Abstract
A combination of cultivation-based methods with a molecular biological approach was used to investigate whether planktonic bacteria with identical 16S rRNA gene sequences can represent distinct eco- and genotypes. A set of 11 strains of Brevundimonas alba were isolated from a bacterial freshwater community by conventional plating or by using a liquid most-probable-number (MPN) dilution series. These strains had identical 16S rRNA gene sequences and represented the dominant phylotype in the plateable fraction, as well as in the highest positive dilutions of the MPN series. However, internally transcribed spacer and enterobacterial repetitive intergenic consensus PCR fingerprinting analyses, as well as DNA-DNA hybridization analyses, revealed great genetic diversity among the 11 strains. Each strain utilized a specific combination of 59 carbon substrates, and the niche overlap indices were low, suggesting that each strain occupied a different ecological niche. In dialysis cultures incubated in situ, each strain had a different growth rate and cell yield. We thus demonstrated that the B. alba strains represent distinct populations with genetically determined adaptations and probably occupy different ecological niches. Our results have implications for assessment of the diversity and biogeography of bacteria and increase the perception of natural diversity beyond the level of 16S rRNA gene sequences.
Analysis of 16S rRNA gene sequences has become the primary approach for studying the natural occurrence and distribution of bacteria in a culture-independent manner (3). The vertical and seasonal distributions of distinct 16S rRNA gene sequences (phylotypes) within one ecosystem have been used to infer the ecological niches of bacteria (40, 56). This approach is especially valuable if the physiology of bacteria that have not been cultured yet is to be elucidated.
In many cases phylogenetically closely related bacteria (whose 16S rRNA sequences differ by between 2.7 and 0.3%) have been detected in the same freshwater, marine, or soil habitat (19, 32, 43, 56). According to macroecological principles of competitive exclusion, physiologically similar microorganisms should not co-occur in nutrient-poor systems which are dominated by physical and chemical fluctuations (40). Accordingly, phylogenetically closely related bacteria coexisting in the same habitat occupy distinct ecological niches (19, 32, 43, 56).
For pathogenic bacteria it is well established that even phylogenetically identical strains or species can exhibit distinct ecophysiological properties. Certain serovars of Mycobacterium intracellulare (10), serovars of Ochrobactrum anthropi (27), strains of Yersinia pestis and Yersinia pseudotuberculosis (55), or strains of Bacillus anthracis and Bacillus cereus (4) contain identical 16S rRNA gene sequences. These phylogenetically identical organisms are also genetically highly similar based on DNA-DNA hybridization data (Table 1) but clearly represent different ecotypes based on their virulence properties or host ranges. Often, phenotypic differences can be traced back to the presence of plasmids, as in B. anthracis, in which the major virulence determinants are encoded by the 181-kb plasmid pXO1 and the 95-kb plasmid pXO2 not present in B. cereus (44).
TABLE 1.
DNA-DNA similarity and physiological differences for strains with identical 16S rRNA gene sequencesa
| Strain pair | DNA-DNA similarity (%) | Size of fragment (bp) | Physiological differences | Reference(s) |
|---|---|---|---|---|
| Escherichia coli K-12 and O157:H7 | ND | 1,542b | Virulence factors for hemorrhagic colitis | 9, 39 |
| Nodularia sp. strains GR8b and BY1 | ND | 1,445 | Formation of gas vesicles | 28 |
| Bacillus cereus NCTC11143 (= DSMZ4312) and Bacillus anthracis Sterne | ND | 1,446 | Pathogenicity; toxin and capsule formation | 4 |
| Mycobacterium intracellulare ATCC 35762 and ATCC 35772 | 88 | 1,493 | Serological properties | 10, 18 |
| Pelczaria aurantia DSM 12801T and Kocuria rosea DSM 20447T | 87.1 | 1,502 | Oxidase reaction | 49 |
| Yersinia pestis 1122 and Yersinia pseudotuberculosis P62 /P3 | 84-100 | 1,497 | Y. pestis-specific virulence plasmids pFra and pPla; motility at 28°C; urease; rhamnose fermentation; requirement for amino acids; bubonic/pneumonic plague versus mesenteric lymphademitis; transmission route, flea bite versus fecal-oral route | 8, 55 |
| Ochrobactrum anthropi 1a and LMG 5140 | 84.5 | 1,307 | BIOLOG GN test | 27 |
| Aeromonas salmonicida NCIMB2020T and Aeromonas sp. strain CIP7340 | 50 | 1,502 | ND | 15 |
| Brevundimonas alba H12C5 and 18 | 73.5 | 1,361 | Carbon substrates; oxidase | This study |
| Brevundimonas alba H12C5 and 24 | 86.5 | 1,361 | Carbon substrates; oxidase | This study |
| Brevundimonas alba H12C5 and 20 | 30.1 | 1,361 | Carbon substrates; oxidase; in situ growth | This study |
Only strains with identical 16S rRNA gene sequences are shown. ND, not determined.
rrsE gene.
However, the genomes of certain phylogenetically identical strains exhibit profound differences. Escherichia coli K-12 and O157:H7 differ not only in genome size (by 0.89 Mb) but also in a considerable number of chromosomal genes. Twenty-five percent of the genes present in the enterohemorrhagic organism E. coli O157:H7 are not found in the nonpathogenic organism E. coli K-12, whereas 12% of the genes in the latter organism are absent in the former organism (9, 39). Nevertheless, some of the 16S rRNA gene sequences (e.g., the two rrsE genes) are identical in the two organisms. Similarly, genomic fingerprinting (48, 50, 57) and analysis of fosmid libraries of DNA fragments from marine samples (6) have indicated that nonpathogenic bacteria with identical 16S rRNA gene sequences but distinctly different genomes coexist in natural ecosystems (46). The term microdiversity has been used to describe the phenomenon of phylogenetically closely related but physiologically distinct bacterial populations (32). In order to assess the extent of microdiversity present in a natural habitat, the niche separation between the different genotypes with identical 16S rRNA genes, and finally the potential limitations of 16S rRNA-based methods, more information about the genetic and ecophysiological differences of such bacteria is required.
In the present study, 11 different Brevundimonas alba strains with identical 16S rRNA gene sequences were isolated from the same sample obtained from a freshwater lake. The strains were subsequently analyzed with respect to their genomic and physiological diversities, their growth in situ, and their ecological niches.
MATERIALS AND METHODS
Isolation of strains.
Bacterial strains were isolated from Zwischenahner Meer, a shallow eutrophic lake in northern Germany (5). On 20 May 1998, water samples were obtained from a depth of 0.3 m from the head of a 30-m-long pier located on the east shore of the lake. Samples were sequentially filtered through a 20-μm-mesh plankton net and then an 8-μm-pore-size cellulose nitrate filter to remove algae and larger filamentous cyanobacteria. Chemoheterotrophic bacteria were isolated from the highest positive dilutions of liquid most-probable-number (MPN) series in synthetic freshwater medium supplemented with YPG (0.075% yeast extract, 0.15% peptone, 0.15% glucose) (5). For purification of bacterial strains, these liquid samples were streaked on solid YPG media (1.2% agar). In parallel, 10-fold serial dilutions of water samples were plated directly onto solid YPG medium. After 3 weeks of incubation at 20°C, all colonies (a total of 34 colonies) from the last two positive dilutions were picked from the YPG medium plates and restreaked until pure cultures were obtained. The purity of all strains isolated was examined microscopically.
Microscopy.
Wet mounts for light microscopy were prepared on agar-coated slides (41) and observed with a Leitz DMR microscope (Leitz, Wetzlar, Germany) at a magnification of ×1,000.
DNA extraction.
A lake water subsample used for extraction of genomic DNA was concentrated by tangential flow (21), and genomic DNA was extracted by a modified phenol-chloroform extraction method with subsequent ethanol precipitation (53). Cells of pure bacterial cultures were lysed by freeze-thawing (21) prior to 16S ribosomal DNA (rDNA) amplification and enterobacterial repetitive intergenic consensus (ERIC)-PCR fingerprinting. For dot blot hybridization, the genomic DNA of bacterial strains was extracted by bead beating (33) and was purified by a standard phenol-chloroform extraction and ethanol precipitation technique (47).
PCR, DGGE, and sequencing.
ERIC-PCR fingerprinting of the bacterial strains was performed as described previously (36). For internally transcribed spacer (ITS) fingerprinting, primers G1 (5′-GAAGTCGTAACAAGG-3′) (23) and 23SR (5′-GGGTTBCCCCATTCRG-3′) (11) were used. Amplification products were digested with AluI and MboI (Stratagene, Amsterdam, The Netherlands), and the fragments were separated on 3% small-DNA-agarose (Biozym, Hessisch Oldendorf, Germany) gels. For denaturing gradient gel electrophoresis (DGGE) (35), a fragment of the 16S rDNA between Escherichia coli positions 341 and 907 was amplified by using primer 341f with a GC clamp (36). As the template, 50 ng of extracted DNA or 1 μl of a freeze-thawed DNA preparation was used. Amplification was performed by using a step-down PCR (37), which included 10 cycles at an annealing temperature of 62°C, followed by 20 cycles at 57°C. Each cycle started with a melting step at 96°C and ended with an extension step at 72°C.
Amplification products were separated by DGGE in 6% (wt/vol) polyacrylamide gels containing a linear 30 to 70% denaturant gradient. DNA fingerprints were visualized by ethidium bromide staining of the gels.
For sequencing, a DNA template of the 16S rRNA gene was generated with primers 8f and 1492r (25) by using the conditions described previously (36). PCR products were separated from free PCR primers with a QIAquick spin kit (QIAGEN, Hilden, Germany) and were sequenced directly with a SequiTerm Excel II LC DNA sequencing kit (Epicentre Technologies, Madison, Wis.). An automated infrared laser fluorescence sequencer (model 4000 DNA sequencer; Li Cor, Lincoln, Nebr.) was used for sequencing. The sequencing primers were primers 8f, 341f, 530f, 907r, 926f, 1055r, and 1492r (3, 25). The sequences obtained were ≥1,361 bp long and, compared to the E. coli sequence, had several deletions. Hence, the sequences obtained in the present study spanned E. coli positions 14 through 1465.
Phylogenetic analysis.
The SIMILARITY_RANK tool of the Ribosomal Database Project (30) and the BLASTN 2.0.6 algorithm of GenBank (2) were employed to search for close evolutionary relatives of the 16S rRNA gene sequences of our 11 bacterial isolates. Alignments were generated with ClustalW (54), and base positions which were identical in less than 50% of the sequences were masked (36). Two different methods were used for construction of phylogenetic trees by employing programs of the PHYLIP 3.57c package (16). First, a distance matrix was calculated with the algorithm of Jukes and Cantor (24), and a phylogenetic tree was inferred by using the algorithm of Fitch and Margoliash (17). Second, a phylogenetic tree was constructed by using the maximum-likelihood method (DNAML) of the PHYLIP package.
Dot blot hybridization.
Probes specific for strain H12C5 were prepared from the genomic DNA by using a DIG High Prime DNA labeling and detection kit (Roche). For blotting, DNA of all strains was treated first with DNase-free RNase (50 μg · ml−1; 45 min at 37°C) and then with proteinase K (50 μg · ml−1; 30 min at 37°C). After phenol-chloroform extraction and ethanol precipitation (47), dilution series were prepared in TE buffer (10 mM Tris-HCl, 0.1 mM Na4-EDTA [pH 8]), and the DNA was denatured for 10 min at 95°C and vacuum blotted onto positively charged nylon membranes. Each membrane was baked for 30 min at 120°C and prehybridized for 30 min in 15 ml of DIG Easy Hyb buffer at 62°C. Hybridization was carried out for 18 h in 12 ml of prewarmed Easy Hyb buffer containing 2 μg of denatured strain H12C5 probes. After this, the blots were washed twice for 5 min with 2× SSC (1× SSC is 150 mM NaCl plus 15 mM sodium citrate [pH 7.0]) containing 0.1% sodium dodecyl sulfate at room temperature and then twice under highly stringent conditions (30 min in 0.1× SSC-0.1% sodium dodecyl sulfate at 82°C). The hybridization signal was detected with a DIG luminescence detection kit (Roche) and Lumi-film (Roche) used according to the instructions of the manufacturer. The exposure time was 50 min. Compared to standard DNA-DNA hybridization, the procedure described above allowed better differentiation of closely related strains. For comparison, standard DNA-DNA hybridization (13) was performed for four strains with different levels of genetic relatedness. In this case, the degree of DNA-DNA binding was determined spectophotometrically from the renaturation rates of individual DNA types and of pairwise mixtures of genomic DNA from the different strains.
Physiological characterization.
Based on the substrates detected in natural lake water (29, 34) and the different pathways of substrate utilization, 59 different carbon substrates were chosen for growth tests. Synthetic freshwater medium was added to polystyrene microtiter plates, and each well was supplemented with a different substrate. The polymeric substrates used were peptone (0.05%, wt/vol), casein hydrolysate (0.05%), yeast extract (0.005%), cellulose (15 mm2 of filter paper), starch (0.1%), chitin (0.05%), xylene (0.05%), and laminarin (0.05%). The sugars and sugar derivatives used were glucose, fructose, rhamnose, mannose, arabinose, xylose, sucrose, cellobiose, maltose, trehalose, mannitol, gluconate, and glucosamine (final concentration of each, 5 mM). The carboxy acids and other acids were formate (2.5 mM), acetate (5 mM), propionate (1 mM), butyrate (2.5 mM), valerate (0.5 mM), capronate (0.5 mM), caprylate (0.5 mM), crotonate (0.2 mM), malonate (5 mM), succinate (10 mM), fumarate (5 mM), malate (5 mM), tartrate (2 mM), glycolate (5 mM), pyruvate (5 mM), lactate (10 mM), 2-oxoglutarate (5 mM), and citrate (2 mM). The alcohols tested were methanol (2 mM), ethanol (5 mM), propanol (5 mM), butanol (5 mM), glycol (5 mM), glycerol (5 mM), and Tween 80 (0.001%, vol/vol). In addition, the amino acids alanine (2 mM), arginine (2 mM), asparagine (2 mM), cysteine (2 mM), glutamine (2 mM), isoleucine (2 mM), tyrosine (0.5 mM), tryptophan (1.25 mM), and proline (2 mM) were employed, and betaine (2 mM), benzoate (2 mM), salicylate (2 mM), and nicotinate (2 mM) were tested.
For growth tests, each microtiter well received 180 μl of medium and was inoculated with 20 μl of a suspension (optical density at 436 nm, 0.12) of bacterial cells which had been washed in synthetic freshwater medium to remove residual substrates. The plates were incubated for 5 weeks at 20°C, and growth was monitored by determining the turbidity. The catalase activities of the strains were tested by addition of 3% H2O2 to colonies on glass slides. Oxidase activity was tested as described by Tarrand and Gröschel (52).
The niche overlap index (NOI) between two strains, strains A and B, was calculated by determining the ratio of the number of substrates utilized by both strains, (NA∩B) to the total number of substrates utilized by either of the two (Ntot) (58): NOI = NA∩B/Ntot. The physiological similarity of the 11 strains was determined by cluster analysis. A matrix with a binary code for the presence or absence of each phenotypic trait of the isolates was constructed. The Dice coefficient (14) for all pairs of strains was calculated by using the SIMQUAL similarity program for qualitative data of the NTSYS-pc numerical taxonomy computer package (45). Cluster analysis was performed with the SAHN program of the NTSYS-pc package and by employing the unweighted pair group method with arithmetic average.
In situ growth experiments.
Four of the strains were precultured in liquid YPG medium and diluted to a final titer of 105 cells · ml−1 in prefiltered (pore size, 0.1 μm) lake water. For each strain, six dialysis bags (molecular mass cutoff, 12,000 Da; cellulose tubing; diameter, 1.5 cm; Sigma, Deisenhofen, Germany) were filled aseptically with 10 ml of the diluted culture, sealed with Spectra/Por closures (Spectrum, Houston, Tex.), and attached to a stainless steel incubation rack. The incubation rack was incubated at the original sampling depth in the lake. Dialysis tubes were recovered at regular intervals, the contents were fixed with glutardialdehyde (final concentration, 2% [vol/vol]), and the cell titer was determined by 4′,6-diamidino-2-phenylindole (DAPI) counting (42).
Nucleotide sequence accession numbers.
The 16S rRNA gene sequences of strains 1 through 11 have been deposited in the GenBank database under accession numbers AF296678 to AF296688.
RESULTS AND DISCUSSION
One phylotype dominates the culturable bacterial fraction.
Zwischenahner Meer is a shallow eutrophic lake (mean depth, 3.3 m) with a simple hydrodynamic structure, since its entire water column mixes frequently even during the summer (22). Therefore, our data should be representative of the entire water column.
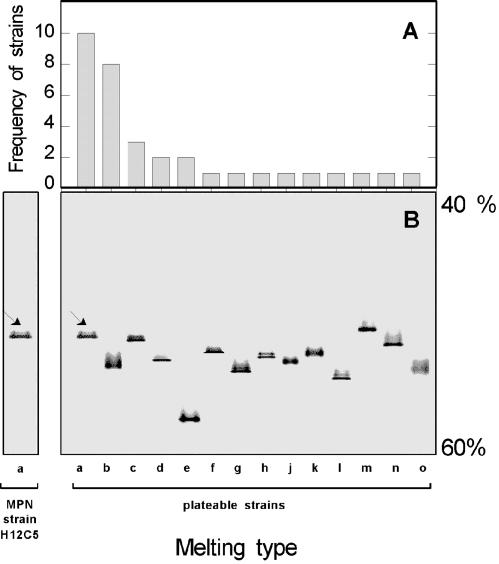
Only 0.03% of the total number of bacterial cells (9.5 × 107 cells · ml−1, as counted by DAPI staining) formed colonies on solid YPG media. This finding is in line with the generally low culturability of planktonic bacteria (3). Thirty-four strains were isolated from the highest positive dilutions on solid YPG media. DGGE analysis of 16S rRNA gene fragments of all strains revealed the same melting behavior for 10 of the strains (strains 8, 14, 18, 20, 24, 25, 26, 30, 31, and 33) (Fig. 1). The same melting behavior was also detected for strain H12C5 isolated from the highest positive dilutions of the MPN series (Fig. 1). Identical melting behaviors during DGGE theoretically could result from sequence identity of the 16S rRNA genes. Indeed, our sequence analysis revealed that the 10 melting type a strains, as well as strain H12C5, had identical sequences for all 1,361 bp sequenced (Fig. 2).
FIG. 1.
Analysis of 16S rRNA gene fragments of all bacterial isolates by DGGE. (A) Frequencies of different melting types (melting types a to o). (B) Different melting types. The arrows indicate the position of the fragments of strains 8, 14, 18, 20, 24, 25, 26, 30, 31, and 33 which were identical to the position of the fragment from strain H12C5. The negative image of an ethidium bromide-stained gel is shown.
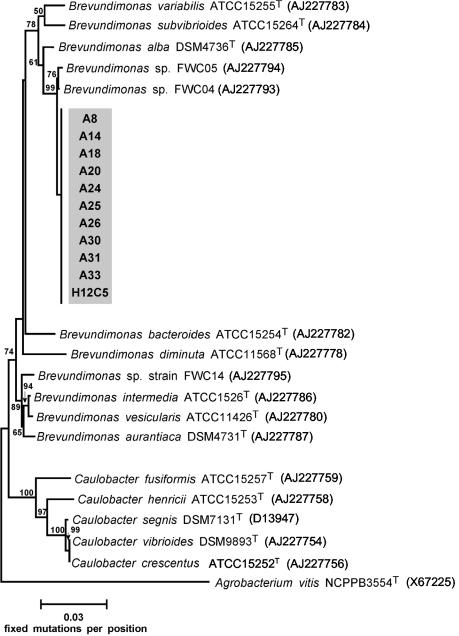
FIG. 2.
Phylogenetic affiliations of 11 strains with the same 16S rDNA melting type. The tree was constructed from evolutionary distances by using the algorithm of Fitch and Margoliash. Accession numbers are indicated in parentheses. The numbers at the nodes are bootstrap values based on 100 resamplings (values that are less than 50 are not shown).
Potentially, there could have been sequence variability in the unsequenced 3′ end of the 16S rRNA gene, and thus this variability could have remained undetected. However, the sequence variability at the end of the 16S rRNA gene is lower than that in the regions upstream (51). It is therefore unlikely that there were significant sequence differences among the strains analyzed in the present study. Another concern could be the potential sequence polymorphism of different copies of the ribosomal operon. Therefore, we compared the 16S rRNA gene sequences of all rrn operons of the closest relative of our isolates. The most closely related bacterium for which data are available is Caulobacter crescentus (GenBank accession no. AE005930 and AE006011), which contains two different rrn operons. A sequence comparison revealed that the 16S rRNA gene sequences of the two operons were identical. Based on these data, sequence polymorphism of the different rrn operons thus appears to be rather unlikely in the Caulobacter-Brevundimonas group.
The 16S rRNA gene sequences of our isolates were 98.4% similar to that of B. alba DSM4736T. This phylogenetic assignment was fully confirmed by the tree constructed by the maximum-likelihood method (data not shown).
Genetic diversity within the dominant culturable phylotype.
Theoretically, the 11 strains with identical phylotypes could be members of one population, in which case they should be genetically and physiologically very similar. However, an especially low rate of evolution of the 16S rRNA gene has been detected in Bacillus and Mycobacterium species (38). Also, the two gram-negative bacteria Aeromonas caviae and Aeromonas trota differ at only one nucleotide position in their 16S rRNA genes, whereas the level of DNA-DNA similarity is ≤30% (31). Therefore, an alternative explanation could be that the strains isolated in the present study may represent members of distinct and coexisting populations with different, genetically determined physiological adaptations and ecological niches.
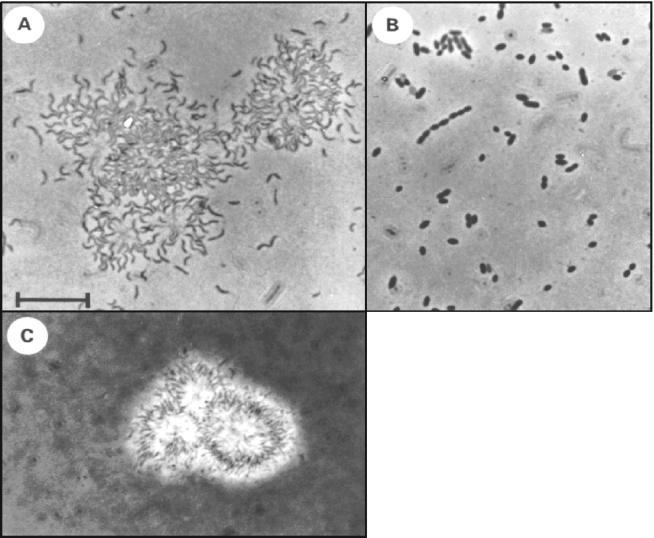
Microscopic inspection revealed two distinct morphologies among the 11 strains. Seven strains (strains H12C5, 8, 14, 18, 24, and 31) had a thin spirilloid morphotype (Fig. 3A) and tended to form aggregates of cells embedded in a slime matrix (Fig. 3C). Four strains (strains 20, 25, 30, and 33) had a rod-shaped morphotype (Fig. 3B). Three different ITS fingerprint patterns (Fig. 4A) were detected, and they largely correlated with morphology: all spirilloid strains had identical ITS fingerprints, whereas two different patterns were generated for the rod-shaped strains. By comparison, ERIC-PCR fingerprinting (Fig. 4B) provided a higher resolution and suggested that strains with identical ITS fingerprints were genetically divergent. Nine different ERIC-PCR fingerprint patterns were detected among the 11 strains, revealing the highest genetic diversity within one 16S rRNA phylotype known so far. In previous studies, a maximum of three different genotypes with identical 16S rRNA gene sequences were recovered from one environment (27, 48).
FIG. 3.
Phase-contrast photomicrographs of B. alba strains. (A) Morphotype of strain H12C5 in squeezed aggregates. (B) Morphotype of strain 20. (C) Extracellular slime layers of aggregates of strain H12C5 as visualized with India ink.
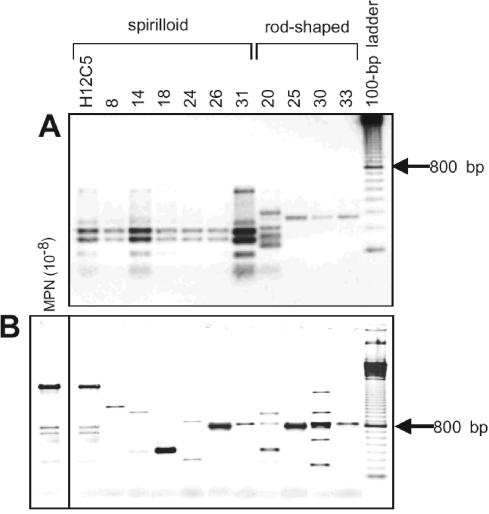
FIG. 4.
ITS fingerprints (A) and ERIC-PCR fingerprints (B) of the 11 B. alba strains. A comparison with ERIC-PCR fingerprints generated directly from the highest positive MPN dilution revealed the dominance of strain H12C5 in the sample. The image is a negative image of an ethidium bromide-stained gel.
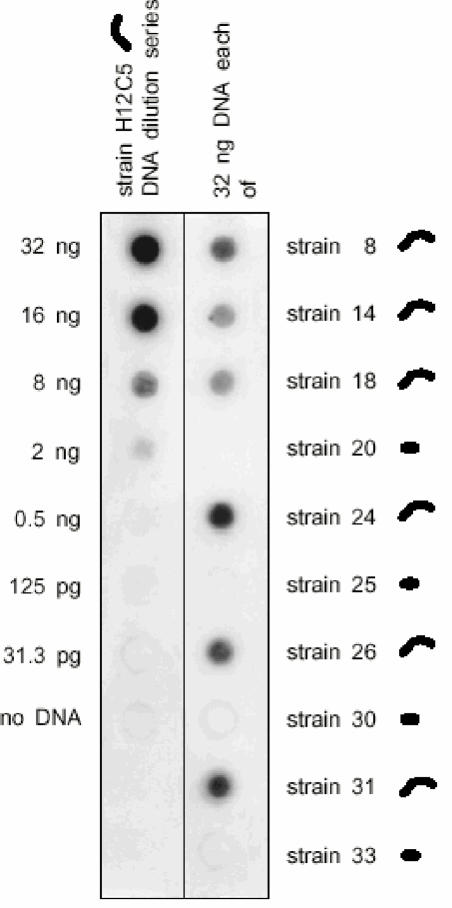
Since ERIC-PCR fingerprinting targets only a small portion of the bacterial genome, we used dot blot hybridization as a second, independent technique to more accurately determine the genetic diversity among the 11 strains. Most notably, the hybridization results correlated with the morphotypes of the strains. Under our highly stringent hybridization conditions, genomic DNA of rod-shaped strains 20, 25, 30, and 33 did not bind detectable amounts of DNA of strain H12C5 (Fig. 5), while the spirilloid strains exhibited much higher DNA-DNA hybridization values. Standard DNA-DNA hybridization fully confirmed the results of our dot blot procedure: strain H12C5 exhibited DNA-DNA similarity of only 30.1% with rod-shaped strain 20 but exhibited 73.5 and 86.5% DNA-DNA similarity with spirilloid strains 18 and 24, respectively (Table 1). So far, low DNA-DNA hybridization values (30%) have been reported to occur only between different bacterial species (51). According to the results of our dot blot hybridization analysis, the rod-shaped strains thus form a separate species. Although low DNA-DNA hybridization values (values as low as 30%) are known to occur rarely among strains with a few different nucleotide positions in their 16S rRNA gene sequences (4, 7, 18, 31, 51), this is, to the best of our knowledge, the first time that such a high level of divergence at the genomic level has been documented for strains with identical 16S rRNA gene sequences (Table 1).
FIG. 5.
Genomic relatedness of the 10 B. alba strains isolated from agar plates to strain H12C5. The corresponding morphotypes are shown next to the strain designations.
To date, niche differentiation of genetically closely related (DNA-DNA similarity, >70%) bacteria with identical phylotypes has only been documented for a few clinically relevant strains (Table 1). As indicated by their genetic heterogeneity, the 11 B. alba strains represent different populations, and even species, with the same phylotype. From an ecological point of view, the persistence of these different populations in the same habitat can be understood only if they occupy distinct ecological niches.
Niche differentiation among strains with identical 16S rRNA gene sequences.
Although strain H12C5 dominated the culturable fraction, it could not grow directly on agar plates. In contrast, none of the other 10 strains could be isolated from the liquid media. These observations alone indicate that the different strains have different physiologies. Consequently, their potential ecological niches were assessed based on their carbon substrate utilization patterns.
Each strain used a unique combination of the 59 carbon substrates (Table 2). While the strains isolated directly from agar plates were able to utilize 3 to 37 substrates, strain H12C5 had the lowest metabolic versatility of all strains and grew only on 2 substrates, peptone and laminarin. Furthermore, H12C5 did not exhibit oxidase activity like the other strains did. The NOI calculated for all pairs of spirilloid strains indicated that they occupy different ecological niches (mean NOI for all strain pairs, 0.27 ± 0.16). Likewise, the NOI for spirilloid and rod-shaped strains was low (mean, 0.26 ± 0.12). However, the NOI for the four rod-shaped B. alba strains was significantly (P < 0.01) higher (0.48 ± 0.11). In the phyllosphere, resource partitioning among different bacterial species with NOI values of 0.25 to 0.59 allows stable coexistence (59), whereas catabolically identical strains (NOI, 1.0), even if they belong to different species, cannot coexist (58).
TABLE 2.
Substrate utilization pattern of the 11 strains of Brevundimonas sp.a
| Substrate | Utilization by:
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| H12C5 | 8 | 14 | 18 | 20 | 24 | 25 | 26 | 30 | 31 | 33 | |
| Sugars and derivatives | |||||||||||
| Glucose | − | + | + | − | − | − | − | − | + | − | − |
| Fructose | − | + | − | + | − | − | + | − | + | − | − |
| Mannose | − | + | − | + | − | − | − | − | − | − | − |
| Rhamnose | − | + | − | + | − | − | + | − | − | − | − |
| Arabinose | − | − | − | − | − | − | + | − | + | − | + |
| Trehalose | − | − | − | − | + | − | − | − | + | − | − |
| Mannitol | − | − | − | − | − | − | − | − | + | − | − |
| Gluconate | − | − | − | − | + | − | − | − | − | − | − |
| Glucosamine | − | − | − | − | − | − | − | − | + | − | − |
| Laminarin | + | + | + | + | + | + | + | + | + | + | + |
| Organic acids | |||||||||||
| Acetate | − | − | − | − | + | − | − | − | + | − | − |
| Propionate | − | − | − | − | + | − | − | − | + | − | − |
| Lactate | − | − | − | − | + | − | + | + | + | − | + |
| Butyrate | − | − | + | + | − | − | − | − | + | − | + |
| Crotonate | − | − | + | − | − | − | − | − | + | − | − |
| Valerate | − | − | + | + | − | − | − | − | + | − | − |
| Capronate | − | − | + | + | − | − | − | − | − | − | − |
| Caprylate | − | − | + | + | − | − | − | − | + | − | − |
| Malonate | − | − | − | + | − | − | − | − | − | − | − |
| Succinate | − | − | − | + | + | − | − | − | + | − | − |
| Fumarate | − | − | − | + | − | − | − | − | − | − | − |
| Malate | − | − | − | + | + | − | − | − | + | − | − |
| Tartrate | − | − | − | + | + | − | − | − | − | − | − |
| Pyruvate | − | − | − | + | − | − | − | − | − | − | − |
| 2-Oxoglutarate | − | − | − | + | − | − | + | − | + | − | + |
| Citrate | − | − | − | − | + | − | − | + | + | − | + |
| Alcohols | |||||||||||
| Methanol | − | − | − | + | − | − | − | − | − | − | − |
| Ethanol | − | − | − | + | − | − | − | − | − | − | − |
| Propanol | − | − | − | + | − | − | − | − | − | − | − |
| Butanol | − | − | − | + | − | − | − | − | − | − | − |
| Glycol | − | − | − | + | − | − | − | − | − | − | − |
| Glycerol | − | − | − | + | − | − | − | − | + | − | + |
| Amino acids | |||||||||||
| Alanine | − | − | + | + | + | − | + | − | + | − | + |
| Arginine | − | − | − | + | − | − | + | − | + | − | + |
| Asparagine | − | + | + | + | − | − | − | − | − | − | + |
| Glutamine | − | + | + | + | + | + | + | + | + | − | + |
| Isoleucine | − | − | − | − | + | − | + | + | + | − | + |
| Proline | − | + | − | + | − | − | + | + | + | − | + |
| Tyrosine | − | + | − | + | + | − | + | + | + | − | + |
| Tryptophan | − | + | − | + | − | − | − | − | − | − | − |
| Polymers | |||||||||||
| Starch | − | − | − | + | + | + | + | + | + | + | + |
| Xylan | − | − | − | − | + | + | + | − | − | − | − |
| Complex substrates | |||||||||||
| Casamino Acids | − | + | + | + | − | − | − | − | − | − | − |
| Yeast extract | − | + | + | + | + | + | + | + | + | − | + |
| Others | |||||||||||
| Tween 80 | − | − | + | + | + | − | − | − | + | − | + |
| Betaine | − | + | − | + | − | − | + | + | + | − | + |
| Benzoate | − | + | − | + | − | − | − | + | + | − | + |
| Salicylate | − | + | − | + | − | − | + | − | − | − | − |
| Nicotinate | − | + | − | − | − | − | − | − | − | − | − |
Peptone was utilized by all strains. The following compounds were tested but were not utilized by any of the strains: xylose, sucrose, maltose, cellobiose, cellulose, chitin, formate, glycolate, and cysteine.
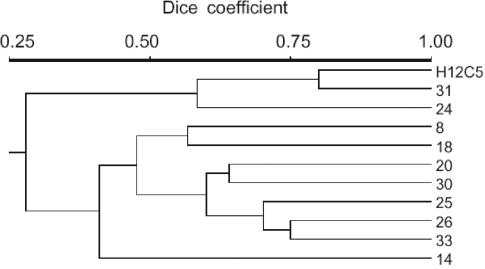
The similarity analysis of the data (Fig. 6) demonstrated that all strains exhibited distinct substrate utilization patterns. Spirilloid strains H12C5 and 31 showed the closest similarity (Dice coefficient, 0.8), whereas the similarity values for all other strains were ≤0.75.
FIG. 6.
Physiological similarity of the 11 Brevundimonas strains as assessed by cluster analysis based on the substrate utilization patterns.
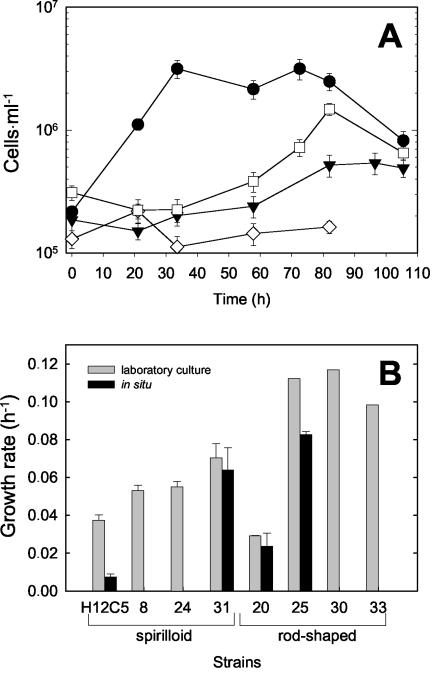
Additional experiments were conducted in order to compare the growth rates of the different strains in our dilute freshwater medium (Fig. 7B). The growth rates of four spirilloid and four rod-shaped strains growing in our liquid YPG medium varied considerably, confirming their genetic and physiological heterogeneity. Most notably, the low growth rate of strain 20 corresponds to its unique ITS and ERIC fingerprints, whereas the genetically more similar strains 25, 30, and 33 also had similar growth rates. Our data indicate that there was considerable diversification and niche differentiation of the Brevundimonas strains despite their identical phylotypes.
FIG. 7.
(A) Growth of two spirilloid strains (open symbols) and two rod-shaped strains (solid symbols) in dialysis cultures incubated in Zwischenahner Meer. ⋄, strain H12C5; □, strain 31; ▾, strain 20; •, strain 25. (B) Growth rates of the 11 B. alba strains in pure laboratory cultures growing in artificial freshwater medium with YPG as the substrate and growing in dialysis bags in situ in Zwischenahner Meer. The error bars indicate one standard deviation.
A parallel analysis (22) revealed that α-proteobacteria represented over 50% of the isolates. These organisms comprised the dominant Brevundimonas phylotype and a second phylotype related to Pedomicrobium (22). Based on a culture-independent analysis of the phylogenetic composition by fluorescence in situ hybridization (12), however, α-proteobacteria represent only between 2.6 and 3.2% of the bacterioplankton cells in Zwischenahner Meer during the spring. Among the 11 strains, strain H12C5 was obtained from the highest positive dilutions and hence should be the most frequent strain in the natural bacterioplankton community. Therefore, genomic DNA of strain H12C5 was quantified by employing a highly stringent dot blot hybridization protocol and using the other 10 Brevundimonas strains as negative controls. Based on this analysis, strain H12C5 accounted for only 0.07% of all genomic DNA in the bacterioplankton community of Zwischenahner Meer.
Growth experiments in situ.
NOIs determined in laboratory growth experiments do not necessarily reflect the actual competition as it occurs in the natural environment (1). Furthermore, their low abundance in situ (see above) theoretically could indicate that the isolated strains are not capable of significant growth in the natural environment.
In order to assess the potential for growth and the competition of the different Brevundimonas strains, two rod-shaped and two spirilloid strains with very different growth rates were selected for in situ growth experiments in dialysis cultures (Fig. 7A). In two cases (strains 20 and 31), the in situ growth rates did not differ significantly from those in laboratory media, indicating that our artificial freshwater medium with diluted complex substrates mimicked the conditions in Zwischenahner Meer rather well (Fig. 7B). Two other strains, however, grew significantly more slowly in dialysis cultures.
It therefore has to be concluded that at least some of the Brevundimonas strains are capable of appreciable growth in situ (the doubling time was as short as 8.7 h for strain 25 [Fig. 7B]). Our data also demonstrate that the differences between the strains are even more pronounced than the differences indicated by laboratory experiments. The different growth rates could be caused by the different substrate utilization patterns of the strains, but they could also be the result of other influences (e.g., temperate phages).
Evidently, the low abundance of our isolates in the bacterioplankton of Zwischenahner Meer is caused by factors other than a low growth rate. It is feasible that the Brevundimonas strains are more sensitive to loss processes like protozoan grazing or viral lysis than the other members of the bacterioplankton assemblage (20).
Conclusions
The 11 Brevundimonas strains investigated in the present study were retrieved from the same lake water sample. Therefore, a hitherto unknown multitude of ecotypes must thrive in the same habitat. Based on our results, it has to be concluded that the extent of genomic and physiological diversity masked by identical 16S rRNA sequences is much larger than has been assumed previously and that this so-called microdiversity has ecological relevance. If a bacterial species is defined as a “monophyletic and genomically coherent cluster of individual bacteria that show a high degree of overall similarity in many independent characteristics ” (46), then bacterial diversity may indeed exceed present estimates by several orders of magnitude, as previously suggested (15).
In addition, our data imply that, at least in some cases, only limited conclusions regarding the biogeography and ecophysiology of uncultured bacteria can be drawn from the mere presence or absence of their 16S rRNA sequences in an ecosystem. In cases like the present one, molecular methods with higher resolution, like analyses of ITS sequences, of fosmid libraries (6), or of chromosomal painting (26), are more appropriate for culture-independent analyses of bacterioplankton community structure.
Acknowledgments
We thank Alke Bruns for analysis of the ITS fingerprint patterns. Standard DNA-DNA hybridization was done by the Deutsche Sammlung von Mikroorganismen und Zellkulturen, Braunschweig, Germany.
This study was supported by grants 0311098 and 0311949 from the Bundesministerium für Bildung, Wissenschaft, Forschung und Technologie to Heribert Cypionka and Jörg Overmann.
REFERENCES
- 1.Abrams, P. A. 1980. Some comments on measuring niche overlap. Ecology 61:44-49. [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amann, R., W. Ludwig, and K.-H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ash, C., J. A. E. Farrow, M. Dorsch, E. Stackebrandt, and M. D. Collins. 1991. Comparative analysis of Bacillus anthracis, Bacillus cereus, and related species on the basis of reverse transcriptase sequencing of 16S rRNA. Int. J. Syst. Bacteriol. 41:343-346. [DOI] [PubMed] [Google Scholar]
- 5.Bartscht, K., H. Cypionka, and J. Overmann. 1999. Evaluation of cell activity and of methods for the cultivation of bacteria from a natural lake community. FEMS Microbiol. Ecol. 28:249-259. [Google Scholar]
- 6.Béjà, O., E. V. Koonin, L. Aravind, L. T. Taylor, H. Seitz, J. L. Stein, D. C. Bensen, R. A. Feldman, R. V. Swanson, and E. F. DeLong. 2002. Comparative genomic analysis of archaeal genotypic variants in a single population and in two different oceanic provinces. Appl. Environ. Microbiol. 68:335-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bennasar, A., R. Rosselló-Mora, J. Lalucat, and E. R. B. Moore. 1996. 16S rRNA gene sequence analysis relative to genomovars of Pseudomonas stutzeri and proposal of Pseudomonas balearica sp. nov. Int. J. Syst. Bacteriol. 46:200-205. [DOI] [PubMed] [Google Scholar]
- 8.Bercovier, H., H. H. Mollaret, J. M. Alonso, J. Brault, G. R. Fanning, A. G. Steigerwalt, and D. J. Brenner. 1980. Intra- and interspecies relatedness of Yersinia pestis by DNA hybridization and its relationship to Yersinia pseudotuberculosis. Curr. Microbiol. 4:225-229. [Google Scholar]
- 9.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode,G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1462. [DOI] [PubMed] [Google Scholar]
- 10.Böddinghaus, B., J. Wolters, W. Heikens, and E. C. Böttger. 1990. Phylogenetic analysis and identification of different serovars of Mycobacterium intracellulare at the molecular level. FEMS Microbiol. Lett. 70:197-204. [DOI] [PubMed] [Google Scholar]
- 11.Borneman, J., and E. W. Triplett. 1997. Molecular microbial diversity in soils from eastern Amazonia: evidence for unusual microorganisms and microbial population shifts associated with deforestation. Appl. Environ. Microbiol. 63:2647-2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bruns, A., U. Nübel, H. Cypionka, and J. Overmann. 2003. Effect of signal compounds and incubation conditions on the culturability of freshwater bacterioplankton. Appl. Environ. Microbiol. 69:1980-1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeLey, J., H. Cattoir, and A. Reynaerts. 1970. The quantitative measurement of DNA hybridization from renaturation rates. Eur. J. Biochem. 12:133-142. [DOI] [PubMed] [Google Scholar]
- 14.Dice, L. R. 1945. Measures of the amount of ecologic association between species. Ecology 26:297-302. [Google Scholar]
- 15.Dykhuizen, D. E. 1998. Santa Rosalia revisited: why are there so many species of bacteria? Antonie Leeuwenhoek 73:25-33. [DOI] [PubMed] [Google Scholar]
- 16.Felsenstein, J. 1989. PHYLIP, phylogenetic inference package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 17.Fitch, W. M., and E. Margoliash. 1967. Construction of phylogenetic trees: a method based on mutation distances as estimated from cytochrome c sequences is of general applicability. Science 155:279-284. [DOI] [PubMed] [Google Scholar]
- 18.Fox, G. E., J. D. Wisotzkey, and P. Jurtshuk. 1992. How close is close: 16S rRNA sequence identity may not be sufficient to guarantee species identity. Int. J. Syst. Bacteriol. 42:166-170. [DOI] [PubMed] [Google Scholar]
- 19.Gray, N. D., and I. M. Head. 2001. Linking genetic identity and function in communities of uncultured bacteria. Environ. Microbiol. 3:481-492. [DOI] [PubMed] [Google Scholar]
- 20.Hahn, M. W., and M. G. Höfle. 2001. Grazing of protozoa and its effect on populations of aquatic bacteria. FEMS Microbiol. Ecol. 35:113-121. [DOI] [PubMed] [Google Scholar]
- 21.Jaspers, E., and J. Overmann. 1997. Separation of bacterial cells by isoelectric focusing, a new method for analysis of complex microbial communities. Appl. Environ. Microbiol. 63:3176-3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jaspers, E., K. Nauhaus, H. Cypionka, and J. Overmann. 2001. Multitude and temporal variability of ecological niches as indicated by the diversity of cultivated bacterioplankton. FEMS Microbiol. Ecol. 36:153-164. [DOI] [PubMed] [Google Scholar]
- 23.Jensen, M. A., J. A. Webster, and N. Straus. 1993. Rapid identification of bacteria on the basis of polymerase chain reaction-amplified ribosomal DNA spacer polymorphism. Appl. Environ. Microbiol. 59:945-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jukes, T. H., and C. R. Cantor. 1969. Evolution of protein molecules, p. 21-132. In H. N. Munro (ed.), Mammalian protein metabolism Academic Press, New York, N.Y.
- 25.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley, Chichester, United Kingdom.
- 26.Lanoil, B. D., and S. J. Giovannoni. 1997. Identification of bacterial cells by chromosomal painting. Appl. Environ. Microbiol. 63:1118-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lebuhn, M., W. Achouak, M. Schloter, O. Berge, H. Meier, A. Hartmann, and T. Heulin. 2000. Taxonomic characterization of Ochrobactrum sp. isolates from soil samples and wheat roots, and description of Ochrobactrum tritici sp. nov. and Ochrobactrum grignonense sp. nov. Int. J. Syst. Evol. Microbiol. 50:2207-2223. [DOI] [PubMed] [Google Scholar]
- 28.Lehtimäki, J., C. Lyra, S. Suomalainen, P. Sundman, L. Rouhiainen, L. Paulin, and M. Salkinoja-Salonen. 2000. Characterization of Nodularia strains, cyanobacteria from brackish waters, by genotypic and phenotypic methods. Int. J. Syst. Evol. Microbiol. 50:1043-1053. [DOI] [PubMed] [Google Scholar]
- 29.Lock, M. A. 1994. The dynamics of dissolved and particulate organic material over the substratum of water bodies, p. 137-160. In R. S. Wotton (ed.), The biology of particles in aquatic systems, 2nd ed. Lewis Publishers, Boca Raton, Fla.
- 30.Maidak, B. L., G. J. Olsen, N. Larsen, R. Overbeek, M. J. McCaughney, and C. R. Woese. 1997. The Ribosomal Database Project. Nucleic Acids Res. 25:109-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martínez-Murcia, A. J., S. Benlloch, and M. D. Collins. 1992. Phylogenetic interrelationships of members of the genera Aeromonas and Plesiomonas as determined by 16S ribosomal DNA sequencing: lack of congruence with results of DNA-DNA hybridizations. Int. J. Syst. Bacteriol. 42:412-421. [DOI] [PubMed] [Google Scholar]
- 32.Moore, L. R., G. Rocap, and S. W. Chisholm. 1998. Physiology and molecular phylogeny of coexisting Prochlorococcus ecotypes. Nature 393:464-467. [DOI] [PubMed] [Google Scholar]
- 33.Moré, M. I., J. B. Herrick, M. C. Silva, W. C. Ghiorse, and E. L. Madsen. 1994. Quantitative cell lysis of indigenous microorganisms and rapid extraction of microbial DNA from sediment. Appl. Environ. Microbiol. 60:1572-1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Münster, U., and R. J. Chróst. 1990. Origin, composition, and microbial utilization of dissolved organic matter, p. 8-46. In J. Overbeck and R. J. Chróst (ed.), Aquatic microbial ecology. Springer, New York, N.Y.
- 35.Muyzer, G., S. Hottenträger, A. Teske, and C. Waver. 1995. Denaturing gradient gel electrophoresis of PCR-amplified 16S rDNA—a new molecular approach to analyse the genetic diversity of mixed microbial communities, p. 3.4.4.1-3.4.4.22. In A. D. L. Akkermans, J. D. Van Elsas, and F. J. de Bruijn (ed.), Molecular microbial manual, 2nd ed. Kluwer, Dordrecht, The Netherlands.
- 36.Overmann, J., and C. Tuschak. 1997. Phylogeny and molecular fingerprinting of green sulfur bacteria. Arch. Microbiol. 167:302-309. [DOI] [PubMed] [Google Scholar]
- 37.Overmann, J., M. J. L. Coolen, and C. Tuschak. 1999. Specific detection of different phylogenetic groups of chemocline bacteria based on PCR and denaturing gradient gel electrophoresis of 16S rRNA gene fragments. Arch. Microbiol. 172:83-94. [DOI] [PubMed] [Google Scholar]
- 38.Palys, T., E. Berger, I. Mitrica, L. K. Nakamura, and F. M. Cohan. 2000. Discovery and classification of ecological diversity in the bacterial world: the role of DNA sequence data. Int. J. Syst. Ecol. Microbiol. 50:1021-1028. [DOI] [PubMed] [Google Scholar]
- 39.Perna, N. T., G. Plunkett III, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 40.Pernthaler, J., F.-O. Glöckner, S. Unterholzner, A. Alfreider, R. Psenner, and R. Amann. 1998. Seasonal community and population dynamics of pelagic bacteria and archaea in a high mountain lake. Appl. Environ. Microbiol. 64:4299-4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pfennig, N., and S. Wagener. 1986. An improved method of preparing wet mounts for photomicrographs of microorganisms. J. Microbiol. Methods 4:303-306. [Google Scholar]
- 42.Porter, K. G., and Y. S. Feig. 1980. Use of DAPI for identifying and counting aquatic microflora. Limnol. Oceanogr. 25:943-948. [Google Scholar]
- 43.Postius, C., and A. Ernst. 1999. Mechanisms of dominance: coexistence of picocyanobacterial genotypes in a freshwater ecosystem. Arch. Microbiol. 172:69-75. [DOI] [PubMed] [Google Scholar]
- 44.Read, T. D., S. N. Peterson, N. Tourasse, L. W. Baillie, I. T. Paulsen, K. E. Nelson, H. Tettelin, D. E. Fouts, J. A. Eisen, S. R. Gill, E. K. Holtzapple, O. A. Okstad, E. Helgason, J. Rilstone, M. Wu, J. F. Kolonay, M. J. Beanan, R. J. Dodson, L. M. Brinkac, M. Gwinn, R. T. DeBoy, R. Madpu, S. C. Daugherty, A. S. Durkin, D. H. Haft, W. C. Nelson, J. D. Peterson, M. Pop, H. M. Khouri, D. Radune, J. L. Benton, Y. Mahamoud, L. Jiang, I. R. Hance, J. F. Weidman, K. J. Berry, R. D. Plaut, A. M. Wolf, K. L. Watkins, W. C. Nierman, A. Hazen, R. Cline, C. Redmond, J. E. Thwaite, O. White, S. L. Salzberg, B. Thomason, A. M. Friedlander, T. M. Koehler, P. C. Hanna, A. B. Kolsto, and C. M. Fraser. 2003. The genome sequence of Bacillus anthracis Ames and comparison to closely related bacteria. Nature 423:81-86. [DOI] [PubMed] [Google Scholar]
- 45.Rohlf, F. J. 1993. NTSYSpc numerical taxonomy and multivariate analysis system for the IBM PC microcomputer (and compatibles). Applied Biostatistics, Setauket, N.Y.
- 46.Rosselló-Mora, R., and R. Amann. 2001. The species concept for prokaryotes. FEMS Microbiol. Rev. 25:39-67. [DOI] [PubMed] [Google Scholar]
- 47.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 48.Sass, H., E. Wieringa, H. Cypionka, H.-D. Babenzien, and J. Overmann. 1998. High genetic and physiological diversity of sulfate-reducing bacteria isolated from an oligotrophic lake sediment. Arch. Microbiol. 170:243-251. [DOI] [PubMed] [Google Scholar]
- 49.Schumann, P., B. J. Tindall, U. Mendrock, I. Kramer, and E. Stackebrandt. 2000. Pelczaria aurantia ATCC 49321T (= DSM 12801T) is a strain of Kocuria rosea (Flugge 1886) Stackebrandt et al. 1995. Int. J. Syst. Evol. Microbiol. 50:1421-1424. [DOI] [PubMed] [Google Scholar]
- 50.Sikorski, J., M. Möhle, and W. Wackernagel. 2002. Identification of complex composition, strong strain diversity and directional selection in local Pseudomonas stutzeri populations from marine sediment and soils. Environ. Microbiol. 4:465-476. [DOI] [PubMed] [Google Scholar]
- 51.Stackebrandt, E., and B. M. Goebel. 1994. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44:846-849. [Google Scholar]
- 52.Tarrand, J. J., and D. H. M. Gröschel. 1982. Rapid, modified oxidase test for oxidase-variable bacterial isolates. J. Clin. Microbiol. 16:772-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Teske, A., N. B. Ramsing, K. Habicht, M. Fukui, J. Küver, B. B. Jørgensen, and Y. Cohen. 1998. Sulfate-reducing bacteria and their activities in cyanobacterial mats of Solar Lake (Sinai, Egypt). Appl. Environ. Microbiol. 64:2943-2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thompson, J. D., D. G. Higgins, T. J. Gibson. 1994. Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Trebesius, K., D. Harmsen, A. Rakin, J. Schmelz, and J. Heesemann. 1998. Development of rRNA-targeted PCR and in situ hybridization with fluorescently labeled oligonucleotides for detection of Yersinia species. J. Clin. Microbiol. 36:2557-2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ward, D. M., M. J. Ferris, S. C. Nold, and M. M. Bateson. 1998. A natural view of microbial biodiversity within hot spring cyanobacterial mat communities. Microbiol. Mol. Biol. Rev. 62:1353-1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wieringa, E., J. Overmann, and H. Cypionka. 2000. Detection of abundant sulphate-reducing bacteria in marine oxic sediment layers by a combined cultivation and molecular approach. Environ. Microbiol. 2:417-427. [DOI] [PubMed] [Google Scholar]
- 58.Wilson, M., and S. E. Lindow. 1994. Ecological similarity and coexistence of epiphytic ice-nucleating (Ice+) Pseudomonas syringae strains and a non-ice-nucleating (Ice−) biological control agent. Appl. Environ. Microbiol. 60:3128-3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wilson, M., and S. E. Lindow. 1994. Coexistence among epiphytic bacterial populations mediated through nutritional resource partitioning. Appl. Environ. Microbiol. 60:4468-4477. [DOI] [PMC free article] [PubMed] [Google Scholar]