Abstract
Background. KRAS mutation is an important predictive and prognostic factor for patients receiving anti-EGFR therapy. An expanded KRAS, NRAS, BRAF, PIK3CA mutation analysis provides additional prognostic information, but its role in predicting bevacizumab efficacy is unclear. The aim of our study was to evaluate the incidence of KRAS, NRAS, BRAF and PIK3CA mutations in metastatic colorectal cancer patients receiving first line oxaliplatin based chemotherapy with or without bevacizumab and to evaluate their prognostic and predictive significance.
Methods. 55 patients with the first-time diagnosed CRC receiving FOLFOX ± bevacizumab were involved in the study. Tumour blocks were tested for KRAS mutations in exons 2, 3 and 4, NRAS mutations in exons 2, 3 and 4, BRAF mutation in exon 15 and PIK3CA mutations in exons 9 and 20. The association between mutations and clinico-pathological factors, treatment outcomes and survival was analyzed.
Results. KRAS mutations were detected in 67.3% of the patients, BRAF in 1.8%, PIK3CA in 5.5% and there were no NRAS mutations. A significant association between the high CA 19–9 level and KRAS mutation was detected (mean CA 19–9 levels were 276 and 87 kIU/l, respectively, p = 0.019). There was a significantly higher response rate in the KRAS, NRAS, BRAF and PIK3CA wild type cohort receiving bevacizumab compared to any gene mutant type (100 and 60%, respectively, p = 0.030). The univariate Cox regression analysis did not confirm KRAS and other tested mutations as prognostic factors for PFS or OS.
Conclusions. Our study revealed higher KRAS and lower NRAS, BRAF and PIK3CA mutation rates in the Lithuanian population than those reported in the literature. KRAS mutation was associated with the high CA 19–9 level and mucinous histology type, but did not show any predictive or prognostic significance. The expanded KRAS, NRAS, BRAF and PIK3CA mutation analysis provided additional significant predictive information.
Keywords: KRAS, NRAS, BRAF, PIK3CA, colorectal cancer, bevacizumab
Abstract
KRAS, NRAS, BRAF IR PIK3CA MUTACIJŲ REIKŠMĖ METASTAZAVUSIU STOROSIOS ŽARNOS VĖŽIU SERGANČIUS PACIENTUS GYDANT CHEMOTERAPIJOS IR BEVACIZUMABO DERINIU: ĮSTAIGOS PATIRTIS
Santrauka
Įvadas. Pacientams, kuriems taikomas gydymas anti- EGFR terapija, KRAS mutacijos – svarbus predikcinis ir prognozinis veiksnys. Išplėstinė KRAS, NRAS, BRAF ir PIK3CA analizė suteikia papildomos prognozinės informacijos, tačiau jos reikšmė nuspėjant gydymo bevacizumabu efektyvumą neaiški. Tyrimo tikslas – ištirti ir įvertinti KRAS, NRAS, BRAF ir PIK3CA mutacijų prognozinę bei predikcinę reikšmę pacientams, kuriems taikoma pirmos eilės chemoterapija oksaliplatinos pagrindu su bevacizumabu.
Metodai. Tyrime dalyvavo 55 pacientai, jiems dėl metastazavusios ligos skirtas pirmos eilės gydymas FOLFOX4 schema su arba be bevacizumabo. Naviko medžiagoje, gautoje iš parafininių blokų, tirtos KRAS 2, 3 ir 4 egzono, NRAS 2, 3 ir 4 egzono, BRAF 15 egzono ir PIK3CA 9 ir 20 egzono mutacijos. Vertintas šių mutacijų ryšys su klinikinėmis ir patologinėmis charakteristikomis, atsaku į gydymą bei išgyvenamumu.
Rezultatai. KRAS mutacijų nustatyta 67,3 %, BRAF – 1,8 %, PIK3CA – 5,5 % pacientų ir nė vienam neaptikta NRAS mutacijų. Pastebėtas reikšmingas ryšys tarp KRAS mutacijos ir CA 19–9 lygio (vidutinė CA 19–9 reikšmė buvo 276 kIU/l KRAS mutuotų pacientų grupėje, palyginti su 87 kIU/l laukinio tipo grupėje, p = 0,019). Nustatytas statistiškai geresnis atsakas pacientams, gydytiems chemoterapija su bevacizumabu, jiems nenustatyta jokių tirtųjų mutacijų, palyginti su tais, kuriems aptikta bent vieno tirtojo geno mutacija (atsako dažnis atitinkamai buvo 100 ir 60 %, p = 0,030). KRAS ar kitų mutacijų prognozinė reikšmė išgyvenamumui be ligos progresijos bei bendrajam išgyvenamumui atlikus vienamatę Cox regresijos analizę nebuvo patvirtinta.
Išvados. Tyrimo metu nustatytas KRAS mutacijos dažnis yra didesnis, o NRAS, BRAF ir PIK3CA – mažesnis nei skelbiama literatūroje. KRAS mutacija buvo susijusi su didesniu CA 19–9 lygiu bei mucininio tipo navikais, tačiau neturėjo predikcinės ar prognozinės reikšmės. Išplėstinė KRAS, NRAS, BRAF ir PIK3CA mutacijų analizė suteikė reikšmingos papildomos predikcinės informacijos gydant FOLFOX4 ir bevacizumabo deriniu.
Raktažodžiai: KRAS, NRAS, BRAF, PIK3CA, storosios žarnos vėžys, bevacizumabas
INTRODUCTION
Colorectal cancer (CRC) is the third most common cancer type worldwide. Globally, it accounts for 1.2 million of new diagnoses and 600,000 deaths every year (1). The five-year survival is about 50–59% and depends on the geographic region and economic development of the country. In Lithuania CRC is the second most common cancer type with 3–6% increasing morbidity each year (2). According to the EUROCARE-5 data CRC survival rates in Lithuania are much worse than the European average (3).
Despite high morbidity, a survival improvement tendency is noticed worldwide over the past 10 years. It is associated with new active chemotherapeutic drugs and targeted agents. Doublet or triplet combinations of chemotherapy agents and biologics increase survival of metastatic CRC to 30 months. Unfortunately, new anticancer agents increase toxicity and treatment costs and not all the patients benefit from these treatments. Understanding biology and molecular mechanisms of disease and drug resistance could help in predicting treatment efficacy.
RAS/RAF/MAPK and PI3K/AKT/MTOR are two major intracellular signaling pathways involved in proliferation, adhesion, angiogenesis, migration and survival. Activation of these pathways is common in CRC and mostly associated with KRAS, NRAS, BRAF and PIK3CA mutations (4, 5). Several studies revealed KRAS as an independent predictor of relapse and death (6–9). BRAF mutation was associated with a distinct tumour phenotype and more aggressive disease (10, 11).
KRAS and NRAS mutations were associated with a worse response to anti-EGFR therapy and treatment outcomes (12–14). Also they have been investigated as potential predictive markers of the response to bevacizumab or oxaliplatin, but results are controversial (8, 9, 15–17).
Recently, it was reported that the KRAS, BRAF, NRAS and PIK3CA mutation analysis gives additional prognostic information. According to the mutation status patients were divided into 4 groups, with the worse prognosis in the BRAF and KRAS mutation group and the best prognosis in all genes wild type group (18). This kind of the expanded mutation analysis also provides an additional predictive value for anti-EGFR therapy (19). There is limited information regarding the role of the mentioned mutations in predicting bevacizumab or oxaliplatin efficacy.
So far, KRAS and NRAS mutations are the only approved predictive markers for metastatic colorectal cancer. These mutations predict efficacy of anti- EGFR therapy, but still there are no validated predictive markers for one of the most common treatment combinations of oxaliplatin based chemotherapy and bevacizumab.
The aim of our study was to evaluate the incidence of KRAS, NRAS, BRAF and PIK3CA mutations in metastatic colorectal cancer patients receiving first line oxaliplatin based chemotherapy with or without bevacizumab and to evaluate their prognostic and predictive significance.
MATERIALS AND METHODS
Patients
55 patients with first-time diagnosed metastatic colorectal cancer participated in a prospective observational study conducted in the National Cancer Institute (Lithuania) in 2011–2014. All the patients had histological confirmed adenocarcinoma, tumour samples were obtained by a primary tumour removal operation or biopsy before starting chemotherapy. The patients received FOLFOX4 chemotherapy (oxaliplatin 85 mg/m2 iv infusion on day 1, calcium folinate 200 mg/m2 iv infusion on days 1–2, 5-fluorouracil 400 mg/m2 bollus on days 1–2 and 5-fluorouracil 600 mg/m2 22-hour continuous iv infusion on days 1–2; repeated every 2 weeks) with or without bevacizumab (5 mg/kg iv infusion every 2 weeks) until disease progression or an unacceptable toxicity according to the institutional guidelines. Treatment efficacy was evaluated every 2 months by a CT (computer tomography) scan according to the RECIST 1.1 criteria. After completing the treatment, patients were followed up for progression or survival every 3 months.
The study has been approved by the Regional Biomedical Research Ethics Committee and performed in accordance with the Helsinki Declaration. All patients signed an informed consent before entering the study. Tumour samples were analyzed in the National Pathology Center, Affiliate of Vilnius University Hospital Santariškių Clinics (Lithuania) and the Laboratory of Molecular Medicine of Hematology, Oncology and Transfusiology Center of Vilnius University Hospital Santariškių Clinics (Lithuania), and all were blinded to treatment allocation and outcomes.
DNA extraction and mutation analysis
Formalin-fixed paraffin-embedded (FFPE) tumour tissue blocks were selected by a histopathologist ensuring the presence of at least 50% tumour cells. From the selected FFPE tumour block 4–5 sections of 5 µm thickness were obtained and processed for genomic DNA extraction using the Maxwell® 16 FFPE Plus LEV DNA Purification Kit (Promega). KRAS, NRAS, BRAF and PIK3CA mutations were analysed using PCR (Maxima Hot Start PCR Master Mix (2X) kits according to manufacturer’s protocols). The primers sequences used for PCR are presented in Table 1. The purified PCR products were sequenced using the BigDye® Terminator v1.1 Cycle Sequencing Kit and analysed by an ABI PRISM® 3100 Genetic Analyzer, ContigExpress (Vector NTI).
Table 1.
The primers sequences used for PCR
| Gene | Exon | Primer 5’-3’ | |
|---|---|---|---|
| Forward | Reverse | ||
| KRAS | 2 | GGTACTGGTGGAGTATTTGATAGTGT | GCAGGACCATTCTTTGATACAGA |
| 3 | CTTTGGAGCAGGAACAAT GTCT | GGGGAGGGCTTTCTTTGTGTA | |
| 4 | GTGTTACTAAT GACT GTGCTATAAC | GATTAAGAAGCAATGCCCTCTC | |
| NRAS | 2 | AT GTGGCT CGCCAAT TAACC | TCCGACAAGT GAGAGACAGGA |
| 3 | CACACCCCCAGGATTCTTACA | TCCTTT CAGAGAAAATAATGCTCCT | |
| 4 | CCCGTTTTTAGGGAGCAGA | GAATAT GGAT CACATCTCTACCAGAG | |
| PIK3CA | 9 | CCTGTCTCTGAAAATAAAGTCTTGC | AAAAGCATTTAATGTGCCAACGACC |
| 20 | TCGACAGCATGCCAAT CTCT | CT GAGAGTTAT TAACAGTGCAGT G | |
| BRAF | 15 | TCTTCATAAT GCTT GCTCT GATAGGA | CCCTGAGATGCTGCTGAGTT |
CEA and CA 19–9 analysis
Blood samples were taken before starting chemotherapy. The level of CEA and CA 19–9 was evaluated by enzyme-linked immunosorbent assay (ELISA) using CUSABIO (China) kits according to the manufacturer’s recommendations. Normal CEA value ranges were considered less than 5 µg/l and for CA 19–9 less than 37 kIU/l.
Statistical analysis
Descriptive statistics were used to describe demographic characteristics. A non-parametric Wilcoxon test was used to evaluate the differences between the two independent data sets because data was not normally distributed. The differences between the two independent qualitative data groups were evaluated by a Chi-square or Fisher exact test. Risk factors for PFS and OS were assessed by a Cox regression analysis. Survival trends were evaluated by the Kaplan–Meier method. A log-rank test was used to evaluate the difference between Kaplan–Meier curves. Progression free survival (PFS) was calculated as the time from the first day of treatment to the first date of disease progression or the day of a confirmed new tumour or death. Overall survival (OS) was calculated as the time from the first day of treatment to death. If during the last visit to the clinician there was no evidence of disease progression or a new tumour, the date was confirmed as censored. A two-tailed p-value less than 0.05 was considered to be significant. A statistical analysis was performed using the Statistical Analysis System (SAS) package version 9.2.
RESULTS
During 2011–2014, 55 patients with first-time diagnosed metastatic colorectal cancer were included into the study. The median age was 63 years (range 44–76). There were 29 (52%) males and 26 (48%) females. 35 (64%) of tumours were located in the colon and 20 (36%) in the rectum. The histological type in 44 (80%) of the cases was adenocarcinoma, and in 11 (20%) it was adenocarcinoma with mucinous differentiation. 49 (89%) of the tumours were medium grade, 1 (2%) were low grade and 5 (9%) were high grade. All the patients had metastases in the liver, and for 21 (38%) of the patients it was the only site of the metastases. 38 (69%) of the patients had synchronous metastases, and in 49 (89%) a primary tumour was removed. 14 (25%) of the patients underwent liver resection. The median number of chemotherapy cycles was 8. Bevacizumab was administered to 29 (53%) of the patients.
Distribution according to the site of mutations and their incidence is presented in Table 2. KRAS mutations were detected in 37 (67.3%) of the patients, exon 2 in all cases, with codon 12 as the most frequent site. One patient had simultaneous KRAS mutations in exon 2 (G13D) and exon 3 (R68S), it accounted for 1.8% prevalence of exon 3 mutations. Other mutations were less frequent: 1 BRAF exon 15 mutation (1.8%), 3 PIK3CA mutations all were detected in codon 9 (5.5%) and there were no NRAS mutations. Two of the patients with PIK3CA mutations also had KRAS codon 12 mutations, BRAF and KRAS mutations were mutually exclusive. Taken together, it accounted for 16 (29.1%) of multigene wild type patients.
Table 2.
Frequency and types of tested mutations
| Gene | Status | Exon | Codon | Number | % | % |
|---|---|---|---|---|---|---|
| G12A | 3 | 5.5 | ||||
| G12C | 3 | 5.5 | ||||
| 2 | G12D | 10 | 18.2 | |||
| Mutant | G12S | 4 | 7.3 | 67.3 | ||
| KRAS | G12V | 11 | 20.0 | |||
| G13D | 6 | 10.9 | ||||
| 3 | R68S | 1 | 1.8 | |||
| 4 | 0 | 0 | ||||
| Wild Type | 18 | 32.7 | 32.7 | |||
| 2 | 0 | 0 | ||||
| NRAS | Mutant | 3 | 0 | 0 | 0 | |
| 4 | 0 | 0 | ||||
| Wild Type | 55 | 100 | 100 | |||
| BRAF | Mutant | 15 | V600 | 1 | 1.8 | 1.8 |
| Wild Type | 54 | 98.2 | 98.2 | |||
| PIK-3CA | Mutant | 9 | E545K | 3 | 5.5 | 5.5 |
| 20 | 0 | 0 | ||||
| Wild Type | 52 | 94.5 | 94.5 | |||
Mucinous differentiation of adenocarcinoma was significantly associated with the KRAS mutant type (p = 0.010). More KRAS mutated tumours were detected in the colon (especially the right side), but the difference was not significant. There was a non-significant association between 12 codon mutations and lung metastases: 42% of patients with codon 12 mutations and no patients with codon 13 mutations developed lung metastases. There were no significant associations with other clinical and pathological features (Table 3).
Table 3.
The association of KRAS and all gene (KRAS, BRAF, NRAS, PIK3CA) mutations with clinical and pathological characteristics
| Characteristic | N | KRAS | All genes | ||||
|---|---|---|---|---|---|---|---|
| MT | WT | P | MT | WT | P | ||
| Total | 55 | 37 | 18 | 39 | 16 | ||
| Age | |||||||
| <65 | 29 | 20 | 9 | 1.000 | 22 | 7 | 0.393 |
| ≥65 | 26 | 17 | 9 | 17 | 9 | ||
| Gender | |||||||
| Female | 26 | 20 | 6 | 0.166 | 20 | 6 | 0.352 |
| Male | 29 | 17 | 12 | 19 | 10 | ||
| Location | |||||||
| C18-19 | 35 | 25 | 10 | 0.358 | 26 | 9 | 0.466 |
| C20 | 20 | 12 | 8 | 13 | 7 | ||
| Side | |||||||
| Right | 12 | 10 | 2 | 0.289 | 10 | 2 | 0.284 |
| Left | 43 | 27 | 16 | 29 | 14 | ||
| Type of adenocarcinoma | |||||||
| Adenocarcinoma | 44 | 26 | 18 | 0.010 | 28 | 16 | 0.023 |
| Mucinous | 11 | 11 | 0 | 11 | 0 | ||
| Tumour budding | |||||||
| Yes | 21 | 15 | 6 | 0.606 | 16 | 5 | 0.498 |
| No | 34 | 22 | 12 | 23 | 11 | ||
| Extrahepatic metastases | |||||||
| Yes | 34 | 23 | 11 | 0.938 | 25 | 9 | 0.586 |
| No | 21 | 14 | 7 | 14 | 7 | ||
| Lung metastases | |||||||
| Yes | 20 | 13 | 7 | 0.786 | 15 | 5 | 0.614 |
| No | 35 | 24 | 11 | 24 | 11 | ||
| Peritoneal metastases | |||||||
| Yes | 8 | 5 | 3 | 0.756 | 6 | 2 | 0.783 |
| No | 47 | 32 | 15 | 33 | 14 | ||
| Lymphnode metastases | |||||||
| Yes | 12 | 7 | 5 | 0.455 | 8 | 4 | 0.714 |
| No | 43 | 30 | 13 | 31 | 12 | ||
| Treatment arm | |||||||
| FOLFOX4 | 26 | 19 | 7 | 0.389 | 19 | 7 | 0.738 |
| FOLFOX4 + bevacizumab | 29 | 18 | 11 | 20 | 9 | ||
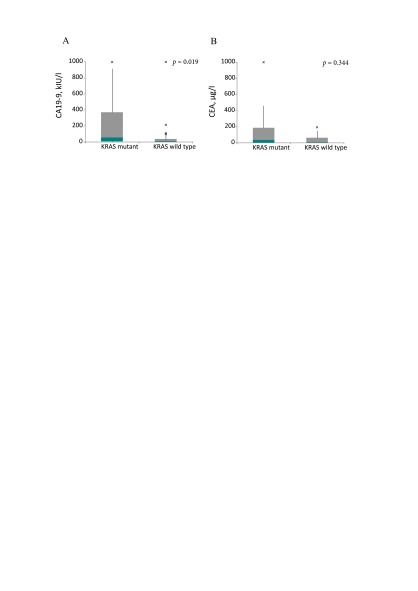
A significant association between the high CA 19–9 level and KRAS mutation was detected (Fig. 1). The mean CA 19–9 level in the KRAS mutant patients’ group was 276 kIU/l compared to 87 kIU/l in the KRAS wild type patients’ group, p = 0.019). The mean CEA level in KRAS mutant patients’ group was 235 µg/l compared to 37 µg/l in the KRAS wild type patients’ group, but the difference was not significant, p = 0.344, because data was not normally distributed.
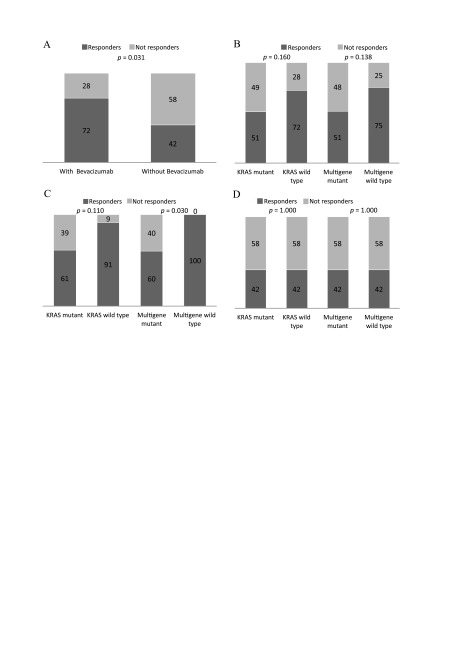
Based on the response to the treatment patients were divided into 2 groups: responders (complete and partial response; 32 patients, 58%) and non-responders (stable and progressive disease; 23 patients, 41%). The response rate was counted as a percentage of the patients that achieved a partial or complete response. The patients with KRAS wild type tumours had better response rates (percentage of patients with achieved complete and partial response) compared to the patients with KRAS mutant tumours (72 and 51%, respectively, p = 0.160) (Fig. 2A, B). Similar results were obtained in all gene (KRAS, BRAF and PIK3CA) wild type group compared to any gene mutant (75 and 51%, respectively, p = 0.138). Both results were not significant.
A trend toward better response in bevacizumab receiving KRAS wild type patients compared to MT was observed (91 and 61%, respectively, p = 0.11). All gene wild type patients had a significantly better response than any gene mutant patients (100 vs 60%, respectively, p = 0.030) (Fig. 2C, D). There were no differences in the rates of response to FOLFOX4 regarding KRAS and any gene mutations.
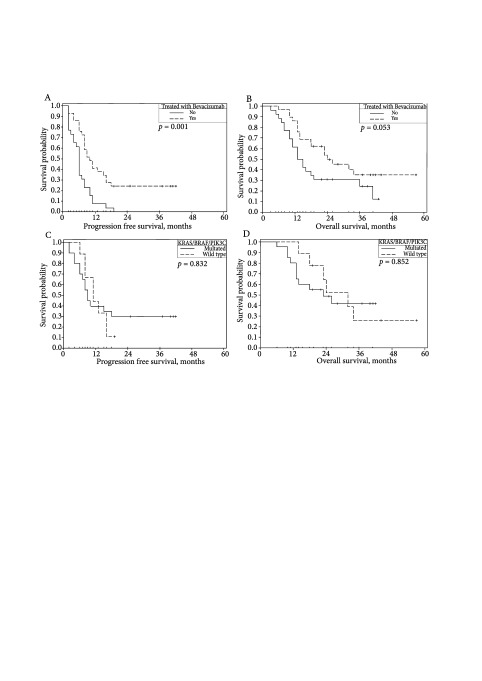
The median observation time was 18 months (range 3–57 months) for all patients, PFS was 8 months (95% CI 6–10 months), and OS was 18 months (95% CI 18–26 months). The Kaplan– Meier analysis revealed that bevacizumab significantly prolonged PFS an OS. The median PFS in the group of patients receiving bevacizumab was 10 months (95% CI 7–13 months) compared to 6 months (95% CI 5–7 months) in the group of patients not receiving bevacizumab, p = 0.001. The median OS was 24 months (95% CI 15– 33 months) and 13 months (95% CI 9–17 months), respectively, p = 0.053 (Fig. 3A, B). Neither KRAS nor other mutations did influence the progression free survival (PFS) or the overall survival (OS) irrespectively of the treatment arm (Fig. 3C, D).
Fig 1.
Differences of CA 19–9 and CEA levels depending on the KRAS status. CA 19–9 level depending on the KRAS status (A); CEA level depending on the KRAS status (B)
Fig 2.
Overall response rates according to the mutation status and treatment group. Response rates in all study population according to the treatment arm (A); Response rates in all study population according to the KRAS and multigene (KRAS, NRAS, BRAF, PIK3CA) mutation status (B); Response rates in the patients’ group receiving bevacizumab according to the KRAS and multigene (KRAS, NRAS, BRAF, PIK3CA) mutation status (C); Response rates in the patients’ group not receiving bevacizumab according to the KRAS and multigene (KRAS, NRAS, BRAF, PIK3CA) mutation status (A)
The univariate Cox regression analysis revealed that bevacizumab was associated with longer PFS (p = 0.0016, HR 2.575, 95% CI 1.431–4.636) and OS (p = 0.0624, HR 0.539, 95% CI 0.282–1.032). The multivariate analysis confirmed bevacizumab as an independent prognostic factor for better PFS (p = 0.0163, HR 2.081, 95% CI 1.144–3.783), but not OS. The univariate Cox regression analysis did not confirm KRAS and other tested mutations as prognostic factors for PFS or OS.
DISCUSSION
RAS/RAF/MAPK and PI3K/AKT/MTOR are two major intracellular signaling pathways involved in proliferation, adhesion, angiogenesis, migration and survival. Activation of these pathways is common in CRC and mostly associated with KRAS, NRAS, BRAF and PIK3CA mutations. The reported KRAS mutation prevalence in metastatic CRC patients is 40–55%, with the following distribution observed: KRAS exon 2 (43%), KRAS exon 3 (4%), KRAS exon 4 (6%) (18, 20). The reported incidence of NRAS mutations is 3–5%, BRAF 5–15%, and PIK3CA 15–20% (18).
We have determined KRAS mutations in 67.3% of cases, all in exon 2, with the dominant 12 codon (84% of all KRAS mutations). One patient had simultaneous KRAS mutations in exon 2 (G13D) and exon 3 (R68S) and it accounted for 1.8% prevalence of exon 3 mutations. We did not detect any KRAS mutation in exon 4. The determined prevalence of KRAS mutations is much higher than that reported in the literature and distribution of the mutations is slightly different. Such a high incidence of the KRAS mutation could be influenced by the selected patient population with advanced disease and liver metastases. Absence of KRAS exon 4 and NRAS mutation as well as a low BRAF and PIK3CA mutations rate are contradictory. On the one hand, it might have been influenced by the high KRAS mutation rate, taken into account that NRAS, BRAF and KRAS mutations are mutually exclusive. On the other hand, it could be influenced by a small sample size, the peculiarity of the Lithuanian population or methodological issues (lower sensitivity of Sanger sequencing compared to other methods, such as pyrosequencing, locked nucleic acid (LNA) PCR assay or mutant-enriched PCR, for low frequency RAS mutations) (21–23).
Fig 3.
Kaplan–Meier plots of PFS and OS according to the treatment arm and KRAS, NRAS, BRAF and PIK3CA gene status. PFS according to the treatment arm: the median PFS in the group of patients receiving bevacizumab was 10 months (95% CI 7–13 months) compared to 6 months (95% CI 5–7 months) in the group of patients not receiving bevacizumab, p = 0.001 (A). The median OS in the group of patients receiving bevacizumab was 24 months (95% CI 15–33 months) compared to 13 months (95% CI 9–17 months) in the group of patients not receiving bevacizumab, p = 0.053 (B). The median PFS of bevacizumab receiving patients in the multigene wild type group was 11 months (95% CI: 7–15) compared to 9 months (95% CI: 7–11) in the multigene mutant group, p = 832 (C). The median OS of bevacizumab receiving patients in the multigene wild type group was 32 months (95% CI: 20–44) compared to 23 months (95% CI: 8–38) in the multigene mutant group, p = 852 (D)
As reported in the literature (24), in our study KRAS mutations were more frequently observed in the tumours with mucinous differentiation. Also there were significantly higher levels of CA 19–9 in KRAS mutant patients. CEA levels in KRAS patients were also higher, but the difference was not significant. There were only a couple of similar reports published so far. Narita et al. reported that KRAS and BRAF mutations in the tumour tissue are associated with elevated CA 19–9 levels (25). Trevisiol et al. indicated that the serum KRAS gene mutation status was significantly associated with preoperative CA 19–9 levels (26). Data regarding the association between KRAS mutations and the elevated CEA level is still controversial (27, 28). The pattern of possible influence of the KRAS mutation on CA 19–9 excretion has not been well established yet. Considering the negative prognostic value of mucinous hystology and high CEA and CA 19–9 levels, once again we show the association of KRAS mutation with a more aggressive tumour phenotype.
RAS/RAF/MAPK and PI3K/AKT/MTOR path- ways are involved in angiogenesis (29), and this gives the potential of RAS, BRAF and PIK3CA mutations to predict the efficacy of antiangiogenetic therapy. It is supposed that the KRAS mutation might activate angiogenesis in several ways: by increasing production of VEGF and CXCL-8 (proangiogenetic interleukin 8) and by activating growth of the tumour cells (30, 31). Petrelli et al. published a metha-analysis of 12 studies involving 2,266 patients (9) revealing a negative impact of KRAS mutation on the response rate (48.3 vs 54.8%, HR, 1.42; 95% CI, 1.05–1.92; p = 0.02), progression free survival (9.42 vs 11.8 months, HR, 0.85; 95% CI, 0.74–0.98; p = 0.02) and overall survivall (20.2 vs 24.5 months, HR, 0.65; 95% CI, 0.46–0.92; p = 0.01). Researches concluded that the absence of KRAS mutation could be considered as a predictor of better response (9). On the other hand, a retrospective analysis of the Czech population registry CORRECT did not show any influence of KRAS exon 2 mutations on the results of first line bevacizumab combinations with chemotherapy (32). Bruera et al. have reported the possible impact of the KRAS genotype on angiogenesis and a significantly shorter survival of the patients with G12D mutation receiving bevacizumab (33).
We have determined a better response rate in the FOLFOX4 and bevacizumab group compared to that in the FOLFOX4 group. The results were not significant, but they are in concordance with the metha-analysis reported by Petrelli. We did not find any influence of any specific codon mutation on the outcome. Taking in consideration data regarding the additional predictive value of other mutations (BRAF and PIK3CA) to anti-EGFR therapy (19), we have analysed their prediction and the prognostic value in our population. We have determined that 29.1% of the study population did not have mutations in any of the tested genes (KRAS, NRAS, BRAF or PIK3CA) and all these patients in the bevacizumab group achieved a partial or complete response. The difference in response rates in FOLFOX4 plus bevacizumab compared to only the FOLFOX group was significant (p = 0.03). We suppose that our results could show the potential predictive value of the extended mutation analysis of KRAS, NRAS, BRAF and PIK3CA genes, but also this hypothesis should be tested on larger patient population.
The prognostic role of KRAS mutations remains controversial. More than 20 reported trials did not show any prognostic significance, but some have the established negative prognostic value of KRAS mutations on the progression free survival (6, 8, 9), overall survival (6, 9) or early recurrence after liver surgery for metastases (34). BRAF mutations are not only associated with a more aggressive tumour phenotype, but are also found to be an independent negative prognostic marker for the overall survival (10, 11, 35).
Foltran et al. (18) reported that simultaneous testing for KRAS, BRAF, NRAS and PIK3CA mutations can help more accurately predict survival. They devided patients into four groups with significantly different survival results: the worse prognosis in the BRAF and KRAS mutation group (OS median 7.6 months) and the best prognosis in the all genes wild type group (OS median 27.7 months).
In our study, despite the longer median overall survival of bevacizumab receiving patients in the all genes wild type group compared to that of any gene mutant group (23 months, 95% CI: 8–38; and 32 months, 95% CI: 20–44, respectively), the difference was not significant. We did not confirm the prognostic value of the tested mutations irrespectively of the treatment arm.
There were several methodological limitations of the present study. First, we have selected patients with liver metastases, some of them did undergo liver resection but some also had extrahepatic disease, which could determine differences in survival. Second, a small sample size and single-center population might have also influenced the results. Our analysis was explorative and hypothesis generating and should be analyzed in a larger cohort.
CONCLUSIONS
Our study revealed higher KRAS and lower NRAS, BRAF and PIK3CA mutation rates in the Lithuanian population than those reported in the literature. The KRAS mutation was associated with the high CA 19–9 level and mucinous histology type, but did not show any predictive or prognostic significance. The expanded KRAS, NRAS, BRAF and PIK3CA mutation analysis provided additional significant predictive information.
ACKNOWLEDGEMENTS
The authors greatfully acknowledge the mutation testing assistance of Rimvydas Norvilas and other co-workers in the Laboratory of Molecular Medicine of Hematology, Oncology and Transfusiology Center of Vilnius University Hospital Santariškių Clinics.
Edita Baltruškevičienė, Ugnius Mickys, Tadas Žvirblis, Rokas Stulpinas, Teresė Pipirienė Želvienė, Eduardas Aleknavičius
References
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011; 61: 69–90. [DOI] [PubMed] [Google Scholar]
- Smailytė G, Aleknavičienė B.. Vėžys Lietuvoje 2012 metais. Nacionalinis vėžio institutas, Vėžio registras; 2015. [Google Scholar]
- De Angelis R, Sant M, Coleman MP, Francisci S, Baili P, Pierannunzio D, et al.. Cancer survival in Europe 1999–2007 by country and age: results of EUROCARE-5 – a population-based study. Lancet Oncol. 2014; 15: 23–34. [DOI] [PubMed] [Google Scholar]
- Fodde R, Smits R, Clevers H.. APC, signal transduction and genetic instability in colorectal cancer. Nat Rev Cancer. 2001; 1: 55–67. [DOI] [PubMed] [Google Scholar]
- Danielsen SA, Eide PW, Nesbakken A. Portrait of the PI3K/AKT pathway in colorectal cancer. Biochim Biophys Acta. 2015; 1855: 104–21. [DOI] [PubMed] [Google Scholar]
- Andreyev HJN, Norman AR, Clarke PA, Cunningham D, Oates JR.. Kirsten ras mutations in patients with colorectal cancer: the multicenter “RASCAL” study. J Natl Cancer Inst. 1998; 90: 675–84. [DOI] [PubMed] [Google Scholar]
- Andreyev H, Norman AR, Cunningham D, Oates J, Dix BR, Iacopetta BJ, et al.. Kirsten ras mutations in patients with colorectal cancer: the ‘RASCAL II’ study. Br J Cancer. 2001; 85: 692–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richman SD, Seymour MT, Cjambers P, Elliott F, Daly CL, Meade AM, et al.. KRAS and BRAF mutations in advanced colorectal cancer are associated with poor prognosis but not preclude benefit from oxaliplatin or irinotecan: results from MRC FOCUS trial. J Clin Oncol. 2009; 27: 5931–7. [DOI] [PubMed] [Google Scholar]
- Petrelli F, Coinu A, Cabiddu M, Ghilardi M, Barni S.. KRAS as prognostic biomarker in metastatic colorectal cancer patients treated with bevacizumab: a pooled analysis of 12 published trials. Med Oncol. 2013; 30: 650. [DOI] [PubMed] [Google Scholar]
- Clancy C, Burke JP, Kalady MF.. BRAF mutation is associated with distinct clinicopathological characteristics in colorectal cancer: a systematic review and meta-analysis. Colorectal Dis. 2013; 15(12): e711–8. [DOI] [PubMed] [Google Scholar]
- Samowitz WS, Sweeney C, Herrick J, Albertsen H, Levin TR, Murtaugh MA, et al.. Poor survival associated with the BRAF V600E mutation in microsatellite- stable colon cancers. Cancer Res. 2005; 65(14): 6063–9. [DOI] [PubMed] [Google Scholar]
- Douillard JY, Siena S, Cassidy J, Tabernero J, Burkes R, Barugel M, et al.. Randomized, phase III trial of panitumumab with infusional fluorouracil, leucovorin, and oxaliplatin (FOLFOX4) versus FOLFOX4 alone as first-line treatment in patients with previously untreated metastatic colorectal cancer: the PRIME study. J Clin Oncol. 2010; 28(31): 4697–705. [DOI] [PubMed] [Google Scholar]
- Bokemeyer C, Van Cutsem E, Rougier P, Ciardiello F, Heeger S, Schlichting M, et al.. Addition of cetuximab to chemotherapy as first-line treatment for KRAS wild-type metastatic colorectal cancer: pooled analysis of the CRYSTAL and OPUS randomised clinical trials. Eur J Cancer. 2012; 48: 1466–75. [DOI] [PubMed] [Google Scholar]
- Maughan TS, Adams RA, Smith CG, Meade AM, Seymour MT, Wilson RH, et al.; MRC COIN Trial Investigators. Addition of cetuximab to oxaliplatin based first-line combination chemotherapy for treatment of advanced colorectal cancer: results of the randomised phase 3 MRC COIN trial. Lancet. 2011; 377: 2103–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurwitz HI, Yi J, Ince W, Novotny WF, Rosen O.. The clinical benefit of bevacizumab in metastatic colorectal cancer is independent of KRAS mutation status: analysis of phase III sudy of bevacizumab with chemotherapy in previously untreated metastatic colorectal cancer. Oncologist. 2009; 14: 22–8. [DOI] [PubMed] [Google Scholar]
- Díaz-Rubio E, Gómez-España A, Massutí B, Sastre J, Reboredo M, Manzano JL, et al.. Role of Kras status in patients with metastatic colorectal cancer receiving first-line chemotherapy plus bevacizumab: a TTD group cooperative study. PLoS One. 2012; 7(10): e47345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YL, Liang YH, Tsai JH, Liau JY, Liang JT, Lin BR, et al.. Oxaliplatin-based chemotherapy is more beneficial in KRAS mutant than in KRAS wild type metastatic colorectal cancer patients. PlosOne. 2014; 9(2): e86789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltran L, De Maglio G, Pella N.. Prognostic role of KRAS, NRAS, BRAF and PIK3CA mutations in advanced colorectal cancer. Future Oncol. 2015; 11(4): 629–40. [DOI] [PubMed] [Google Scholar]
- De Roock W, Claes B, Bernasconi D, De Schutter J, Biesmans B, Fountzilas G, et al.. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy- refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol. 2010; 11(8): 753–62. [DOI] [PubMed] [Google Scholar]
- Peeters M, Kafatos G, Taylor A, Gastanaga VM, Oliner KS, Hechmati G, et al.. Prevalence of RAS mutations and individual variation patterns among patients with metastatic colorectal cancer: A pooled analysis of randomised controlled trials. Eur J Cancer. 2015. September; 51(13): 1704–13. [DOI] [PubMed] [Google Scholar]
- Molinari F, Felicioni L, Buscarino M, De Dosso S, Buttitta F, Malatesta S, et al.. Increased detection sensitivity for KRAS mutations enhances the prediction of anti-EGFR monoclonal antibody resistance in metastatic colorectal cancer. Clin Cancer Res. 2011; 17: 4901–14. [DOI] [PubMed] [Google Scholar]
- Dono M, Massucco C, Chiara S, Sonaglio C, Mora M, Truini A, et al.. Low percentage of KRAS mutations revealed by locked nucleic acid polymerase chain reaction: implications for treatment of metastatic colorectal cancer. Mol Med. 2013; 18: 1519–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsiatis AC, Norris-Kirby A, Rich RG, Hafez MJ, Gocke CD, Eshleman JR, Murphy KM.. Comparison of Sanger sequencing, pyrosequencing, and melting curve analysis for the detection of KRAS mutations: diagnostic and clinical implications. J Mol Diagn. 2010. July; 12(4): 425–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Qiu T, Zhi W, Shi S, Zou S, Ling Y, et al.. Colorectal carcinomas with KRAS codon 12 mutation are associated with more advanced tumor stages. BMC Cancer. 2015. May 1; 15: 340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita Y, Taniguchi H, Komori A, Nitta S, Yamaguchi K, Kondo C, et al.. CA19-9 level as a prognostic and predictive factor of bevacizumab efficacy in metastatic colorectal cancer patients undergoing oxaliplatin-based chemotherapy. Cancer Chemother Pharmacol. 2014. February; 73(2): 409–16. [DOI] [PubMed] [Google Scholar]
- Trevisiol C, Di Fabio F, Nascimbeni L, Peloso L, Salbe C, Ferruzzi E, et al.. Prognostic value of circulating KRAS2 gene mutations in colorectal cancer with distant metastases. Int J Biol Markers. 2006. Oct-Dec; 21(4): 223–8. [DOI] [PubMed] [Google Scholar]
- Selcukbiricik F, Erdamar S, Ozkurt CU, Molinas Mandel N, Demirelli F, Ozguroglu M, et al.. The role of K-RAS and B-RAF mutations as biomarkers in metastatic colorectal cancer. J BUON. 2013. Jan-Mar; 18(1): 116–23. [PubMed] [Google Scholar]
- Cho M, Akiba C, Lau C, Smith D, Telatar M, Afkhami M, et al.. Impact of RAS and BRAF mutations on carcinoembryonic antigen production and pattern of colorectal metastases. World J Gastrointest Oncol. 2016. January 15; 8(1): 128–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Custodio A, Barriuso J, de Castro J, Martínez- Marín V, Moreno V, Rodríguez-Salas N, Feliu J.. Molecular markers to predict outcome to antiangiogenic therapies in colorectal cancer: current evidence and future perspectives. Cancer Treat Rev. 2013; 39: 908–24. [DOI] [PubMed] [Google Scholar]
- Rak J, Mitsuhashi Y, Bayko L, Filmus J, Shirasawa S, Sasazuki T, Kerbel RS.. Mutant ras oncogenes upregulate VEGF/VPF expression: implications for induction and inhibition of tumor angiogenesis. Cancer Res. 1995; 55: 4575–80. [PubMed] [Google Scholar]
- Rak J, Yu JL, Kerbel RS, Coomber BL.. What do oncogenic mutations have to do with angiogenesis/ vascular dependence of tumors? Cancer Res. 2002; 62: 1931–4. [PubMed] [Google Scholar]
- Bencsikova B, Bortlicek Z, Halamkova J, Ostrizkova L, Kiss I, Melichar B, et al.. Efficacy of bevacizumab and chemotherapy in the first-line treatment of metastatic colorectal cancer: broadening KRAS-focused clinical view. BMC Gastroenterol. 2015; 15: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruera G, Cannita K, Tessitore A, Russo A, Alesse E, Ficorella C, Ricevuto E.. The prevalent KRAS exon 2 c.35 G>A mutation in metastatic colorectalcancer patients: A biomarker of worse prognosis and potential benefit ofbevacizumab-containing intensive regimens? Crit Rev Oncol Hematol. 2015; 93: 190–202. [DOI] [PubMed] [Google Scholar]
- Nash GM, Gimbel M, Shia J, Nathanson DR, Ndubuisi MI, Zeng ZS, et al.. KRAS mutation correlates with accelerated metastatic progression in patients with colorectal liver metastases. Ann Sur Oncol. 2010; 14: 572–8. [DOI] [PubMed] [Google Scholar]
- Rizzo S, Bronte G, Fanale D, Corsini L, Silvestris N, Santini D, et al.. Prognostic vs predictive molecular biomarkers in colorectal cancer: is KRAS and BRAF wild type status required for anti-EGFR therapy? Cancer Treat Rev. 2010; 36 Suppl 3: S56–61. [DOI] [PubMed] [Google Scholar]