Abstract
Opioids are the primary drugs used in Western medicine for pain management and palliative care. Farming of opium poppies remains the sole source of these essential medicines despite diverse market demands and uncertainty in crop yields due to weather, climate change, and pests. Here, we engineered yeast to produce the selected opioid compounds thebaine and hydrocodone starting from sugar. All work was conducted in a laboratory that is permitted and secured for work with controlled substances. We combined enzyme discovery, enzyme engineering, and pathway and strain optimization to realize full opiate biosynthesis in yeast. The resulting opioid biosynthesis strains required expression of 21 (thebaine) and 23 (hydrocodone) enzyme activities from plants, mammals, bacteria, and yeast itself. This is a proof-of-principle, and major hurdles remain before optimization and scale up could be achieved. Open discussions of options for governing this technology are also needed in order to responsibly realize alternative supplies for these medically relevant compounds.
Opioids are an important class of medicines that include the analgesic morphine and the antitussive codeine. The World Health Organization (WHO) classifies these compounds as essential medicines due to their use in treating severe pain, in pain management, and in palliative care (1). In the developing world there are shortages of painkillers; the WHO has estimated that 5.5 billion people have “low to nonexistent access to treatment for moderate or severe pain” (2).
All natural opiates (e.g. morphine and codeine) and semi-synthetic opioids (e.g. oxycodone, hydrocodone, and hydromorphone) are currently derived from the opium poppy (Papaver somniferum). Approximately 100,000 hectares of opium poppy are cultivated annually to yield poppy straw containing over 800 tons of opiates, primarily morphine and thebaine, to meet licit medical and scientific demand (3). The majority of poppy-derived morphine and thebaine is chemically converted into higher value compounds, including codeine, oxycodone, and hydrocodone. Industrial poppy farming is susceptible to environmental factors such as pests, disease, and climate, which can introduce instability and variability into this geographically-concentrated supply chain and is resulting in growing pressure to diversify supply (4). Despite diverse market demands and increasing supply risks poppy farming remains the sole source of opioids, in part because chemical synthesis of these complex molecules is not commercially competitive. Approximately 30 chemical syntheses of morphine and derivatives have been reported (5) but none are feasible at scale.
A microbial-based manufacturing process for opioids or opioid precursors, which are part of the larger class of benzylisoquinoline alkaloids (BIAs), has the potential to address many of the challenges associated with the poppy-based supply chain. Industrial cultivation of microorganisms, such as the baker’s yeast Saccharomyces cerevisiae, occurs over days versus the annual cultivation of plants. Also, because microbes are grown in closed fermentation vessels, the production process is not susceptible to external environmental factors and could provide greater consistency in product composition and impurity profiles across batches.
In recent years, researchers have engineered yeast to produce a variety of plant-based natural products (6), most notably artemisinic acid, a precursor to the antimalarial drug artemisinin (7). Semi-synthetic production of artemisinin has now reached the market, meeting up to one-third of global need (8, 9). Yeast-based production of artemisinic acid required the introduction of three to six heterologous plant genes and numerous genetic modifications to increase productivity (7, 9). While advances in synthetic biology have increased the complexity of plant pathways that can be reconstructed (6), all efforts to engineer yeast to produce BIAs downstream of (S)-reticuline, including morphinans, have relied on an external supply of BIA precursors (10–15). Escherichia coli (16) and, very recently, yeast (17, 18) have been engineered to produce early BIA intermediates de novo, suggesting that yeast might be capable of synthesizing opioids from simple carbon and nitrogen sources. We and others engineered the first part of the biosynthetic pathway, from tyrosine to (S)-reticuline (17, 18). Separately, we engineered a second part of the pathway, from thebaine to morphine (13) and later others from (R)-reticuline to codeine (15). However, functionally expressing the more than 20 heterologous genes required for complete biosynthesis of these complex molecules has been challenging because of the decreases in titer observed with each additional enzymatic step. Also, key enzyme(s) that epimerize the (S)-benzylisoquinoline scaffold to the (R)-enantiomer, which is the biosynthetic precursor of the promorphinan and morphinan scaffolds, have remained unknown even after decades of study until recently identified by two groups (19, 20) and by our team as described below
A decade ago, when we began work to realize total biosynthesis of opioids in yeast, we were motivated by the many foreseeable benefits yet mindful of potential negative impacts. Most specifically, we were and remain concerned that a yeast-based opioid supply might contribute to opioid abuse (21, 22). Thus, before starting this project, we sought and received permission to carry it out via Stanford University’s institutional research registration with the U.S. Drug Enforcement Agency (DEA). Gaining permission required (i) background screening for researchers handling Schedule II compounds or yeast strains capable of making such compounds; (ii) detailed protocols limiting fermentation volumes and compound concentrations and including provisions for culture and product destruction and disposal immediately following experiments; (iii) increased physical containment for the strains and controlled compounds; (iv) increased laboratory security; and (v) explicit management and reporting. Taken together, these requirements reduce the chance that any compounds or strains generated in our research would directly enable individuals to abuse opioids.
We first built a yeast strain to produce (S)-reticuline, a key biosynthetic intermediate to many downstream BIAs including the morphinans. This strain was built with a new modular genetic design that incorporated modifications designed to divert greater carbon flux through tyrosine to (S)-reticuline. The reticuline biosynthetic pathway was split into four genetic modules that contain the coding sequences for 17 biosynthetic enzymes (Fig. 1A, figs. S1–2, table S1). We selected chromosomal regions from which we expected no growth defect and active expression as integration loci (23–25). A precursor overproduction module (I) designed to increase accumulation of L-tyrosine and 4-hydroxyphenylacetaldehyde (4-HPAA) encodes the overexpression of three or four yeast proteins – mutants of 3-deoxy-D-arabino-2-heptulosonic acid 7-phosphate (DAHP) synthase and chorismate mutase (Aro4pQ166K, Aro7pT226I) that are less inhibited by L-tyrosine, and transketolase (TKL1p) only, or additionally phenylpyruvate decarboxylase (Aro10p). A tetrahydrobiopterin (BH4) module (II) designed to synthesize and recycle this mammalian redox cofactor encodes expression of four proteins from Rattus norvegicus – sepiapterin reductase (SepR), 6-pyruvoyl tetrahydrobiopterin synthase (PTPS), quinonoid dihydropteridine reductase (QDHPR), and pterin carbinolamine dehydratase (PCD). An (S)-norcoclaurine module (III) designed to synthesize the first BIA backbone molecule encodes expression of four proteins – a mutant (R37E, R38E, W166Y) of tyrosine hydroxylase (TyrHWR) that is less inhibited by L-tyrosine and catecholamines and the BH4 salvage enzyme dihydrofolate reductase (DHFR) both from R. norvegicus, DOPA decarboxylase (DoDC) from the bacteria Pseudomonas putida, and norcoclaurine synthase (NCS) from the plant Coptis japonica. An (S)-reticuline module (IV) designed to synthesize the key BIA branchpoint molecule encodes expression of five plant proteins – norcoclaurine 6-O-methyltransferase (6OMT), coclaurine-N-methyltransferase (CNMT), 4’-O-methyltransferase (4’OMT), and cytochrome P450 reductase (CPR) all from P. somniferum, and N-methylcoclaurine hydroxylase (NMCH) from Eschscholzia californica. The enzyme variants were selected on the basis of examined activities in engineered yeast (11, 18) and incorporated the addition of several new activities (i.e. Aro10p, DHFR) to increase flux to reticuline biosynthesis.
Fig 1. Engineered biosynthetic pathway for de novo production of thebaine and hydrocodone and optimization of reticuline-producing platform strains.
(A) Biosynthetic scheme for production of thebaine and hydrocone from sugar. Thebaine is a starting material for many opioid drugs through biosynthetic and semi-synthetic routes. Block arrows indicate enzyme-catalyzed steps. Light grey arrows, unmodified yeast enzymes; dark grey arrows, overexpressed and modified yeast enzymes; purple arrows, mammalian (Rattus norvegicus) enzymes; orange arrows, bacterial (Pseudomonas putida) enzymes; green arrows, plant (Papaver somniferum, Papaver bracteatum, Coptis japonica, Eschscholzia californica) enzymes. Yellow outline indicates DRS-DRR; red outline indicates engineered SalSyn. E4P, erythrose 4-phosphate; PEP, phosphoenolpyruvate; DAHP, 3-deoxy-D-arabino-2-heptulosonic acid 7-phosphate; 4-HPP, 4-hydroxyphenylpyruvate; 4-HPAA, 4-hydroxyphenylacetaldehyde; BH4, 5,6,7,8-tetrahydrobiopterin; Tkl1p, transketolase; CPR, cytochrome P450 reductase; Aro4pQ166K, DAHP synthase; Aro1p, pentafunctional arom enzyme; Aro2p, bifunctional chorismate synthase and flavin reductase; Aro7pT226I, chorismate mutase; Tyr1p, prephenate dehydrogenase; Aro8p, aromatic aminotransferase I; Aro9p, aromatic aminotransferase II; Aro10p, phenylpyruvate decarboxylase; TyrHWR, feedback inhibition-resistant tyrosine hydroxylase (R37E, R38E, W166Y); DoDC, L-dopa decarboxylase; NCS, (S)-norcoclaurine synthase; 6OMT, norcoclaurine 6-O-methyltransferase; CNMT, coclaurine N-methyltransferase; NMCH, N-methylcoclaurine hydroxylase; 4’OMT, 3’-hydroxy-N-methylcoclaurine 4’-O-methyltransferase; DRS-DRR, 1,2-dehydroreticuline synthase-1,2-dehydroreticuline reductase; SalSyn, salutaridine synthase; SalR, salutaridine reductase; SalAT, salutaridinol 7-O-acetyltransferase; T6ODM, thebaine 6-O-demethylase; morB, morphinone reductase. Details on the BH4 biosynthesis, recycling, and salvage pathway, conversion of (S)-norcoclaurine to (S)-reticuline, and genetic pathway modules are provided in figs. S1–2. (B) Optimization of the reticuline-producing platform strain through pathway and strain engineering. Reticuline in the growth media was analyzed by LC-MS/MS MRM and quantified with an external standard curve. Error bars represent standard deviation of three biological replicates (C) Chiral analysis of reticuline produced by the platform strain. Reticuline was isolated from the growth media of strain CSY1061 and separated on a chiral column. This chromatogram is one of two similar traces from replicate yeast cultures. The chromatogram was smoothed using a 7-point boxcar moving average.
The BIA modules were integrated into a wild-type haploid CEN.PK2 strain. We assayed reticuline production by growing yeast strains in minimal synthetic complete media supplemented with ascorbic acid without ammonium sulfate for 72 h (fig. S3) and analyzing the growth media for BIA molecules by liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS) (table S2). A minimal reticuline-producing strain (CSY1057, table S3), which incorporated modules II-IV, produced 12.3 µg/L reticuline (Fig. 1B, fig. S4A, table S4). The addition of module I to the strain, increasing BIA precursor supply, resulted in a 1.6-fold improvement to reticuline accumulation with or without Aro10p (CSY1059, 20.0 µg/L; CSY1058, 20.7 µg/L). We observed nearly complete consumption of L-DOPA (90 µg/L, fig S4B, table S4), substantial accumulation of dopamine (10 mg/L, fig. S4C), and accumulation of 3’-hydroxy-N-methylcoclaurine (fig. S4D, E) by LC-MS/MS for the strain harboring modules I–IV (CSY1059). We hypothesized that increasing expression of (i) NCS would increase conversion of dopamine and the native yeast metabolite 4-HPAA to norcoclaurine and downstream products, (ii) TyrHWR would replenish the supply of dopamine, and (iii) 4’OMT would reduce accumulation of 3’-hydroxy-N-methylcoclaurine and increase flux to reticuline. Thus, we designed a bottleneck module (V), which encodes the overexpression of three proteins – TyrHWR, 4’OMT, and NCS – by incorporation of additional gene copies. The module was integrated into CSY1059 and designed to knockout the native ZWF1 gene (zwf1Δ; CSY1061) or to integrate into a separate locus (CSY1060). A yeast platform strain with the addition of module V into the zwf1 locus resulted in a further 4-fold improvement to reticuline accumulation (82 µg/L, Fig. 1B, table S4) and a corresponding 2-fold decrease in accumulated 3’-hydroxy-N-methylcoclaurine relative to CSY1059 (fig. S4D).
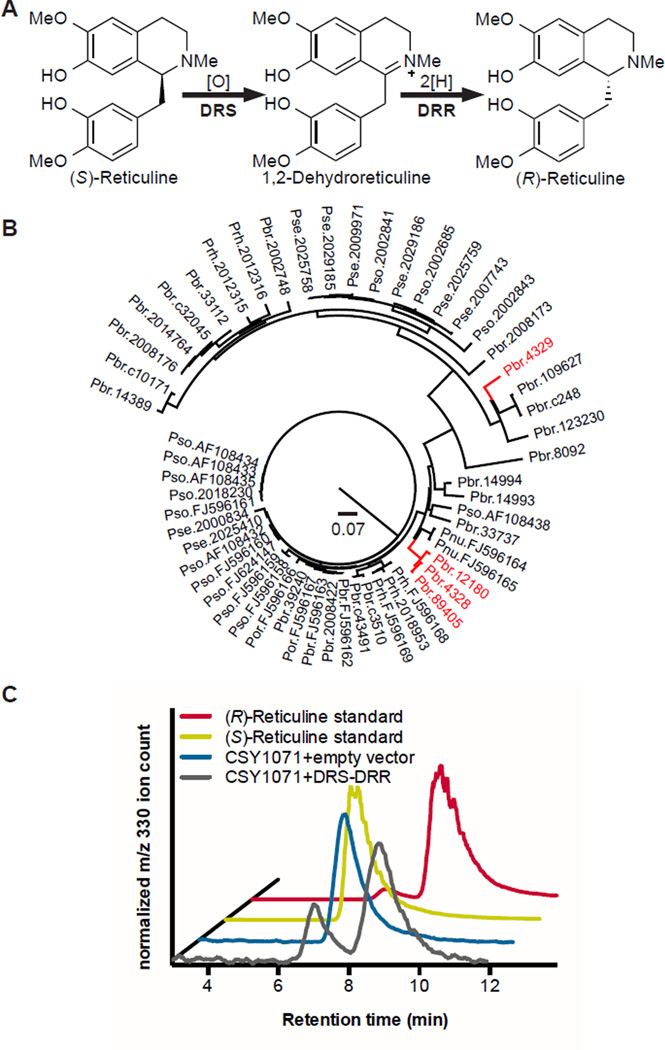
Reticuline produced by the yeast platform strain CSY1061 was isolated by reverse phase HPLC and analyzed by chiral LC-MS. The chiral analysis indicated that the majority of the reticuline produced is the (S)-enantiomer (Fig. 1C), as was expected due to the stereospecificity of the NCS-catalyzed condensation. The production of primarily the (S)-enantiomer in our platform strain corroborates similar observations from other bacteria and yeast engineered with the three methyltransferase enzymes, even when fed racemic substrates (10, 15–17). Opium poppy has the unique ability to convert (S)-reticuline to (R)-reticuline, from which the morphinan alkaloids are derived (26, 27). Although the epimerase activity has been characterized by extensive isotope feeding and biochemical studies to proceed via oxidation to a Schiff base intermediate and stereospecific reduction (Fig. 2A), the 1,2-dehydroreticuline synthase (DRS) and 1,2-dehydroreticuline reductase (DRR) enzyme(s) had not been isolated and sequenced when we began our study (27–29). While we were preparing this manuscript for submission, one group reported the discovery of this enzyme in P. somniferum by characterizing mutant alleles from chemically mutagenized opium poppy plants (19). While our manuscript was under review, another group reported using plant transcriptome databases to identify candidates and then cloned the gene from P. somniferum cDNA (20). Our approach instead leveraged plant transcriptome databases, DNA synthesis, and the engineered (S)-reticuline-producing yeast strains, thus not requiring access to physical plant material.
Fig. 2. DRS-DRR converts (S)-reticuline to (R)-reticuline.
(A) Biosynthetic scheme for the reaction catalyzed by DRS-DRR. (B) Identification of DRS-DRR via bioinformatic analysis of COR-like sequences. Bioinformatic query was COR VIGS sequence and subject sequences were the P. bracteatum PhytoMetaSyn transcriptome; P. bracteatum P. setigerum, P. somniferum, P. rhoeas 1000 Plants Project transcriptomes; and all deposited sequences in Genbank belonging to Papaveraceae. The scale bar indicates amount of genetic change in amino acid substitutions per site. Branches highlighted in red indicate sequences containing both CYP and COR-like domains. Phylogenetic tree was generated using ClustalX bootstrap NJ tree with 1000 trials. (C) Chiral analysis of reticuline produced by yeast strains expressing and not expressing DRS-DRR. Chiral analysis of reticuline accumulated in the growth media of strain CSY1071 with empty vector or DRS-DRR (pCS3301) was performed as described in Fig. 1C. This chromatogram is one of two similar traces from replicate yeast cultures. The chromatogram was smoothed using a 7-point boxcar moving average.
More specifically, we noted that two independent plant gene silencing studies found that codeinone reductase (COR) knockdown results in reticuline accumulation, and in one case specifically (S)-reticuline accumulation (30, 31). We hypothesized that a COR-like enzyme may catalyze the stereospecific reduction and used the published virus-induced gene silencing sequence as a BLAST query against Papaver species in the 1000 Plants Project (32) and PhytoMetaSyn (33, 34) transcriptome databases. Hit identity was determined by a reverse BLAST search against sequences deposited in GenBank. Of the 38 COR-like sequences identified that were also greater than 300 nts in length, four had a cytochrome P450 oxidase 82Y1-like domain and a COR-like domain in a single open reading frame (DRS-DRR, Fig. 2B). We considered that this natural fusion protein could catalyze both the oxidation and reduction necessary for (S)-reticuline epimerization. We propose that the oxidation to 1,2-dehydroreticuline may occur via either a carbinolamine or enamine intermediate, and that 1,2-dehydroreticuline is then stereospecifically reduced to (R)-reticuline by the COR-like DRR domain (fig. S5). To identify additional variants of this coding sequence and determine how widespread it is in nature, we used the amino acid sequence from P. bracteatum DRS-DRR (Pbr.89405) to search both databases by translated BLAST nucleotides (tBLASTn), and identified a total of five apparent full-length and 10 partial unique sequences that harbored both domains (fig. S6), which originated from P. somniferum (opium poppy), Papaver setigerum (poppy of Troy), P. bracteatum (Iranian poppy), or Chelidonium majus (greater celandine), despite searching all sequences in the PhytoMetaSyn (67 plant species) and the 1000 Plants Project transcriptome databases (1328 assemblies derived from a few hundred plant species). From this secondary search (fig. S6), we identified a P. somniferum DRS-DRR sequence of interest, Pso.2062398, which was a full-length sequence that had consensus with several individual transcriptome hits.
To determine whether the identified DRS-DRR enzyme possesses epimerase activity, we characterized the DRS-DRR enzyme in the context of a yeast strain engineered to produce (S)-reticuline from fed rac-norlaudanosoline (CSY1071, fig. S1C) (11). In preliminary experiments, we screened the three variants from P. bracteatum that clustered together in the initial search – Pbr.89405, Pbr.12180, and Pbr.4328 – in strain CSY1071 with low-copy plasmids harboring expression cassettes for yeast codon-optimized DRS-DRR and yeast codon-optimized P. somniferum salutaridine synthase (yPsSalSyn). Codon optimization of all synthetic genes was performed by Life Technologies (35). When fed 1 mM norlaudanosoline, only the strain with DRS-DRR variant Pbr.89405 produced substantial salutaridine. We cultured strain CSY1071 containing the low-copy plasmid harboring this DRS-DRR (Pbr.89405, pCS3301) with 1 mM rac-norlaudanosoline for 72 h, and isolated reticuline from the growth media for chiral LC-MS analysis. In strains expressing the DRS-DRR, over half of the reticuline produced was the (R)-enantiomer, whereas exclusively (S)-reticuline was detected in strains lacking the DRS-DRR (Fig. 2C).
We next examined the activity of DRS-DRR variants in the context of the downstream conversion steps to thebaine, the first morphinan alkaloid in opiate biosynthesis. In preliminary experiments, yeast codon-optimized salutaridine reductase (SalR) variants from P. bracteatum and P. somniferum and site-directed mutants that were reported to reduce substrate inhibition and increase Vmax (36, 37) were examined for their ability to catalyze conversion of salutaridine to thebaine with yeast codon-optimized salutaridinol acetyltransferase (SalAT) variants from P. somniferum, P. bracteatum, and P. orientale. The P. bracteatum SalR (PbSalR) and P. somniferum SalAT (PsSalAT) exhibited the highest activities in yeast (fig. S7A, B). A yeast artificial chromosome (YAC, pCS3308) encoding expression cassettes for yPsSalSyn, PbSalR, and PsSalAT was assembled into strain CSY1071, and DRS-DRR variants were expressed from low-copy plasmids (pCS3300–3305). The resulting strains were assayed by feeding 1 mM rac-norlaudanosoline for 72 h and the growth media was analyzed for thebaine production. P. bracteatum and P. somniferum DRS-DRR (Pbr.89405, Pso.2062398) resulted in similar thebaine production (fig. S7C), and the P. bracteatum DRS-DRR (PbDRS-DRR) was used in subsequent experiments. Expression cassettes for the four genes were assembled into a YAC (pCS3309) in strain CSY1071, and the resulting strain was assayed for thebaine production from fed rac-norlaudanosoline. This strain produced thebaine at a concentration of 17 µg/L when cultured with 1 mM rac-norlaudanosoline for 96 h (Fig. 3C, table S5). However, substantial accumulation of the intermediate reticuline (~660 µg/L) was observed.
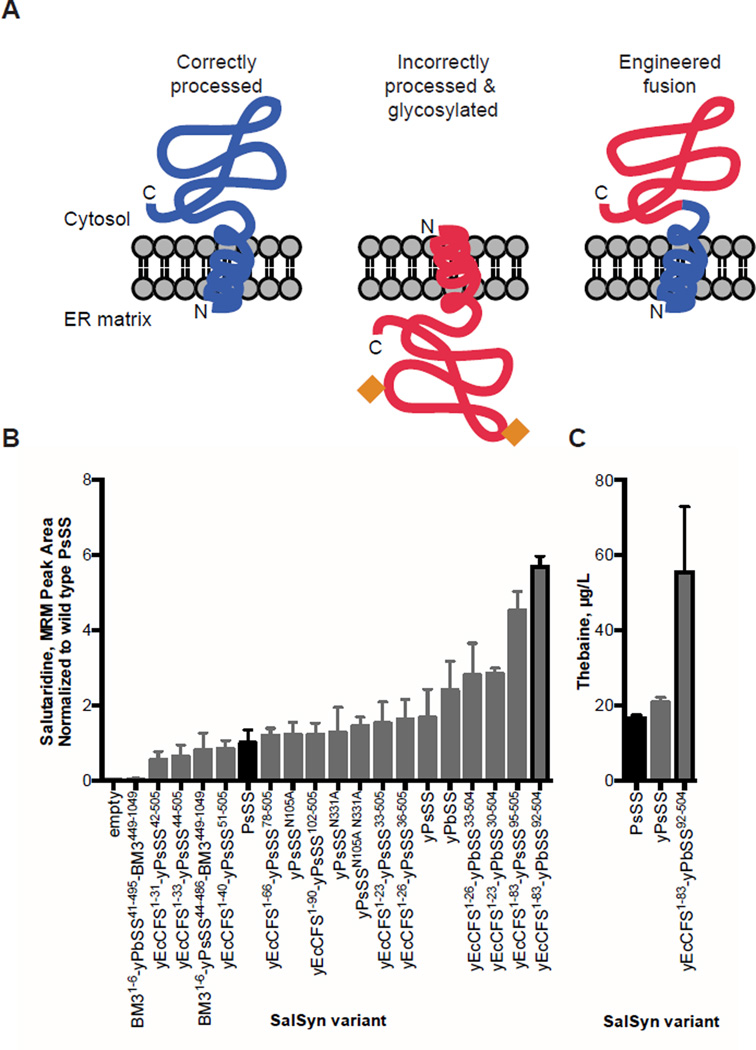
Fig. 3. Engineered SalSyn chimeras improve conversion of (R)-reticuline to salutaridine.
(A) Schematic of the chimeric SalSyn engineering strategy to address incorrect processing and glycosylation of the wild-type SalSyn in yeast. Orange diamonds represent glycosylation. (B) Comparison of salutaridine produced from SalSyn variants, site-directed glycosylation mutants, and engineered fusions in yeast. Yeast strains expressing the indicated SalSyn variant were fed 10 µM (R)-reticuline, and the growth media was analyzed by LC-MS/MS MRM. Peak areas were normalized to wild-type SalSyn (black). (C) Comparison of thebaine produced from SalSyn variants in yeast. Yeast strains were fed 1 mM rac-norlaudanosoline, and thebaine in the growth media was quantified by LC-MS/MS MRM with an external standard curve. Bars outlined in black denote wild-type and best engineered variant. Error bars are standard deviation of at least three biological replicates.
As DRS-DRR is fairly efficient in the conversion of (S)- to (R)-reticuline (Fig. 2C), the accumulation of reticuline indicated that the conversion of (R)-reticuline to salutaridine, catalyzed by SalSyn, warranted further optimization. Western blot analysis indicated that yeast-expressed SalSyn was present as three distinct apparent molecular weight forms, while SalSyn transiently expressed in Nicotiana benthamiana (tobacco) was present primarily at the lowest apparent molecular weight (fig. S8A). Site-directed mutagenesis of three potential N-linked glycosylation sites (N-X-T/S) indicated that the three bands arose from glycosylation of the protein at two sites, N105 and N331. N-linked glycosylation in yeast is indicative of incorrect N-terminal sorting of the nascent SalSyn transcript to the lumen of the endoplasmic reticulum (ER), where it is N-glycosylated rather than anchoring the N-terminus in the outer ER membrane and maintaining the catalytic domain in the cytosol, as is typical of microsomal CYPs (Fig. 3A, fig. S8B) (38, 39). We hypothesized that this mis-processing reduced SalSyn activity in yeast. However, modifying the glycosylation pattern of SalSyn by mutating the glycosylation sites reduced conversion of (R)-reticuline to salutaridine relative to the wild-type yeast codon-optimized enzyme (Fig. 3B).
We performed protein engineering to correct N-terminal sorting of the nascent SalSyn transcript, prevent N-linked glycosylation, and improve the enzyme’s activity in yeast. Cheilanthifoline synthase (CFS) is a plant cytochrome P450 that is 61–68% identical to SalSyn, exhibits high activity when expressed in yeast (14) and is not glycosylated in yeast despite having one N-X-T/S site identical to the SalSyn sequence (fig. S8C). We designed yeast codon-optimized coding sequences for chimeric proteins with one or more N-terminal alpha-helices from CFS replacing those of SalSyn variants from P. somniferum and P. bracteatum, with junction points for the fusions selected on the basis of amino acid alignments and/or protein secondary structure motifs. Western blot analysis of the chimeric proteins indicated that several of the engineered SalSyn enzymes were present as a single band in yeast, similar to the expression pattern observed for the plant-expressed parent enzyme (fig. S8D). The data indicated that the mis-processing of the nascent protein in yeast that resulted in N-linked glycosylation was repaired by the engineered fusions. As an alternative strategy, the coding sequence for the SalSyn CYP domain was cloned in place of the CYP domain in the cytosolic Bacillus megaterium P450 monooxygenase CYP102A1 (BM3), resulting in a chimeric protein with fused CYP and cytochrome P450 reductase domains. The chimeric SalSyn proteins were expressed from a low-copy plasmid in CSY1071 and assayed for salutaridine production from fed (R)-reticuline. Several of the engineered SalSyn variants exhibited improved activity relative to both the wild-type and codon-optimized enzymes, with the engineered P. bracteatum variant yEcCFS1–83-yPbSalSyn92–504 exhibiting ~6-fold greater conversion of (R)-reticuline to salutaridine (Fig. 3B) and over 3-fold greater conversion of rac-norlaudanosoline to thebaine (60 µg/L; Fig. 3C, table S5) than wild-type PsSalSyn.
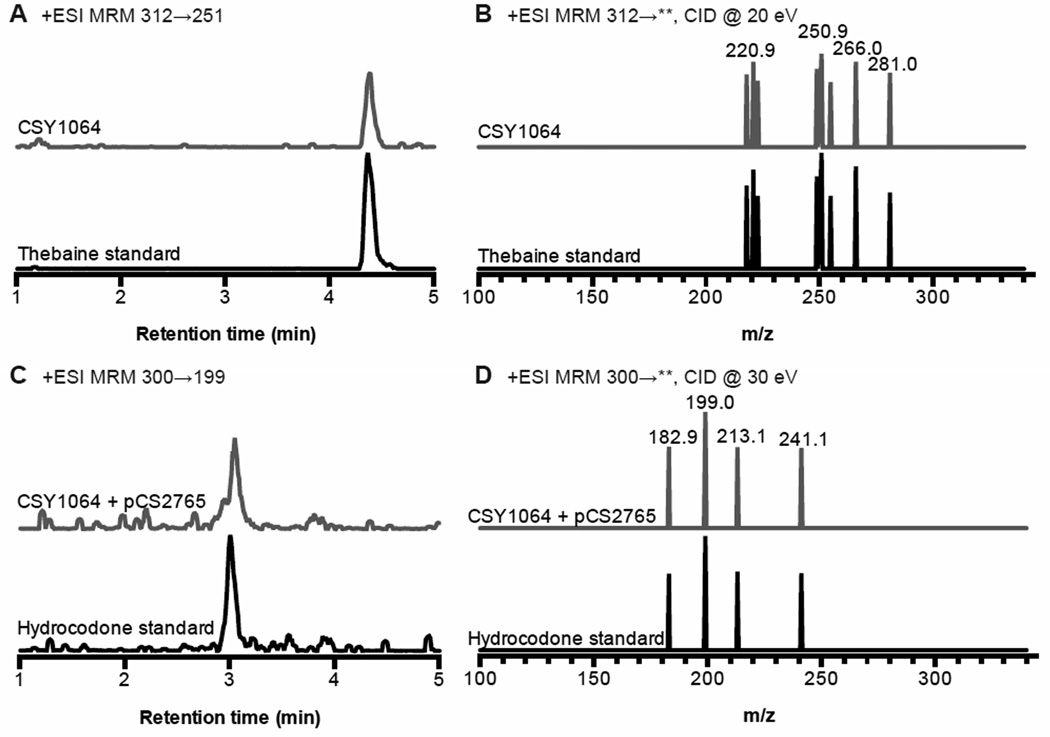
To engineer a yeast strain that produces thebaine from simple carbon and nitrogen sources we designed a thebaine module (VI) that encodes the expression of the best enzyme variants identified in our work – PbDRS-DRR, yEcCFS1–83-yPbSalSyn92–504, PbSalR, and PsSalAT – to convert (S)-reticuline to the morphinan alkaloid thebaine. Thebaine is extracted from opium poppy for use in semi-synthesis of a number of opioids. This module was added to the reticuline producing platform strain (CSY1060) as a chromosomal integration (CSY1064). The resulting strains were cultured in minimal media for 120 h and the growth media assayed for thebaine (Fig. 4A, B). These strains containing 21 heterologous enzymes, overexpression of 2 native enzymes, and inactivation of one native enzyme produced 6.4 ± 0.3 µg/L thebaine (table S6). We further extended the reconstructed biosynthetic pathway to a downstream opioid drug hydrocodone (Fig. 1A), which is a main component in the second most dispensed prescription medicine in the United States (40). A hydrocodone module (VII), which encodes the expression of thebaine 6-O-demethylase (T6ODM) from P. somniferum and morphine reductase (morB) from P. putida M10 (13), was introduced as a YAC (pCS2765) into the thebaine producing strain CSY1064. The resulting strain was cultured in minimal media with 50 mM 2-oxoglutarate to support T6ODM activity for 120 h and the growth media assayed for opioid compounds (Fig. 4C, D, table S6). The engineered yeast were able to produce low levels of hydrocodone, ~0.3 µg/L.
Fig. 4. Complete biosynthesis of the opiate thebaine and semi-synthetic opioid drug hydrocodone in yeast.
(A) Chromatograms of thebaine detected in CSY1064 media and 7.8 µg/L (25 nM) thebaine standard. (B) Spectra of eight multiple reaction monitoring (MRM) transitions of thebaine produced by engineered yeast and standard. (C) Chromatograms of hydrocodone detected in CSY1064+pCS2765 media and 0.3 µg/L (1 nM) hydrocodone standard. (D) Spectra of four MRM transitions of hydrocodone produced by engineered yeast and standard. Growth media was analyzed for opioids by LC-MS/MS MRM. Traces are representative of four biological replicates.
This work represents the complete biosynthesis of opiates in a heterologous host starting from central metabolism. Through our synthetic approach we validated the capability of DRS-DDR variants from different plants to catalyze the (S)- to (R)-epimerization of reticuline in the context of the heterologous opiate biosynthetic pathway. The engineering of yeast able to convert central metabolites to the complex pentacyclic morphinan scaffold required enzyme engineering to correct the processing and increase the activity of the key pathway cytochrome P450 leading to the promorphinan scaffold (SalSyn), and pathway and strain optimization, including the expression of 21 heterologous enzymes from plants, mammals, bacteria, and yeast, overexpression of 2 native yeast enzymes, and deletion of one native yeast gene. The current report represents a proof-of-principle for generating morphinan scaffolds de novo in yeast, and opens the possibility of derivatizing these and other molecules by new biosynthetic or semi-synthetic routes to improve their therapeutic properties.
Over a 100,000-fold improvement in fermentation titer (i.e., to ~5 g/L) would be required for yeast-based production of opioids to be a feasible alternative to poppy farming for commercial production. Thus, the strains reported here would not be suitable for commercial scale up. Future engineering efforts could take advantage of the pathway’s role as an electron sink for fermentative production to direct greater electron and carbon flux to the opiates rather than ethanol or other fermentation products. At commercial productivities, ~5 mL of yeast grown over several days would provide one dose of pain medication, which is currently sourced from 0.2 m2 of poppy field land over the course of a year; sourcing opiates from sugar and yeast instead of opium poppy could decrease the overall land area required for production by over 500-fold.
There is some concern that biosynthesis of opioids in yeast may soon lead to ‘home brew’ opiates (41). The production levels achieved here under controlled fermentation conditions do not enable home brew of these drugs and also are not economically competitive with poppy farming for supplying either licit or illicit markets. Specifically, at the titers reported here (<1 µg/L), a single dose of hydrocodone, as used in Vicodin® (5 mg), would require thousands of liters of fermentation broth, which no home brewer would reasonably pursue. Such improvements are not merely a matter of fermentation scale up and would require additional research to achieve the necessary strain and pathway improvements. Thus, the work reported here does not provide a ‘recipe’ for making opioid drugs in a manner that directly undermines public health or security. Nevertheless, as a prepositioned safeguard of future public health, our yeast strains only produce opioids (e.g., hydrocodone, thebaine) with reduced potential for diversion to illicit markets due to the added steps and cost of chemically converting these specific compounds to heroin. While such strains could potentially be further engineered to produce morphine directly, prior work to convert thebaine to morphine only realized a 1.5% yield (13). Thus, a strain that converts sugar to morphine would require over a 7 million-fold improvement in overall yield relative to the work reported here.
Nevertheless, substantially improved production of opioids via yeast should be expected in the next several years. More broadly, our work highlights the potential of yeast as a chassis for bio-based production of many complex chemicals and materials. Synthetic biology is poised to replace or supplement many supply chains with advanced bio-based manufacturing. A greatly expanded capacity to build with biology will contribute to changes in land and natural resource use, employment, and policy. Practical strategies that address concerns while enabling innovation and the realization of benefits must be developed now in order to secure our future bioeconomy. Given the complexity and diversity of both the potential concerns and possible benefits we would strongly endorse an open deliberative process that develops options for the governance (42) of medicinal compound biosynthesis before economically competitive processes are realized.
Supplementary Material
Acknowledgments
We thank Stanford Cell Sciences Imaging Facility for providing fluorescence microscopy access (Leica SP5, NIH grant SIG number: 1S10RR02557401) and training; Agilent Technologies for an award through their Global Academic Research Support Program; T. Kutchan, P. Facchini for contributing Papaver samples to the 1KP and PMS transcriptome projects, respectively; W. Lau of the Sattely laboratory for training, growing plants, and helpful discussions regarding plant experiments; Y. Li, Y-H. Wang, A.P. Klein, D. Endy for valuable feedback in the preparation of the manuscript. This work was supported by the National Institutes of Health (grant to C.D.S., AT007886), National Science Foundation (fellowship to S.G., I.J.T.), ARCS Foundation (fellowship to I.J.T.), and Stanford University (fellowship to S.G., REU fellowship to M.F.I.). Stanford University has a pending patent application on this work on which SG, KT, IJT, and CDS are inventors. CDS, KT, IJT are co-founders of Antheia, Inc.
References and Notes
- 1.World Health Organization. Geneva, Switzerland: 2013. 18th WHO essential medicines list. [Google Scholar]
- 2.Seya MJ, Gelders SF, Achara OU, Milani B, Scholten WK. A first comparison between the consumption of and the need for opioid analgesics at country, regional, and global levels. J Pain Palliat. Care Pharmacother. 2011;25:6–18. doi: 10.3109/15360288.2010.536307. [DOI] [PubMed] [Google Scholar]
- 3.International Narcotics Control Board (INCB) Narcotic drugs: Estimated world requirements for 2015 - Statistics for 2013. 2014 [Google Scholar]
- 4.Bradsher K. Shake-up on opium island. New York Times. 2014 [Google Scholar]
- 5.Reed JW, Hudlicky T. The quest for a practical synthesis of morphine alkaloids and their derivatives by chemoenzymatic methods. Acc. Chem. Res. 2015;48:674–687. doi: 10.1021/ar500427k. [DOI] [PubMed] [Google Scholar]
- 6.Siddiqui MS, Thodey K, Trenchard I, Smolke CD. Advancing secondary metabolite biosynthesis in yeast with synthetic biology tools. FEMS Yeast Res. 2012;12:144–170. doi: 10.1111/j.1567-1364.2011.00774.x. [DOI] [PubMed] [Google Scholar]
- 7.Ro DK, et al. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature. 2006;440:940–943. doi: 10.1038/nature04640. [DOI] [PubMed] [Google Scholar]
- 8.PATH. Stabilizing the antimalarial drug supply. 2014 [Google Scholar]
- 9.Paddon CJ, Keasling JD. Semi-synthetic artemisinin: A model for the use of synthetic biology in pharmaceutical development. Nat. Rev. Microbiol. 2014;12:355–367. doi: 10.1038/nrmicro3240. [DOI] [PubMed] [Google Scholar]
- 10.Minami H, et al. Microbial production of plant benzylisoquinoline alkaloids. Proc. Natl. Acad. Sci. USA. 2008;105:7393–7398. doi: 10.1073/pnas.0802981105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hawkins KM, Smolke CD. Production of benzylisoquinoline alkaloids in Saccharomyces cerevisiae. Nat. Chem. Biol. 2008;4:564–573. doi: 10.1038/nchembio.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fossati E, et al. Reconstitution of a 10-gene pathway for synthesis of the plant alkaloid dihydrosanguinarine in Saccharomyces cerevisiae. Nat. Commun. 2014;5:3283. doi: 10.1038/ncomms4283. [DOI] [PubMed] [Google Scholar]
- 13.Thodey K, Galanie S, Smolke CD. A microbial biomanufacturing platform for natural and semisynthetic opioids. Nat. Chem. Biol. 2014;10:837–844. doi: 10.1038/nchembio.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trenchard IJ, Smolke CD. Engineering strategies for the fermentative production of plant alkaloids in yeast. Metab. Eng. 2015;30:96–104. doi: 10.1016/j.ymben.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fossati E, Narcross L, Ekins A, Falgueyret JP, Martin VJ. Synthesis of morphinan alkaloids in Saccharomyces cerevisiae. PLoS One. 2015;10:e0124459. doi: 10.1371/journal.pone.0124459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakagawa A, et al. A bacterial platform for fermentative production of plant alkaloids. Nat. Commun. 2011;2:326. doi: 10.1038/ncomms1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DeLoache WC, et al. An enzyme-coupled biosensor enables (S)-reticuline production in yeast from glucose. Nat. Chem. Biol. 2015;11:465–471. doi: 10.1038/nchembio.1816. [DOI] [PubMed] [Google Scholar]
- 18.Trenchard IJ, Siddiqui MS, Thodey K, Smolke CD. De novo production of the key branch point benzylisoquinoline alkaloid reticuline in yeast. Metab. Eng. 2015 doi: 10.1016/j.ymben.2015.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Winzer T, et al. Morphinan biosynthesis in opium poppy requires a P450-oxidoreductase fusion protein. Science. 2015 doi: 10.1126/science.aab1852. [DOI] [PubMed] [Google Scholar]
- 20.Farrow SC, Hagel JM, Beaudoin GAW, Burns DC, Facchini PJ. Stereochemical inversion of (S)-reticuline by a cytochrome P450 fusion in opium poppy. Nat. Chem. Biol. 2015 doi: 10.1038/nchembio.1879. [DOI] [PubMed] [Google Scholar]
- 21.Gomes T, et al. The burden of premature opioid-related mortality. Addiction. 2014;109:1482–1488. doi: 10.1111/add.12598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reuben DB, et al. National Institutes of Health Pathways to Prevention Workshop: The role of opioids in the treatment of chronic pain. Ann. Intern. Med. 2015;162:295–300. doi: 10.7326/M14-2775. [DOI] [PubMed] [Google Scholar]
- 23.Dean EJ, Davis JC, Davis RW, Petrov DA. Pervasive and persistent redundancy among duplicated genes in yeast. PLoS Genet. 2008;4:e1000113. doi: 10.1371/journal.pgen.1000113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nogae I, Johnston M. Isolation and characterization of the ZWF1 gene of Saccharomyces cerevisiae, encoding glucose-6-phosphate dehydrogenase. Gene. 1990;96:161–169. doi: 10.1016/0378-1119(90)90248-p. [DOI] [PubMed] [Google Scholar]
- 25.Flagfeldt DB, Siewers V, Huang L, Nielsen J. Characterization of chromosomal integration sites for heterologous gene expression in Saccharomyces cerevisiae. Yeast. 2009;26:545–551. doi: 10.1002/yea.1705. [DOI] [PubMed] [Google Scholar]
- 26.Battersby AR, Foulkes DM, Binks R. Alkaloid biosynthesis VIII. Use of optically active precursors for investigations on biosynthesis of morphine alkaloids. J Chem. Soc. 1965:3323–3332. [PubMed] [Google Scholar]
- 27.Hirata K, Poeaknapo C, Schmidt J, Zenk MH. 1,2-Dehydroreticuline synthase, the branch point enzyme opening the morphinan biosynthetic pathway. Phytochemistry. 2004;65:1039–1046. doi: 10.1016/j.phytochem.2004.02.015. [DOI] [PubMed] [Google Scholar]
- 28.Beaudoin GA, Facchini PJ. Benzylisoquinoline alkaloid biosynthesis in opium poppy. Planta. 2014;240:19–32. doi: 10.1007/s00425-014-2056-8. [DOI] [PubMed] [Google Scholar]
- 29.Deeknamkul W, Zenk MH. Purification and properties of 1,2-dehydroreticuline reductase from Papaver somniferum seedlings. Phytochemistry. 1992;31:813–821. [Google Scholar]
- 30.Allen RS, et al. RNAi-mediated replacement of morphine with the nonnarcotic alkaloid reticuline in opium poppy. Nat. Biotechnol. 2004;22:1559–1566. doi: 10.1038/nbt1033. [DOI] [PubMed] [Google Scholar]
- 31.Wijekoon CP, Facchini PJ. Systematic knockdown of morphine pathway enzymes in opium poppy using virus-induced gene silencing. Plant J. 2012;69:1052–1063. doi: 10.1111/j.1365-313X.2011.04855.x. [DOI] [PubMed] [Google Scholar]
- 32.Matasci N, et al. Data access for the 1,000 Plants (1KP) project. Gigascience. 2014;3:17. doi: 10.1186/2047-217X-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Facchini PJ, et al. Synthetic biosystems for the production of high-value plant metabolites. Trends Biotechnol. 2012;30:127–131. doi: 10.1016/j.tibtech.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 34.Xiao M, et al. Transcriptome analysis based on next-generation sequencing of non-model plants producing specialized metabolites of biotechnological interest. J Biotechnol. 2013;166:122–134. doi: 10.1016/j.jbiotec.2013.04.004. [DOI] [PubMed] [Google Scholar]
- 35.Fath S, et al. Multiparameter RNA and codon optimization: A standardized tool to assess and enhance autologous mammalian gene expression. PLoS One. 2011;6:e17596. doi: 10.1371/journal.pone.0017596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ziegler J, Brandt W, Geissler R, Facchini PJ. Removal of substrate inhibition and increase in maximal velocity in the short chain dehydrogenase/reductase salutaridine reductase involved in morphine biosynthesis. J Biol. Chem. 2009;284:26758–26767. doi: 10.1074/jbc.M109.030957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Higashi Y, Kutchan TM, Smith TJ. Atomic structure of salutaridine reductase from the opium poppy (Papaver somniferum) J Biol. Chem. 2011;286:6532–6541. doi: 10.1074/jbc.M110.168633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goder V, Spiess M. Topogenesis of membrane proteins: Determinants and dynamics. FEBS Lett. 2001;504:87–93. doi: 10.1016/s0014-5793(01)02712-0. [DOI] [PubMed] [Google Scholar]
- 39.Higy M, Junne T, Spiess M. Topogenesis of membrane proteins at the endoplasmic reticulum. Biochemistry. 2004;43:12716–12722. doi: 10.1021/bi048368m. [DOI] [PubMed] [Google Scholar]
- 40.IMS Institute for Healthcare Informatics. Medicines use and spending shifts: A review of the use of medicines in the U.S. in 2014. 2015 [Google Scholar]
- 41.Oye KA, Lawson JC, Bubela T. Drugs: Regulate 'home-brew' opiates. Nature. 2015;521:281–283. doi: 10.1038/521281a. [DOI] [PubMed] [Google Scholar]
- 42.Garfinkel MS, Endy D, Epstein GL, Friedman RM. Synthetic genomics: Options for governance. Biosecur. Bioterror. 2007;5:359–362. doi: 10.1089/bsp.2007.0923. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.