 V617F knockin heterozygous mice presenting MPN phenotype (d) Co-immunoprecipitation of endogenous STAT5 and p53 in HEL cells, in the presence (18 h pretreatment, 10 μM INCB018424) or absence of JAK2 inhibitor. (e) ChIP of RNA pol II, STAT5B and p53 (*P = 2.7e − 2) in p53 knockdown HEL cells and control cells, and in HEL cells treated with the JAK2 inhibitor INCB018424 or solvent alone (DMSO) (STAT5B: *P = 1.1e − 2; p53: *P = 1.8e − 5). Error bars represent s.d. for the qPCR performed in triplicate on sh-Ctrl and sh-p53 cells, or s.e.m. from two independent experiments performed on HEL cell after treatment by the JAK2 inhibitor qPCR, quantitative PCR; RT–PCR, PCR in reverse transcription. DMSO, dimethyl sulfoxide.
ChIP experiments then showed that STAT5B was still bound to the LPP/miR-28 promoter in the absence of p53 (down-modulated by shRNA against p53), whereas in contrast, the binding of p53 was diminished in the absence of STAT5B binding (inhibited by JAK2 inhibitor, panel 2b) (Figure 2d). This is consistent with a model where recruitment of p53 depends on prior binding of STAT5B to the promoter to drive LPP/miR-28 transcription.
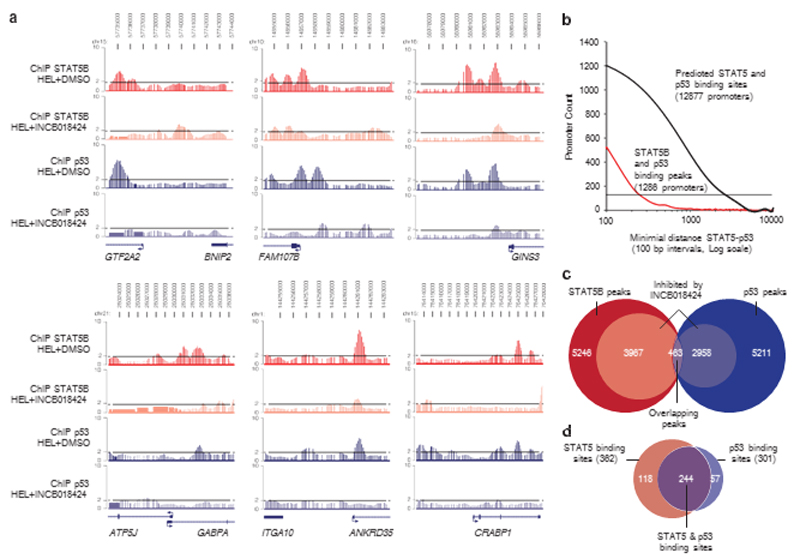
Although in a yeast transcriptional assay the p53 M133K was reported as lacking DNA-binding activity to canonical p53 target genes,13 we tested whether indeed in our system this mutant would not induce canonical targets, and whether it would bind to novel targets having the p53-binding consensus sites, but that are normally not induced by p53. First, we verified that p53 M133K was not bound to p53 canonical targets in HEL cells, which was the case (Supplementary Figure S3). Second, from our bioinformatics analysis, we predicted that p53 and STAT5 binding sites are frequently found close together, in the human genome within a 2.5 kb distance compared with an equiprobable distribution (Po2.2e − 16, chi-squared test) (Figure 3b), suggesting that these two transcription factors may frequently cooperate to regulate transcription.
Figure 3.
STAT5B and p53 ChIP-on-chip binding peaks are frequently co-localized and dependent on STAT5B activation. (a) Enrichment levels of STAT5B or p53 chromatin immunoprecipitations from HEL cells (+DMSO) are represented in red and blue, respectively, and STAT5B and p53 enrichment levels after JAK2 inhibition (+INCB018424) are represented in pink and light blue, respectively. Dotted lines correspond to the twofold enrichment ratio compared with the input samples. (b) The number of promoters is plotted as a function of the relative distances between STAT5B- and p53-binding peaks on promoters (red line) and the relative distance between STAT5 and p53 predicted binding sites in promoters of the human genome (black line). A significant proportion of promoters contain both STAT5- and p53-binding sites within a 2.5 kb distance compared with an equiprobable distribution (dashed line) using the chi-squared test (2.2e− 16). (c) Venn diagram summarizing the total amount of STAT5B-binding peaks (red, 5246) and p53-binding peaks (blue, 5211); STAT5B peaks (3967) and p53 peaks (2958) inhibited by JAK2 inhibition are represented in pink and light blue, respectively. The intersection represents overlapping STAT5B and p53 responsive peaks (purple, 463). (d) From this list of 463 peaks, 362 (78%) contained a consensus STAT5 binding site (>85% similar to the STAT5 consensus site, in red), 301 (65%) contained a p53 consensus binding site (>70% similar to the p53 consensus site, in blue) and 244 (52%) contained both (purple). DMSO, dimethyl sulfoxide.
Next, we studied more in detail the interaction between STAT5 and p53 using: (i) co-immunoprecipitation, (ii) luciferase assays measuring the transcriptional activities of each of the two transcription factors after co-expressing wild-type and mutant form of each.
We tested co-immunoprecipitation between p53 and wild-type STAT5 or truncated mutants where either the N-terminus (residues 1–136, denoted deltaN) or the C-terminus (residues 749–794, denoted deltaC) was missing, thereby reducing their size (Supplementary Figure S1e, upper panel). When each of these mutants were expressed in Ba/F3 cells along with p53 by electroporation, immunoprecipitation with anti-p53 (recognizing the N-terminus of endogenous p53) led to the co-immunoprecipitation of full length STAT5, but also of deltaNSTAT5 and deltaC-STAT5, with the latter being co-precipitated stronger (Supplementary Figure S1e, middle panel). Our assay was not sensitive enough to detect co-immunoprecipitation between endogenous p53 and endogenous STAT5 in Ba/F3 cells, whereas p53 sufficed to co-immunoprecipitate exogenous STAT5 variants (Supplementary Figure S1e, right panel), where again deltaC-STAT5 co-precipitated stronger with p53 than wild type or deltaN-STAT5. Control isotype IgG did not co-precipitate any of the STAT5 proteins. These data and Figures 2c and d would indicate that p53 interacts with a region of STAT5 that is containing the core protein sequence with the DNA-binding domain (between sequences 136 and 749 that are also highly conserved between the two STAT5A/B isoforms). Our data suggest that the C-terminal transactivation domain (that is also a site for tyrosine phosphatase docking and thus determines stability of activation) might exert a negative effect on the interaction. However, these conclusions need validation in a STAT5-deficient background.
In Supplementary Figure S1f, we show that, as described by the Groner lab19 p53 suppresses transcriptional activity of STAT5. In contrast, STAT5 does not inhibit p53 transcriptional activity, as detected by the luciferase reporter PG13-luc (data not shown, and Fritsche et al.19). We used this assay as a read-out for functional interaction and observed that wild-type p53, the M133K and V143A mutants (Supplementary Figure S1f) and to a lower extent the truncated mutant p53 Delta288 of UT7 cells (Supplementary Figure S1g) are all able to reduce transcriptional activity of STAT5 on a luciferase reporter driven by the spi-luc STAT5 responsive promoter. Thus, all such mutants can functionally interact with STAT5, explaining for example why, in UT7 cells, expression of JAK2 V617F induced RNA-polymerase II binding to the LPP promoter (detected by ChIP only after JAK2 V617F expression Figure 2c and data not shown) and induction of LPP gene expression.12 Interestingly, the deltaN-STAT5 mutant that lacks the N-terminus (residues 1–136) remains able to induce transcription and to be inhibited by p53 proteins in gamma2A cells (Supplementary Figure S1f).
To characterize this previously unrecognized STAT5–p53 cooperation for transcriptional activity, we performed ChIP of STAT5B and p53 followed by hybridization to promoter microarrays with or without previous treatment with JAK2 inhibitor in HEL cells, which inhibits the JAK2 V617F mutant, that is homozygously expressed in this cell line. We observed that about 9% peaks corresponding to STAT5B and p53 binding were co-localized on promoters (Figure 3a) and separated by less than 1 kb (Figure 3b). Seventy-five percent (3967/5246) of STAT5Bbinding peaks and fifty-six percent (2958/5211) of p53-binding peaks were diminished by more than twofold after JAK2 V617F inhibition (Figure 3c), suggesting that p53 M133K binding is dependent on STAT5B DNA binding.
Gene ontology analysis showed that unique STAT5 targets were enriched for genes coding for phosphoproteins, serine/threonine kinases, nuclear and cytoplasmic proteins, and were associated with biological activities such as cell cycle, cytoskeleton organization, RNA processing or cytokine production. In contrast, common p53-STAT5B targets are coding for transmembrane proteins, glycoproteins and secreted proteins (Supplementary Figure S4), and are associated with different biological processes, such as regulation of secretion or synaptic transmission. Taken together, these results indicate that the p53–STAT5B complex may bind to genes that contribute to distinct cellular functions from that of STAT5B alone. Such genes might be directly linked to the pathogenesis and phenotypes of malignancies in the hematopoietic lineage or may become candidate markers of disease.
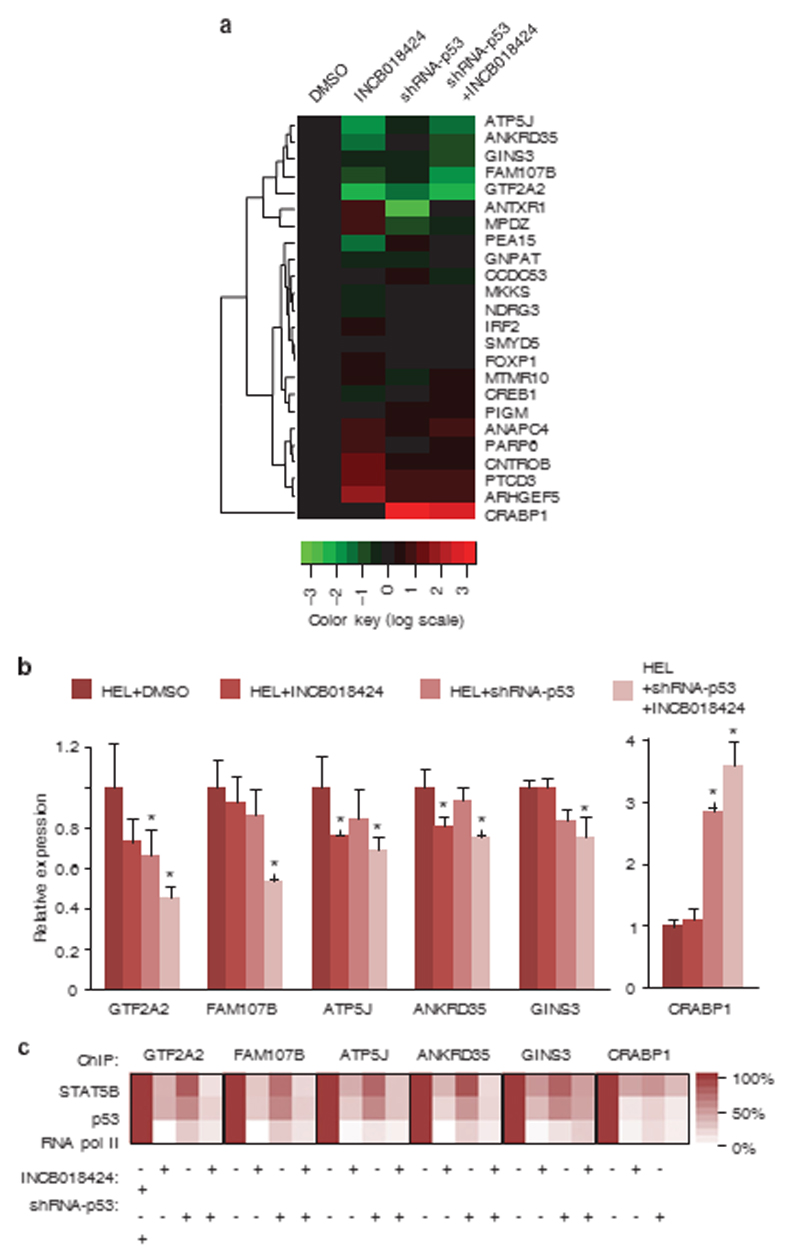
We selected 463 genomic positions where STAT5B- and p53binding peaks overlap and are sensitive to JAK2 inhibition targets shown to be downregulated by JAK2 inhibition.21 We found that 10 genes were upregulated and 9 genes were downregulated after either JAK2 inhibition, p53 M133K knockdown or both (Figure 4a). Among these, GTF2A2, FAM107B, ATP5J, ANKRD35 and GINS3 (Figure 3) were the most significantly downregulated genes and CRABP1 was the most significantly upregulated gene upon inhibition of JAK2/STAT5 phosphorylation, p53 M133K knockdown or both (Figure 4b). Furthermore we confirmed the diminished RNA pol II recruitment at the STAT5B-p53 common peaks’ positions in HEL cells by ChIP (Figure 4c). These data demonstrate that cooperative DNA binding of STAT5B and p53 M133K is involved in the transcriptional regulation of target genes.
Figure 4.
Transcriptional activity of STAT5B and p53 target genes. (a) Heatmap profile summarizing qRT–PCR on STAT5B and p53 target genes in HEL cells (DMSO), HEL cells treated by a JAK2 inhibitor (INCB018424), HEL cells knocked-down for p53 expression (shRNA-p53) and HEL cells treated with the JAK2 inhibitor and p53 shRNA (shRNA-p53+INCB018424). For each gene, red represents the highest and green the lowest expression level. (b) Relative expression of target genes compared with beta-actin expression assessed by qRT–PCR, showing a significant change upon STAT5B and p53 inhibition (*Po0.05). (c) Heatmap profiles of STAT5B, p53 and RNA pol II binding at selected promoters analyzed by chromatin immunoprecipitation using primers reported in Supplementary Table S3. DMSO, dimethyl sulfoxide. Relative expression of target genes compared with beta-actinexpression assessed by qRT–PCR, showing a significant change upon STAT5B and p53 inhibition (*Po0.05). (c) Heatmap profiles of STAT5B, p53 and RNA pol II binding at selected promoters analyzed by chromatin immunoprecipitation using primers reported in Supplementary Table S3. DMSO, dimethyl sulfoxide.
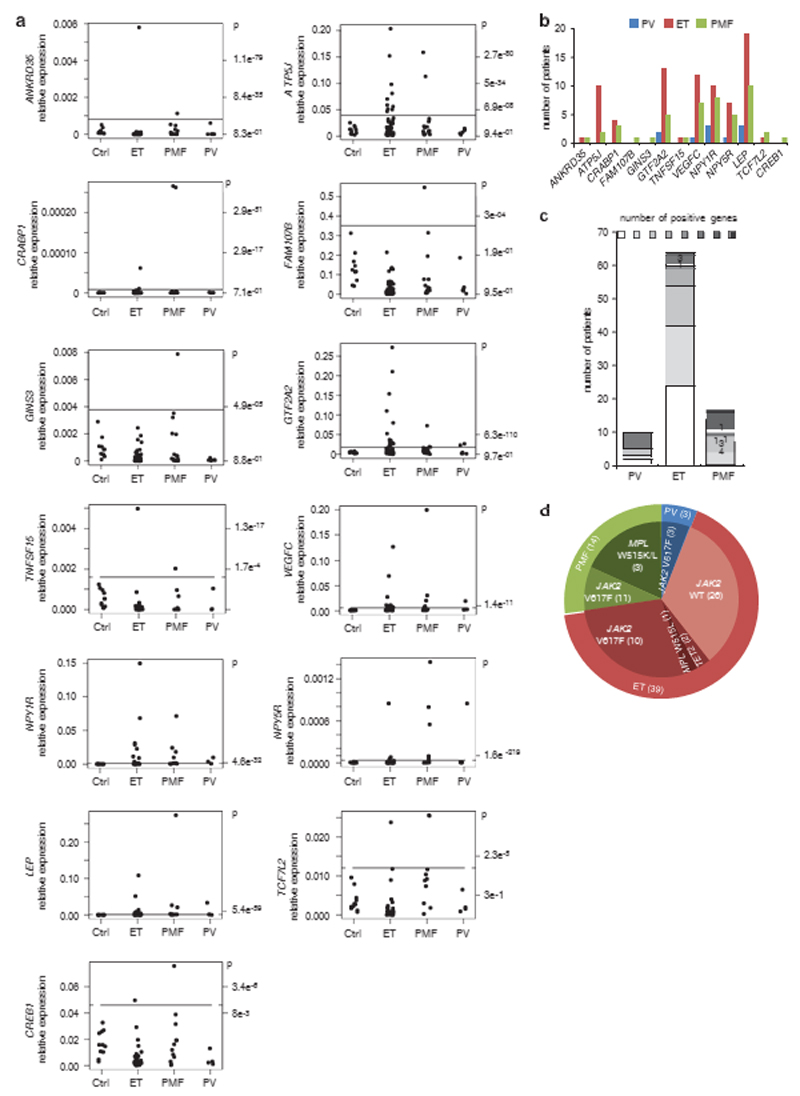
Next, we evaluated the expression levels of the novel STAT5p53 target genes in platelets from the peripheral blood of MPN patients. Platelets are the product of maturation of megakaryocytes in the bone marrow. We tested 11 healthy controls and 86 MPN patients, namely 6 polycythemia vera, 16 primary myelofibrosis and 64 essential thrombocythemia (ET) patients (Figure 5a). Strikingly, 39 out of the 64 ET, 14 out of the 16 primary myelofibrosis and 8 out of the 10 polycythemia vera patient samples overexpressed such genes. These results might be relevant as 50% of ET and primary myelofibrosis were until very recently negative for known pathogenic mutations. LEP, ATP5J, GTF2A2, VEGFC, NPY1R and NPY5R are the genes that were most frequently overexpressed in ET patients (Figure 5b). Overall, 62% ET, 87% primary myelofibrosis and 80% polycythemia vera patients were positive for the increased expression of at least one of these genes (Figure 5c). Some of those patients were carrying JAK2 V617 or MPL (TpoR) W515K/L mutations known to lead to constitutive STAT5 activation (Figure 5d) whereas others did not, suggesting those that overexpress this MPN-disease cluster regulated by STAT5-p53 might harbor STAT5-activating mutations. LEP codes for the hormone leptin, which induces weight loss, a characteristic of MF, and which has been reported to be increased in MF patients.22 GTF2A2 codes for the general initiation factor TFIIA of the RNA polymerase II important for transcriptional activity23 and ATP5J codes for the F6 subunit of the Fo complex of the mitochondrial ATP synthase responsible for ATP synthesis.24 VEGFC codes for the growth factor VEGFC that could be important for bone marrow changes during MPN progression. Another such regulated gene, LPP, hosts miR-28 which induces megakaryocyte differentiation defects.12 Altogether these data suggest that contrary to the cytokine-activated STAT5 in healthy controls, the persistent STAT5 activity in MPN patients may recruit p53 onto promoters of alternative target genes inducing pathological transcription in megakaryocytes that remains detectable in platelets and that might be relevant for disease.
Figure 5.
Increased expression of STAT5B and p53 target genes in the platelets of MPN patients. (a) Relative gene expression levels in the platelets of healthy controls (Ctrl, n = 11), essential thrombocythemia (ET, n = 64), primary myelofibrosis (PMF, n = 16) and polycythemia vera (PV, n = 6) patients. P-values indicate the probability that a given expression level is in the range of the normal distribution measured on control samples. (b) Number of polycythemia vera, essential thrombocythemia and primary myelofibrosis patients with increased gene expression levels (Po1e − 3). (c) Number of patients for whom zero, one, two or more genes were significantly increased. (d) Summary of the molecular alterations for patients positive for STAT5B and p53 target genes expression. Numbers in parenthesis are the number of patients in each category.
Discussion
The data presented here report a novel molecular mechanism of pathologic transcription in myeloid cancers, namely STAT5 transcriptional activation of alternative target genes through p53 recruitment. The constitutive phosphorylation of STAT5 leads to persistence in the nucleus and an increased probability to bind to GAS (gamma interferon activated sites) that are not normally high-affinity targets.9 On the other hand, STAT5 interacts with p53, as shown by co-immunoprecipitation (Figures 2c and d, Supplementary Figure S1e) and this interaction does not require tyrosine phosphorylation of STAT5, as it is maintained in the presence of JAK2 inhibition. However, the effect on LPP transcription requires active tyrosine-phosphorylated STAT5, as show by inhibition of STAT5 chromatin binding and LPP induction after treatment with ruxolitinib (Figures 2b and e) and several other JAK2 inhibitors.12 Both constitutive activation of STAT5 and its ability to associate with p53 appear to stabilize p53 binding on specific new targets in the human genome leading to transcription of a new set of genes, that are dependent on both p53 and STAT5. Several previous studies described interactions between p53 and STAT signaling. p53 inhibits transcriptional activation by STAT5, whereas STAT5 does not inhibit p53 transcriptional activity.19 In Ba/F3 cells transformed by the oncogenic interaction between the gp55 envelope protein of spleen focus-forming virus and the erythropoietin receptor, p53 transcriptional activity is inhibited by constitutive active signaling via the phosphatidylinositol-3’-kinase pathway and not via STAT5 signaling.25 In addition, STAT1 was shown to interact with p53 and enhance DNA-damageinduced apoptosis.26 Moreover, p53 and STAT5 were reported to cooperate in inducing apoptotic sensitization by oncostatin M in osteoblasts.27 Our present co-immunoprecipitation and luciferase transcription assays show that p53 interacts with STAT5 in a manner that does not involve the NH2-terminal tetramerization domain and that is negatively regulated by the COOH-terminal domain. This negative regulation via the C-terminal STAT5A domain could be meaningful, as the C-terminus of STAT5A can be alternatively spliced, proteolytically processed or posttranslationally modified by acetylation, sumoylation or serine phosphorylation and it is the domain for nuclear shuttling associated with Rho/Rac activity in myeloid leukemia cells.28 Thus, whatever modification might exist in the C-terminus of STAT5A or STAT5B, it will result in changed heterodimers and that will allow tailored STAT5-p53-regulated gene transcription.
The observation that the p53 M133K mutant activates LPP/ miR-28, but cannot bind to and activate promoters of canonical p53 target genes13 (Supplementary Figure S3) indicates that mutants of p53 associated with malignancies might preserve their ability to interact functionally with constitutively active STAT5. We show this for both M133K and the endogenous delta288 p53 variant expressed in UT7 cells (Supplementary Figure S2f and g). The inhibitory effect of p53 delta288 was much weaker than that of wild-type p53, in agreement with the requirement of the COOH-terminal sequence of p53 for effective inhibition of STAT5.19
Although, p53 M133K is mutated in its DNA-binding domain in HEL cells, it is still able to bind DNA in cooperation with an activated STAT5. This is compatible with a model, in which the presence of STAT5B increases the DNA-binding affinity of p53. The p53 M133K mutation does not seem to change the DNA-binding site specificity of p53, as we detected a consensus p53-binding site conserved by more than 70% in the DNA sequences of 65% of the p53 M133K binding peaks in HEL cells responding to JAK2 inhibition. The binding of p53 depends on constitutively active STAT5 only on chromatin, as a luciferase reporter driven by the LPP promoter can be induced by p53 alone not only in UT7 (in which active STAT5 is nuclear and bound to chromatin), but also in COS7 cells, which are known to lack STAT5 expression (Figure 1d). Thus, it will be very interesting to determine the mechanisms by which binding of p53 (or p53 mutants) is restricted on LPP promoter in the absence of STAT5.
Interestingly, STAT5-p53 potential target genes code for different classes of proteins that are normally not active in the hematopoietic system. Their activation may be dependent on the level of phosphorylation of STAT5 in the megakaryocytic progenitors of MPN patients. Epigenetic chromatin changes are well documented in MPN and they may be necessary to activate the transcription of these genes, including recruitment of p53. Several genes pathologically overexpressed in the platelets from MPN patients that are induced via the STAT5–p53 cooperation might have a role in MPN progression. This might be the case for leptin, which has been shown to be expressed and secreted by malignant hematopoietic cells and cell lines.22 Leptin signaling changes lipid and glucose metabolism and it can activate STAT5 proteins to induce further LPP/miR-28 to block megakaryocyte differentiation.12 Moreover, high patient leptin levels can contribute to a chronic inflammatory state and they are contributing to weight loss in MPN and other cancer patients, and this weight loss is reversed upon ruxolitinib treatment. The panel of six genes identified in this study and LPP can be used to test whether in MPN patients without known mutations, such genes are pathologically overexpressed in platelets. Positive results for such a gene signature would suggest molecular defects that still induce constitutive activation of STAT5. In that sense it will be important to test whether the newly described calreticulin mutants, which are associated with >70% of ET and MF negative for JAK2/MPL or other known mutations and which seem to activate also JAK229,30 are associated with such pathologic gene induction in platelets of MPN patients.
The constitutive activation of JAK2 leads to nuclear tyrosine phosphorylation of histone H321 and inhibits the PRMT5 methyltransferase activity on arginines R3 of histones H2A and H4.20 Along the same lines it is possible that JAK2 V617F may also induce epigenetic changes to allow STAT5B and p53 to bind to these alternative genomic sites. This is reminiscent of the enhanced repeat-induced variegation observed in Drosophila in presence of a gain-of-function JAK-D mutation, which disrupts the heterochromatic gene silencing, allowing transcriptional activation of genes that are not necessarily direct targets of D-STAT.31 Further work will certainly be necessary to test the possibility of epigenetic changes at the STAT5-p53 targets and how they change chromatin also in light of oncogenic activation of STAT5 and p53-induced DNA damage or apoptosis response upon chemotherapy or radiotherapy.
Materials and Methods
Plasmids
The human genomic sequence corresponding to the LPP/miR-28 alternative promoter (chr3:187871155–187872055)12 cloned in the pGL4–11[luc2P] vector was purchased from SwichGearGenomics (La Hulpe, Belgium). Site-directed mutagenesis was performed with the QuickChange Site-Directed Mutagenesis Kit (Stratagene, Waldbronn, Germany) and primers indicated in Supplementary Table S1. Expression plasmid for wildtype p53 was described previously.32 Retroviral constructs carrying an shRNA-targeting p53 or a nonsilencing control were described previously.33
Cell culture and transfection
Human erythroleukemia (HEL) and UT7 cells were maintained as described.12 Gamma2A and COS7 cells were maintained in Dulbecco’s modified Eagle’s medium with 10% fetal calf serum. Plasmid constructs and the pRL-TK internal control vector (Promega, Leiden, Germany) were transfected with Lipofectamine (Invitrogen, Gent, Belgium) following manufacturer’s instructions. Luciferase activities were assayed 48 h posttransfection with the Dual-Luciferase Reporter system (Promega) using LPP/miR-28-luciferase reporter or the spi-Luc reporter for STAT5 transcriptional activity34 or the PG13-LUC p53 reporter.35
Antibodies and reagents
ChIP grade antibodies against STAT5B, p53 and RNA pol II were purchased from Upstate/Millipore (Billerica, MA, USA, 06–554), active motif (La Hulpe, Belgium, 39553) and Diagenode (Seraing, Belgium, AC-055–100), respectively. JAK inhibitor INCB018424 (Ruxolitinib, INC4424, Novartis, Vilevorde, Belgium) was used at a 1 μM concentration for 16 h.
ChIP
Chromatin cross-linking, cell lysis and DNA shearing were performed as described.12 Immunoprecipitation was performed using the OneDay ChIP kit (Diagenode) following manufacturer’s instructions. Quantification of DNA immunoprecipitation was performed by real-time PCR using proLPP/miR promoter primers: TTGAGCACAGGACAGAGGAA and TTTTA GCCCTGAGCCTTGAA; or beta-actin primers as negative control: CCTGGC ACCCAGCACAAT and GGGCCGGACTCGTCATACT. Immunoprecipitated DNAs and input samples were amplified by two rounds of GenomePlex Complete whole genome amplification (WGA2) (Sigma, Diegem, Belgium) and hybridized on Human ChIP-chip 2.1 M Deluxe Promoter Array (Roche NimbleGen, Waldkraiburg, Germany, HG18 build).
Gene expression analysis
RNAs were extracted with the RNeasy kit (Qiagen, Venlo, Netherlands) and reverse transcribed with the Reverse Transcriptase Core kit (Eurogentec, Seraing, Belgium) and random primers. Real-time PCRs were performed on a StepOne Real-Time PCR system (Applied Biosystems, Gent, Belgium) with the MESA GREEN MasterMix (Eurogentec) and primers reported in Supplementary Table S3.
Microarray and bioinformatics analyses
Data analysis and processing was performed with the R computing software (http://www.r-project.org/). We calculated IP/input log2 ratio for each oligonucleotide from raw fluorescence values provided by Roche NimbleGen. All data set were normalized as previously described.36 For data representation, we smoothed the immunoprecipitation ratios over a 1 kb window using the Ringo package and created wig files to visualize them with the UCSC genome browser (build hg18). To find ChIP peaks, we identified regions with more than six consecutive oligonucleotides with a smoothed log2 ratio >0.5 using the Ringo package. STAT5- and p53-binding sites were predicted by the matchPWM function from the Biostrings package (http://www.bioconductor.org).
The findOverlaps function (GenomicRanges package) was used to identify coordinates of tiled promoters containing co-localized STAT5B and p53 peaks. For each promoter containing both STAT5 and p53 peaks, we calculated the minimal distance between peaks and the number of promoters as a function of minimal peak distances. Gene ontology analyses were performed with the DAVID functional annotation tool.37 Transcription factor binding site predictions were performed with MatInspector (Genomatix software suite, Munich, Germany). The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE38143.
Immunoprecipitation and western blotting
Cells were lysed either after being pretreated or not for 18 h with 10 μM INCB018424 JAK2 inhibitor in lysis buffer containing 1% NP40 and protease inhibitors. Lysates were incubated with anti-p53 (Santa Cruz, Heidelberg, Germany), empty Protein A Sepharose beads or isotype control for the anti-p53 antibody overnight and immunoprecipitates were analyzed in parallel with aliquots of total cell lysates by western blotting using antiSTAT5, anti-pY STAT5 (Cell Signaling Technology, Leiden, Netherlands), anti-p53 (Santa Cruz) or anti-beta-actin (Sigma) antibodies.
Patient samples
Platelets were isolated from MPN patients diagnosed at Hospital Henri Mondor (Paris, France), Saint-Luc Hospital (Brussels, Belgium) and Cambridge Institute for Medical Research (Cambridge University, Cambridge, UK) (Supplementary Table S4). Blood samples were collected with approval of all participating institutions’ ethical committees and informed consent was obtained in accordance with the Declaration of Helsinki. The JAK2, V617F and MPL W515 status was determined by TaqMan PCR and sequencing.12
Statistical analysis
Statistical significance was assessed by a two-tailed paired Student’s t-test unless otherwise specified.
Supplementary Material
Acknowledgements
We thank Guido Bommer for invaluable p53 reagents and extremely useful criticisms and suggestions. We are grateful for generous support to SNC from the FRS-FNRS, Belgium (Mandat d’Impulsion and FRSM), the Salus Sanguinis Foundation, the Action de Recherche Concertée projects MEXP31C1 and ARC10/15–027 of the University catholique de Louvain, Brussels, the Fondation contre le Cancer, Brussels, the PAI Programs BCHM61B5 and Belgian Medical Genetics Initiative (BeMG), Belgium and the Atlantic Philanthropies, New York (for SNC). RM was supported by SFB-F2807/SFBF4707 from the Austrian Science Fund (FWF).
Footnotes
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Constantinescu SN, Girardot M, Pecquet C. Mining for JAK-STAT mutations in cancer. Trends Biochem Sci. 2008;33:122–131. doi: 10.1016/j.tibs.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 2.Yu H, Jove R. The STATs of cancer–new molecular targets come of age. Nat Rev Cancer. 2004;4:97–105. doi: 10.1038/nrc1275. [DOI] [PubMed] [Google Scholar]
- 3.Baxter EJ, Scott LM, Campbell PJ, East C, Fourouclas N, Swanton S, et al. Acquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disorders. Lancet. 2005;365:1054–1061. doi: 10.1016/S0140-6736(05)71142-9. [DOI] [PubMed] [Google Scholar]
- 4.James C, Ugo V, Le Couedic JP, Staerk J, Delhommeau F, Lacout C, et al. A unique clonal JAK2 mutation leading to constitutive signalling causes polycythaemia vera. Nature. 2005;434:1144–1148. doi: 10.1038/nature03546. [DOI] [PubMed] [Google Scholar]
- 5.Kralovics R, Passamonti F, Buser AS, Teo SS, Tiedt R, Passweg JR, et al. A gain-offunction mutation of JAK2 in myeloproliferative disorders. New Engl J Med. 2005;352:1779–1790. doi: 10.1056/NEJMoa051113. [DOI] [PubMed] [Google Scholar]
- 6.Levine RL, Wadleigh M, Cools J, Ebert BL, Wernig G, Huntly BJ, et al. Activating mutation in the tyrosine kinase JAK2 in polycythemia vera, essential thrombocythemia, and myeloid metaplasia with myelofibrosis. Cancer Cell. 2005;7:387–397. doi: 10.1016/j.ccr.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 7.Walz C, Ahmed W, Lazarides K, Betancur M, Patel N, Hennighausen L, et al. Essential role for Stat5a/b in myeloproliferative neoplasms induced by BCR-ABL1 and JAK2(V617F) in mice. Blood. 2012;119:3550–3560. doi: 10.1182/blood-2011-12-397554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yan D, Hutchison RE, Mohi G. Critical requirement for Stat5 in a mouse model of polycythemia vera. Blood. 2012;119:3539–3549. doi: 10.1182/blood-2011-03-345215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moucadel V, Constantinescu SN. Differential STAT5 signaling by ligand-dependent and constitutively active cytokine receptors. J Biol Chem. 2005;280:13364–13373. doi: 10.1074/jbc.M407326200. [DOI] [PubMed] [Google Scholar]
- 10.Nelson EA, Walker SR, Alvarez JV, Frank DA. Isolation of unique STAT5 targets by chromatin immunoprecipitation-based gene identification. J Biol Chem. 2004;279:54724–54730. doi: 10.1074/jbc.M408464200. [DOI] [PubMed] [Google Scholar]
- 11.Casetti L, Martin-Lanneree S, Najjar I, Plo I, Auge S, Roy L, et al. Differential contributions of STAT5A and STAT5B to stress protection and tyrosine kinase inhibitor resistance of chronic myeloid leukemia stem/progenitor cells. Cancer Res. 2013;73:2052–2058. doi: 10.1158/0008-5472.CAN-12-3955. [DOI] [PubMed] [Google Scholar]
- 12.Girardot M, Pecquet C, Boukour S, Knoops L, Ferrant A, Vainchenker W, et al. miR-28 is a thrombopoietin receptor targeting microRNA detected in a fraction of myeloproliferative neoplasm patient platelets. Blood. 2010;116:437–445. doi: 10.1182/blood-2008-06-165985. [DOI] [PubMed] [Google Scholar]
- 13.Kato S, Han SY, Liu W, Otsuka K, Shibata H, Kanamaru R, et al. Understanding the function-structure and function-mutation relationships of p53 tumor suppressor protein by high-resolution missense mutation analysis. Proc Nati Acad Sci USA. 2003;100:8424–8429. doi: 10.1073/pnas.1431692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rajala HL, Eldfors S, Kuusanmaki H, van Adrichem AJ, Olson T, Lagstrom S, et al. Discovery of somatic STAT5b mutations in large granular lymphocytic leukemia. Blood. 2013;121:4541–4550. doi: 10.1182/blood-2012-12-474577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moriggl R, Gouilleux-Gruart V, Jahne R, Berchtold S, Gartmann C, Liu X, et al. Deletion of the carboxyl-terminal transactivation domain of MGF-Stat5 results in sustained DNA binding and a dominant negative phenotype. Mol Cell Biol. 1996;16:5691–5700. doi: 10.1128/mcb.16.10.5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakatake M, Monte-Mor B, Debili N, Casadevall N, Ribrag V, Solary E, et al. JAK2(V617F) negatively regulates p53 stabilization by enhancing MDM2 via La expression in myeloproliferative neoplasms. Oncogene. 2011;31:1323–1333. doi: 10.1038/onc.2011.313. [DOI] [PubMed] [Google Scholar]
- 17.Pecquet C, Diaconu CC, Staerk J, Girardot M, Marty C, Royer Y, et al. Thrombopoietin receptor down-modulation by JAK2 V617F: restoration of receptor levels by inhibitors of pathologic JAK2 signaling and of proteasomes. Blood. 2012;119:4625–4635. doi: 10.1182/blood-2011-08-372524. [DOI] [PubMed] [Google Scholar]
- 18.Stark GR, Darnell JE., Jr The JAK-STAT pathway at twenty. Immunity. 2012;36:503–514. doi: 10.1016/j.immuni.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fritsche M, Mundt M, Merkle C, Jahne R, Groner B. p53 suppresses cytokine induced, Stat5 mediated activation of transcription. Mol Cell Endocrinol. 1998;143:143–154. doi: 10.1016/s0303-7207(98)00140-3. [DOI] [PubMed] [Google Scholar]
- 20.Liu F, Zhao X, Perna F, Wang L, Koppikar P, Abdel-Wahab O, et al. JAK2V617Fmediated phosphorylation of PRMT5 downregulates its methyltransferase activity and promotes myeloproliferation. Cancer Cell. 2011;19:283–294. doi: 10.1016/j.ccr.2010.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dawson MA, Bannister AJ, Gottgens B, Foster SD, Bartke T, Green AR, et al. JAK2 phosphorylates histone H3Y41 and excludes HP1alpha from chromatin. Nature. 2009;461:819–822. doi: 10.1038/nature08448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mouzaki A, Panagoulias I, Dervilli Z, Zolota V, Spadidea P, Rodi M, et al. Expression patterns of leptin receptor (OB-R) isoforms and direct in vitro effects of recombinant leptin on OB-R, leptin expression and cytokine secretion by human hematopoietic malignant cells. Cytokine. 2009;48:203–211. doi: 10.1016/j.cyto.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 23.DeJong J, Bernstein R, Roeder RG. Human general transcription factor TFIIA: characterization of a cDNA encoding the small subunit and requirement for basal and activated transcription. Proc Natl Acad Sci USA. 1995;92:3313–3317. doi: 10.1073/pnas.92.8.3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Higuti T, Tsurumi C, Kawamura Y, Tsujita H, Osaka F, Yoshihara Y, et al. Molecular cloning of cDNA for the import precursor of human coupling factor 6 of H(+)-ATP synthase in mitochondria. Biochem Biophys Res Commun. 1991;178:793–799. doi: 10.1016/0006-291x(91)90178-a. [DOI] [PubMed] [Google Scholar]
- 25.Lin Y, Brown L, Hedley DW, Barber DL, Benchimol S. The death-promoting activity of p53 can be inhibited by distinct signaling pathways. Blood. 2002;100:3990–4000. doi: 10.1182/blood-2002-02-0504. [DOI] [PubMed] [Google Scholar]
- 26.Townsend PA, Scarabelli TM, Davidson SM, Knight RA, Latchman DS, Stephanou A. STAT-1 interacts with p53 to enhance DNA damage-induced apoptosis. J Biol Chem. 2004;279:5811–5820. doi: 10.1074/jbc.M302637200. [DOI] [PubMed] [Google Scholar]
- 27.Chipoy C, Brounais B, Trichet V, Battaglia S, Berreur M, Oliver L, et al. Sensitization of osteosarcoma cells to apoptosis by oncostatin M depends on STAT5 and p53. Oncogene. 2007;26:6653–6664. doi: 10.1038/sj.onc.1210492. [DOI] [PubMed] [Google Scholar]
- 28.Kramer OH, Moriggl R. Acetylation and sumoylation control STAT5 activation antagonistically. JAKSTAT. 2012;1:203–207. doi: 10.4161/jkst.21232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klampfl T, Gisslinger H, Harutyunyan AS, Nivarthi H, Rumi E, Milosevic JD, et al. Somatic mutations of calreticulin in myeloproliferative neoplasms. New Engl J Med. 2013;369:2379–2390. doi: 10.1056/NEJMoa1311347. [DOI] [PubMed] [Google Scholar]
- 30.Nangalia J, Massie CE, Baxter EJ, Nice FL, Gundem G, Wedge DC, et al. Somatic CALR mutations in myeloproliferative neoplasms with nonmutated JAK2. New Engl J Med. 2013;369:2391–2405. doi: 10.1056/NEJMoa1312542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shi S, Calhoun HC, Xia F, Li J, Le L, Li WX. JAK signaling globally counteracts heterochromatic gene silencing. Nat Genet. 2006;38:1071–1076. doi: 10.1038/ng1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baker SJ, Markowitz S, Fearon ER, Willson JK, Vogelstein B. Suppression of human colorectal carcinoma cell growth by wild-type p53. Science. 1990;249:912–915. doi: 10.1126/science.2144057. [DOI] [PubMed] [Google Scholar]
- 33.Bommer GT, Gerin I, Feng Y, Kaczorowski AJ, Kuick R, Love RE, et al. p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Curr Biol. 2007;17:1298–1307. doi: 10.1016/j.cub.2007.06.068. [DOI] [PubMed] [Google Scholar]
- 34.Wood TJ, Sliva D, Lobie PE, Goullieux F, Mui AL, Groner B, et al. Specificity of transcription enhancement via the STAT responsive element in the serine protease inhibitor 2.1 promoter. Mol Cell Endocrinol. 1997;130:69–81. doi: 10.1016/s0303-7207(97)00075-0. [DOI] [PubMed] [Google Scholar]
- 35.Pearson M, Carbone R, Sebastiani C, Cioce M, Fagioli M, Saito S, et al. PML regulates p53 acetylation and premature senescence induced by oncogenic Ras. Nature. 2000;406:207–210. doi: 10.1038/35018127. [DOI] [PubMed] [Google Scholar]
- 36.Borgel J, Guibert S, Li Y, Chiba H, Schubeler D, Sasaki H, et al. Targets and dynamics of promoter DNA methylation during early mouse development. Nat Genet. 2010;42:1093–1100. doi: 10.1038/ng.708. [DOI] [PubMed] [Google Scholar]
- 37.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.