Abstract
The effects of more than 2 years of trichloroethene (TCE) application on community succession and function were studied in two aerobic sequencing batch reactors. One reactor was fed phenol, and the second reactor was fed both phenol and TCE in sequence twice per day. After initiation of TCE loading in the second reactor, the TCE transformation rates initially decreased, but they stabilized with an average second-order rate coefficient of 0.044 liter mg−1 day−1 for 2 years. In contrast, the phenol-fed reactor showed higher and unstable TCE transformation rates, with an average rate coefficient of 0.093 liter mg−1 day−1. Community analysis by terminal restriction fragment length polymorphism (T-RFLP) analysis of the 16S rRNA genes showed that the phenol-plus-TCE-fed reactor had marked changes in community structure during the first 100 days and remained relatively stable afterwards, corresponding to the period of stable function. In contrast, the community structure of the phenol-fed reactor changed periodically, and the changes coincided with the periodicity observed in the TCE transformation rates. Correspondence analysis of each reactor community showed that different community structures corresponded with function (TCE degradation rate). Furthermore, the phenol hydroxylase genotypes, as determined by restriction fragment length polymorphism analysis, corresponded to community structure patterns identified by T-RFLP analysis and to periods when the TCE transformation rates were high. Long-term TCE stress appeared to select for a different and stable community structure, with lower but stable TCE degradation rates. In contrast, the community under no stress exhibited a dynamic structure and dynamic function.
Trichloroethene (TCE) is a potentially carcinogenic, volatile, chlorinated solvent that is one of the most common groundwater pollutants in the United States (1, 10). TCE can be degraded anaerobically by reductive dechlorination, but the degradation is often incomplete, yielding cis-dichloroethene and vinyl chloride, a carcinogen (22, 30). However, TCE can be degraded aerobically by cometabolism, a fortuitous oxidation that does not result in energy or carbon for cell growth (2). Cometabolism of TCE produces an epoxide that is spontaneously converted to innocuous products such as formate and glyoxylate (39, 40), but some epoxide reacts with the catalytic enzyme, destroying the cells (41). Hence, long-term TCE exposure can select against TCE degraders (26) or possibly for a more active TCE-degrading community if active organisms are competitive for formate and glyoxylate.
Cometabolism has been successfully used for degradation of TCE in bioreactors (12, 35) and in groundwater communities (21, 28) provided the appropriate growth substrate. However, the majority of the studies have focused on the kinetics and efficiency of TCE degradation without considering the microbial communities involved. In only one study did the workers investigate the dynamics and diversity of microbial communities undergoing TCE cometabolism in a test aquifer (18, 19). Understanding the structure and stability of microbial communities is important for the success of bioremediation strategies since particular microbial communities can be responsible for the success or failure of the process. Insights into processes such as community succession are necessary to understand and predict future changes in community structure in natural and managed ecosystems. Often, bioreactor systems are operated for extended periods of time with the assumption that the microbial community structure remains unchanged. However, studies performed with a functionally stable methanogenic reactor revealed that microbial communities can be very dynamic, suggesting that stable function is not always correlated with stable community structure (17). Alternatively, perturbation studies with 1,1-dichlorethene, a very toxic compound (5, 15), have suggested that microbial communities can be quite resilient since the community structure recovered to its original state (19). Hence, there is a need to study the dynamics and stability of microbial communities with respect to their function.
In the study reported here, we compared the structures and functions of two reactor communities, one fed phenol only and the other fed phenol plus TCE for over 2 years of continuous operation. Our objectives were to determine the effect of long-term TCE application on community structure, succession, and reactor performance (performance was measured as rates of phenol and TCE degradation). We used sequencing batch reactors (SBRs) so that the phenol consumption phase was temporally separated from the TCE degradation phase to avoid competitive inhibition for the two substrates. Periodic changes in community structure and function were observed in the SBR that received phenol alone. These changes were not observed in the phenol-plus-TCE-fed reactor, in which long-term exposure to TCE selected for a community that was more stable in terms of structure and function.
MATERIALS AND METHODS
Reactor design and operation.
Two aerobic bench-scale SBRs (12) were inoculated from a phenol-fed reactor that showed high and stable phenol and TCE transformation kinetics (36). Each reactor consisted of a 2.2-liter glass vessel and mixer system (M-100 minijar fermentor; Wheaton, Millville, N.J.). The reactor contents were mixed at 180 rpm, except during the settling phase, and the temperature was maintained at 22 ± 1°C by water recirculation from a temperature-controlled water bath through a cooling plate inside the vessel. The reactors were operated aerobically by pumping air at a flow rate of 220 to 280 ml/min. Both vessels were covered to avoid photosynthetic growth. Phenol and TCE solutions were fed by using 60-ml syringes and syringe pumps (Harvard Apparatus Inc., Holliston, Mass.). Supernatant was decanted and mineral medium was added by using peristaltic pumps (Watson-Marlow, Wilmington, Md.). Peristaltic air and syringe pumps were controlled by a programmable timer (Fisher Scientific, Pittsburgh, Pa.). Both reactors were fed phenol for 30 days before TCE was added to one of them. Reactor operation was as follows. Initially, both reactors received 25 ml of a mixture of phenol (7,200 mg/liter) and mineral medium [containing (per liter) 2.13 g of Na2HPO4, 2.04 g of KH2PO4, 1 g of (NH4)2SO4, 0.067 g of CaCl2 · 2H2O, 0.248 g of MgCl2 · 6H2O, 0.5 mg of FeSO4 · 7H2O, 0.4 mg of ZnSO4 · 7H2O, 0.002 mg of MnCl2 · 4H2O, 0.05 mg of CoCl2 · 6H2O, 0.01 mg of NiCl2 · 6H2O, 0.015 mg of H3BO3, and 0.25 mg of EDTA] over a 0.5-h interval to obtain a final phenol concentration of 100 mg/liter. This concentration was selected because it was sufficient for cell growth and fortuitous TCE oxidation in the next stage. Higher concentrations of phenol could have resulted in growth inhibition. Six hours after phenol addition the TCE-fed reactor received 25 ml of TCE diluted with mineral medium (0.2 mg/ml) and additional mineral medium (1 liter) during 1 h so that the final concentration of TCE was 5 mg/liter, while the phenol-fed reactor received mineral medium only. The 5-mg/liter TCE concentration was chosen because pure-culture studies indicated that concentrations between 0.78 and 5.25 mg/liter were sufficient to observe TCE-mediated toxicity and reduced growth rates, while they did not completely kill the cells (24, 26). Three hours later, suspended solids were allowed to settle for 30 min, and the supernatant (∼1.1 liter) was removed before another 12-h cycle began. Suspended solids (200 ml) were removed every other day 1.5 h after phenol injection to maintain a mean cell residence time of 11 days.
TCE and phenol analysis.
Reactor performance was evaluated by calculation of the kinetics for phenol and TCE removal in batch assays. Samples (5 ml) from each reactor were taken 1.5 h after the end of the phenol addition step of the cycle and were transferred to three 20-ml glass vials that were crimp sealed with Teflon-coated butyl rubber stoppers. For TCE removal measurements, different amounts (10 to 50 μl) of TCE in water (∼1,000 mg/liter) were injected into the vials, which were then incubated at 120 rpm in a rotary shaker at room temperature. Periodically, 0.1 ml of the headspace gas was removed with a 0.5 ml Pressure-Lok series A-2 gas syringe (Hamilton Company, Reno, Nev.) and injected into a Hewlett-Packard 5890A gas chromatograph equipped with a capillary column (DB624; 30 m by 0.53 mm [inside diameter]), a flame ionization detector, and an electron capture detector. The gas chromatograph was operated isothermally at 90°C with helium as the carrier gas (flow rate, 12 ml/min), and the injection port temperature was set at 250°C. The temperatures of the flame ionization detector and the electron capture detector were 250 and 350°C, respectively. Second-order rate constants for TCE removal, which normalized for biomass, were calculated from the TCE concentrations as described previously (12). Analyses of the headspace gas from the phenol-plus-TCE-fed reactor indicated that the injected TCE was completely degraded before the settling and decanting phases. For determinations of phenol removal rates, different amounts of phenol were injected into vials, which were then incubated as described above. Periodically, samples were removed with a syringe and filtered through 0.2-μm-pore-size nylon syringe filters (Wheaton). Filtrates (2 ml) were collected to measure the phenol concentrations with a high-performance liquid chromatograph equipped with a C18 column (Waters, Milford, Mass.) and a UV detector set to 235 nm. The solvent used was an acetonitrile-water mixture (60:40) at a flow rate of 1 ml/min. Analyses of samples from both reactors after phenol injection indicated that the phenol concentration decreased to zero after the first 2 h of the phenol recharge stage. Zero-order kinetic transformation coefficients for phenol removal were calculated from the phenol concentrations as described previously (12). The reactor biomass, expressed as dry weight, varied over the entire operational period. The means ± 95% confidence intervals for the phenol-fed and phenol-plus-TCE-fed reactors were 1,490 ± 66 and 1,641 ± 58 mg/liter, respectively. The dry weight of the phenol-fed reactor included approximately 10% extracellular polymeric substances, while the phenol-plus-TCE-fed reactor contained 45% extracellular polymeric substances (7).
Reactor sampling and DNA extraction.
Samples (∼12 ml) of suspended solids were taken for community analysis at the same time that the kinetic assays were performed. The samples were centrifuged for 15 min at 6,000 × g and 4°C, the supernatant fractions were decanted, and the cell pellets were stored at −80°C. No major wall growth was observed in the reactors.
For nucleic acid extraction, cells were resuspended in TE buffer (10 mM Tris [pH 8.0], 1 mM EDTA [pH 8.0]) and ATL lysis buffer (QIAGEN, Chatsworth, Calif.) before they were mixed with a vortex mixer. Each cell suspension was subjected to two freeze-thaw cycles before it was digested with lysozyme (50 μl of a 100-mg/ml solution; Gibco BRL, Gaithersburg, Md.) and achromopeptidase (10 μl of a 25-mg/ml solution; Sigma, St. Louis, Mo.). The cell lysate was digested with RNase (10 mg/ml; Boehringer Mannheim, Indianapolis, Ind.), followed by proteinase K (45 μl of a 25-mg/ml solution; Gibco BRL) for 1 h at 60°C. Nucleic acids were purified by three phenol-chloroform-isoamyl alcohol extractions and were precipitated overnight at −20°C by addition of 0.1 volume of 3 M sodium acetate (pH 5.3) and 0.6 volume of 100% isopropanol. The DNA pellet was resuspended in 100 μl of sterile deionized water. The purity and quantity of the DNA were determined by UV spectrophotometry at 260 and 280 nm. The DNA was stored at −20°C.
PCR primers and conditions.
Amplification of bacterial 16S rRNA genes (rDNAs) from the community DNA was performed with primer 8F (5′-AGAGTTTGATCMTGGCTCAG-3′, where M is A or C) (20), fluorescently labeled at the 5′ end with the phosphoramidite dye 5-hexachlorofluorescein (Operon Inc., Alameda, Calif.), and primer 1392R (5′-ACGGGCGGTGTGTACA-3′) (3). PCRs were carried out as described previously (9) with the following modifications: 20 ng of template DNA was added to each PCR mixture, and 25 cycles of amplification were performed. Amplification of the phenol hydroxylase genes from the community DNA was performed with primers FD2 (5′-GGCATCAARATYNNNGACTGG-3′, where N is a mixture of A, C, G, and T; R is A or G; and Y is C or T) and Phe212 (5′-GTTGGTCAGCACGTACTCGAAGGAGAA-3′) (42). These primers produced a 490-bp PCR product from the large subunit (α subunit) of the multicomponent phenol hydroxylase gene family. Primer FD2 was designed by comparative analysis of amino acid sequences of this protein from Pseudomonas sp. strain CF600, Pseudomonas putida strain H, P. putida P35X, Burkholderia cepacia G4, and Burkholderia sp. strain JS150. Primer specificity was tested by using a collection of isolates known to degrade phenol (7) and B. cepacia G4. The PCR products were sequenced and were found to be highly similar to phenol hydroxylase genes from known phenol degraders. PCR amplification was performed by using the thermal cycler and reaction mixture described previously (9), except that 25 pmol of each primer was used and five replicate reaction mixtures were prepared for each sample. The following touchdown thermal profile was used for amplification: an initial denaturation step of 5 min at 95°C, followed by 10 cycles of 94°C for 30 s, 30 s of annealing (in which the temperature started at 58°C and decreased 1°C every cycle), and 45 s at 72°C. In addition, 25 more cycles were carried out with an annealing temperature of 55°C. The quality and quantity of the PCR products were determined by electrophoresis of an aliquot (10 μl) of each PCR mixture on a 2.0% (wt/vol) agarose gel (Gibco BRL) in 1× Tris-acetate-EDTA.
PCR product purification, restriction digestion, and gel separation.
PCR product purification, restriction digestion, and gel separation for terminal restriction fragment length polymorphism (T-RFLP) analysis of 16S rDNAs were performed as described previously (9) with the HaeIII and CfoI restriction enzymes (Gibco BRL).
For the phenol hydroxylase genes, the PCR products from the five replicate reactions were combined and concentrated by roto-evaporation. The concentrated PCR products were separated in a 2.5% (wt/vol) agarose gel (Gibco BRL) in 1× Tris-acetate-EDTA, and the band of the correct size was excised from the gel and eluted in 60 μl of sterile filtered distilled water by using a QIAquick gel extraction kit (QIAGEN). The purified PCR products were concentrated by roto-evaporation and digested with 15 U of MspI and RsaI restriction enzymes combined (Gibco BRL). The resulting fragments were separated in a 4% (wt/vol) Metaphor agarose gel (FMC, Indianapolis, Ind.) in 1× Tris-borate-EDTA buffer at 140 V for 3.5 h at 4 °C and then stained with ethidium bromide (0.5 μg/ml). Gel images were analyzed by using GelCompar 4.5 (Applied Maths, Kortrijk, Belgium).
Analysis of T-RFLPs.
Electropherograms from each sample from both reactors were manually aligned with each other by using Genotyper 2.5 (Applied Biosystems Instruments, Foster City, Calif.). To avoid primer artifacts, fragments smaller than 35 bp were excluded from the analysis. The alignment resulted in a matrix in which different peaks were considered to be indicative of different ribotypes present in the community, and the peak heights were used as a measure of ribotype abundance. After alignment, samples for which the sum of the peak heights was less than 10,000 U were redigested and separated, since data below this threshold are unreliable for statistical analysis (8). Histograms with the abundance of each peak normalized by the total signal in each sample were used to compare the changes in community structure in each reactor. Similarity between samples from the same reactor or between samples from the two reactors was determined by correspondence analysis (CA) (25, 37) with the software package ADE-4 (http://biomserv.univ-lyon1.fr/ADE-4.html) (38). CA was chosen for the analysis of the T-RFLP profiles because this ordination method recovers the underlying structure of the data by analysis of the proportional changes in species abundance, providing a global view of the community structure without a severe effect due to noise. For CA, peak height was used as described above, and the data analysis was performed in a step-wise process. CA was initially performed on all the samples (from both reactors) to compare the two reactors (between- and within-treatment variability). Subsequently, each reactor data set was analyzed individually to evaluate within-treatment variability.
To evaluate the reproducibility of the T-RFLP methodology, nine replicate samples from each reactor (taken after 811 days of operation) and 32 lanes of an internal standard were analyzed by using two similarity indices. Similarity indices were used because they generate direct sample-to-sample comparisons, providing more precise estimates of the variability. The Sørensen similarity index takes into account the presence or absence of peaks, while the Steinhaus similarity index takes into account the abundance of each peak (25). Similarity matrixes were generated for each set of nine replicates and 32 lanes of the internal standard (Gene scan 2500; Applied Biosystems, Alameda, Calif.) with the SIMIL module of R-package 4.0 (http://www.fas.umontreal.ca/BIOL/legendre/) (25). Means, ±95% confidence intervals, and coefficients of variation were calculated from the similarity values.
RESULTS
TCE transformation rates.
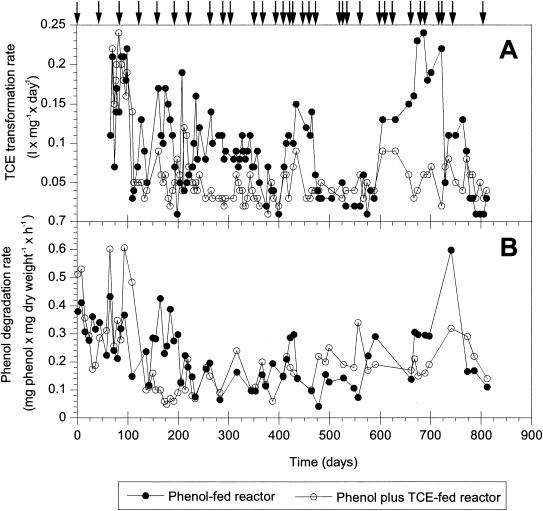
Reactor communities showed different TCE transformation rates after long-term phenol and phenol plus TCE application (Fig. 1A). From day 60 to day 100 the two reactors showed similar, relatively high TCE transformation rates (0.22 and 0.24 liter mg−1 day−1 for the phenol- and phenol-plus-TCE-fed reactors, respectively). By day 109, the TCE transformation rates had decreased to 0.03 liter mg−1 day−1 for the phenol-fed reactor and to 0.05 liter mg−1 day−1 for the phenol-plus-TCE-fed reactor. At this point, the TCE transformation rates of the reactors diverged. The phenol-fed reactor had variable TCE transformation rates that ranged from 0.01 to 0.24 liter mg−1 day−1, with an average TCE transformation rate of 0.082 ± 0.012 liter mg−1 day−1, from day 150 to day 825. In contrast, the TCE transformation rates in the phenol-plus-TCE-fed reactor were less variable, ranging from 0.01 to 0.12 liter mg−1 day−1, and the average TCE transformation rate was 0.044 ± 0.005 liter mg−1 day−1. Visual inspection of the TCE transformation rates in the phenol-fed reactor suggested that changes in TCE kinetics occurred periodically in this reactor (Fig. 1A). Unimodal TCE transformation rates (i.e., a distribution having one distinct peak with a maximum value) were observed approximately between days 60 and 109, 400 and 500, and 575 and 725 for the phenol-fed reactor. In contrast to the phenol-fed reactor, the phenol-plus-TCE-fed reactor did not show periodicity; there were marked changes in transformation rates only during the first 110 days and minor changes after that (Fig. 1A).
FIG. 1.
Long-term functional responses of the phenol-fed and phenol-plus-TCE-fed reactors. (A) TCE transformation rates. (B) Phenol degradation rates. The arrows indicate the sampling days used for the community structure study.
Phenol degradation rates.
The maximum specific rates of phenol degradation for the phenol-fed reactor were variable during the first 200 days of operation, ranging from 0.1 to 0.4 mg of phenol mg (dry weight)−1 h−1 (Fig. 1B). After 200 days of operation, the phenol degradation rates were stable and in the range from 0.1 to 0.3 mg of phenol mg (dry weight)−1 h−1, except for day 747, when for unexplained reasons the rate increased to 0.6 mg of phenol mg (dry weight)−1 h−1. Visual inspections of phenol degradation did not reveal any marked periodicity, such as that observed for the TCE transformation rates (Fig. 1A).
In the phenol-plus-TCE-fed reactor, the phenol degradation rate started at an initial value of 0.5 mg of phenol mg (dry weight)−1 h−1 and then cycled in the early stage, and there were conspicuous decreases (0.19 and 0.22 mg of phenol mg [dry weight]−1 h−1) and increases (0.6 mg of phenol mg [dry weight]−1 h−1) (Fig. 1B). After day 145 the phenol degradation rate did not increase to values as high as those before this time, and the changes that occurred were less pronounced. No major wall growth was observed.
Community structure as determined by T-RFLP analysis.
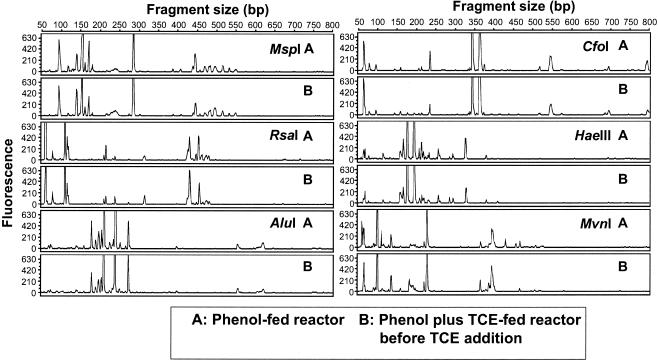
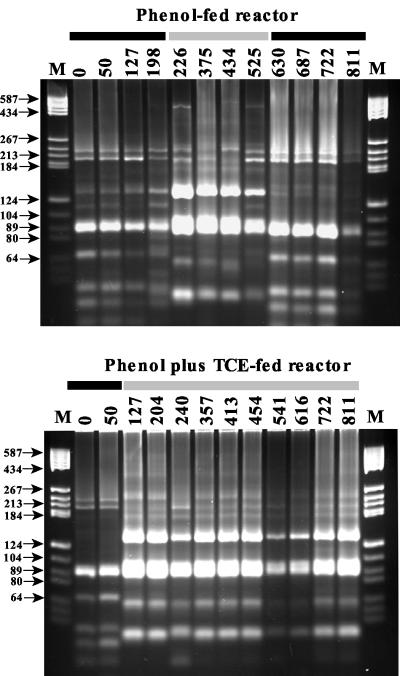
To determine if the two reactor communities were similar after 30 days of operation with phenol and before TCE was added to one of them, T-RFLP analysis was performed with six different tetrameric restriction enzymes (Fig. 2). Each of the enzymes tested produced almost identical patterns for the two reactors, with only minor differences in the heights of some small peaks.
FIG. 2.
T-RFLP electropherograms of PCR-amplified 16S rDNA bacterial genes from replicate reactor communities before TCE application to the phenol-plus-TCE-fed reactor. Samples were digested with six different restriction enzymes individually.
In the T-RFLP procedure, enzyme selection is critical for judging if changes in community structure have occurred. Therefore, to select the best restriction enzymes for the analysis, a subset of all the samples was tested with the six enzymes. Restriction enzymes HaeIII and CfoI were selected for community analysis because they generated the highest number of sharp and distinguishable fragments in all the samples tested. Restriction enzymes such as MspI and RsaI were initially promising (Fig. 2), but in later samples several peaks overlapped.
To assess if the T-RFLP method is a suitable method for monitoring population dynamics, a reproducibility analysis was conducted (Table 1). The indices used measured similarity based on the presence or absence of a fragment (Sørensen index) and the abundance of each fragment (Steinhaus index). To have some reference for comparison, 32 lanes from the DNA marker GS2500 were analyzed. For the two indices the GS2500 standard resulted in the highest values, as expected (Sørensen index, 1.0; Steinhaus index, 0.931), and the phenol- and phenol-plus-TCE-fed reactor T-RFLP patterns showed good reproducibility (Table 1). The variability of the community measurements was lower than the variability in previous studies (9, 16, 33, 34), and the percentages of variation ranged from 3.3 to 6.2%. This is an important finding since in previous studies there were some disagreements concerning the reproducibility of the T-RFLP method (9, 33, 34), which were probably caused by the limited number of samples analyzed. The samples from the phenol-plus-TCE-fed reactor had a notable amount of polymer (7) that could have affected DNA extraction procedures and might account for the slightly higher variability in this reactor. Nevertheless, good reproducibility was observed for the nine replicate samples from each reactor, indicating that the samples were homogeneous and representative of the microbial community in each system.
TABLE 1.
Reproducibility of the T-RFLP dataa
| Sample | n | Sørensen index
|
Steinhaus index
|
Coefficient of variation (%)
|
|||
|---|---|---|---|---|---|---|---|
| Mean | 95% Confidence interval | Mean | 95% Confidence interval | Sørensen index | Steinhaus index | ||
| Phenol-fed reactor | 9 | 0.966 | ±0.012 | 0.921 | ±0.013 | 3.5 | 4.3 |
| Phenol-plus-TCE-fed reactor | 9 | 0.937 | ±0.010 | 0.851 | ±0.018 | 3.3 | 6.2 |
| TAMRA GS2500 standard | 32 | 1 | 0.931 | ±0.003 | 0.0 | 3.4 | |
Means and 95% confidence intervals for Sørensen and Steinhaus similarity indices were calculated from pairwise comparisons. The TAMRA GS2500 standard is a molecular weight marker used to determine the sizes of the fragments in the gels.
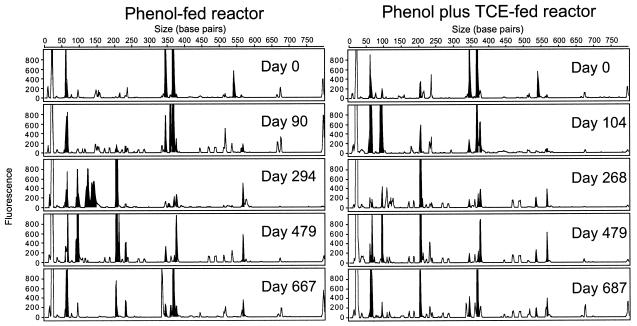
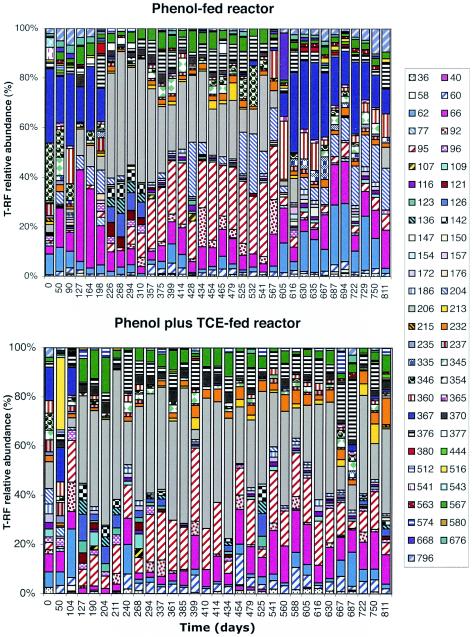
T-RFLP analysis of the two reactors showed that there were marked changes in community structure over time (Fig. 3). Fragments between 340 and 375 bp long had different peak heights for the two reactors. The relative peak heights for other fragments increased over time (200-bp fragment in the phenol-plus-TCE-fed reactor) or remained relatively stable (58-bp fragment in the phenol-fed reactor).
FIG. 3.
Changes in bacterial community structure over time. T-RFLP electropherograms were generated from a CfoI digest of the 16S rDNA PCR product. The shaded peaks indicate dynamic ribotypes.
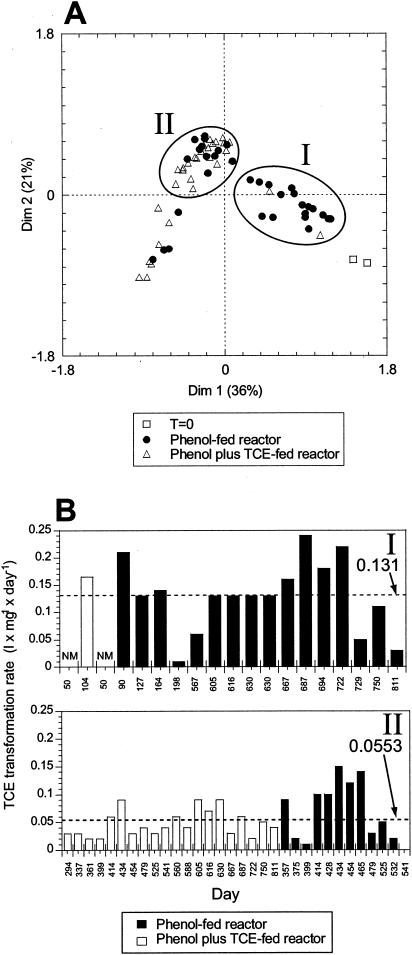
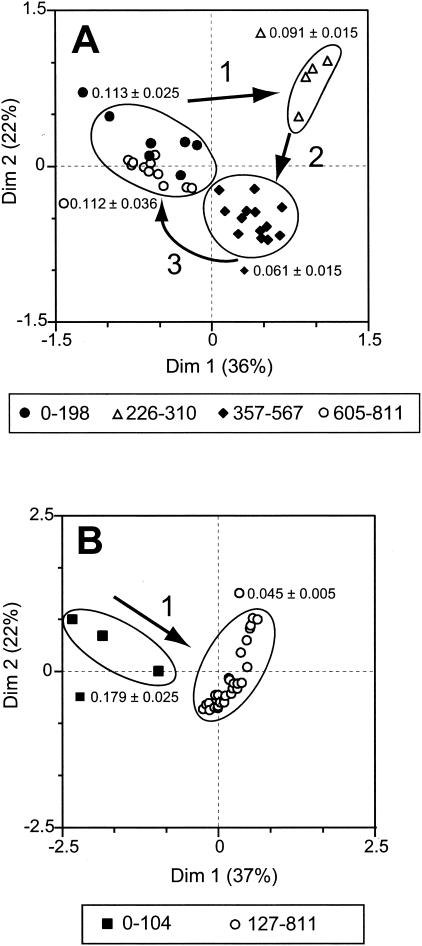
To determine if there were similarities between the two reactor communities, CA was applied to data combined from the reactors. CA produces a graphical representation of samples along two or more axes of reference (dimensions) that contain a fraction of the total variability. Each dimension could represent some of the possible environmental factors that affect species distributions. Samples with similar community structures are closer together in the multidimensional plot. The amount of data scatter in the multidimensional plot represents the variability of the community. CA resulted in data points that formed an arch structure, which is typical of systems undergoing succession (37) (Fig. 4A). CA data from the two reactors before TCE addition (zero time) were nearly identical and different from the data from all the other samples, indicating that the reactor communities were almost identical at the beginning of the experiment and then diverged. Sample data from the phenol-fed reactor were scattered in the plot, which suggests that the phenol-fed community was highly variable. However, the data from the phenol-plus-TCE-fed reactor were more confined to the left arm of the arch, suggesting that the community in the phenol-plus-TCE-fed reactor was more stable than the community in the phenol-fed reactor. The two reactor communities were similar at different periods (Fig. 4A, clusters I and II). Data from days 50 to 104 of the phenol-plus-TCE-fed reactor grouped with data from days 50 to 198 and 605 to 811 of the phenol-fed reactor (Fig. 4A, cluster I). Interestingly, the highest TCE transformation rates were observed in both reactors during the days with similar community structures, suggesting that some community structures were beneficial for high TCE transformation rates (Fig. 4B, graph I). Similarities in community structure between reactors were also observed for days 357 to 541 of the phenol-fed reactor and for days 294 to 361, 399, and 414 to 811 of the phenol-plus-TCE-fed reactor (Fig. 4A, cluster II), which were periods with intermediate to low TCE degradation rates (Fig. 4B, graph II). All these data together indicate that the communities of the two reactors were similar at different times and that similar communities corresponded to similar functions.
FIG. 4.
Community structure and function relationships between reactors. (A) CA of T-RFLP data from reactor communities. Results from independent digests with CfoI and HaeIII were combined for the analysis. Circles and roman numbers indicate periods when the community structures of the two reactors were similar, as follows: cluster I, phenol-fed reactor on days 50 to 198, 567, and 605 to 811 and phenol-plus-TCE-fed reactor on days 50 to 104; cluster II, phenol-fed reactor on days 357 to 541 and phenol-plus-TCE-fed reactor on days 294 to 361, 399, and 414 to 811. T=0, day 0 of each reactor before TCE application to one of them. (B) TCE transformation rates for the two clusters identified by CA. Graph I, cluster I; graph II, cluster II. The dotted lines indicate the average TCE transformation rates (in liter per milligram per day) for the days analyzed. Dim, dimension; NM, not measured.
Changes in community structure can be seen in more detail in histograms that show the relative proportion of each fragment over time (Fig. 5). The appearance and disappearance of fragments indicate that the community in the phenol-fed reactor was dynamic. For example, fragment 367 disappeared or its level was below the detection limit after 198 days, and simultaneously fragment 206 appeared and was present from day 127 until day 616. Fragment 367 reappeared after 605 days of operation. CA of each reactor separately was used to discern temporal patterns in each reactor community individually. CA confirmed that there were differences in community dynamics between the two reactors and revealed a relationship between community structure and function (Fig. 6). In the phenol-fed reactor, the community structure changed at least three times, converging back to its original state (Fig. 6A). The T-RFLP patterns of samples from days 0 to 198 clustered with samples from days 605 to 811, indicating that the communities in these two periods were very similar. The average TCE transformation rates for these two periods were also very similar (0.113 and 0.112 liter mg−1 day−1, respectively). The T-RFLP profiles from days 226 to 310 and from days 357 to 567 formed two distinct clusters. The average TCE transformation rates for these two clusters were sequentially lower, and the values were 0.091 and 0.061 liter mg−1 day−1, respectively. Statistically significant differences in TCE transformation rates were observed between the day 0 to day 198 cluster and the day 357 to day 567 cluster, between the day 226 to day 310 cluster and the day 605 to day 811 cluster, and between the day 357 to day 567 cluster and the day 605 to day 811 cluster (Table 2). All these results suggest that there was a prominent relationship between community structure and function.
FIG. 5.
Relative abundance of bacterial terminal restriction fragments (T-RF) from reactor samples after digestion with CfoI. The numbers next to the symbols on the right indicate the sizes of the T-RFs (in base pairs).
FIG. 6.
CA of T-RFLP data for each reactor community individually. (A) Phenol-fed reactor; (B) phenol-plus-TCE-fed reactor. The numbers next to the symbols under each panel indicate the day periods. The arrows and numbers (1 to 3) in the graphs indicate the order of the changes in community structure. Results from independent digestions with CfoI and HaeIII were combined for the analysis. The circled areas indicate the different clusters. The mean ± 95% confidence interval for the TCE transformation rates for each of the different clusters is indicated next to the circle. Dim, dimension.
TABLE 2.
TCE transformation rate comparisonsa m
| Clusters comparedb | Mean difference | df | P value |
|---|---|---|---|
| P 0-198 and P 226-310 | 0.028 | 17 | 0.1216 |
| P 0-198 and P 357-567 | 0.058 | 25 | 0.0013c |
| P 0-198 and P 605-811 | 0.003 | 19 | 0.8778 |
| P 226-310 and P 357-567 | 0.016 | 17 | 0.1591 |
| P 226-310 and P 605-811 | −0.044 | 14 | 0.0138c |
| P 357-567 and P 605-811 | −0.046 | 19 | 0.0058c |
| PT 0-104 and PT 127-811 | 0.144 | 10 | <0.0001c |
The Student t test was used to determine if there were significant differences between the TCE transformation rates of the different clusters.
Based on CA. P, phenol-fed reactor; PT, phenol-plus-TCE-fed reactor. The numbers indicate the days of operation.
Significant difference at the 0.05% level.
The community in the phenol-plus-TCE-fed reactor was less dynamic than the community in the phenol-fed reactor (Fig. 5). Most of the changes in community structure occurred in the first 100 days of operation. Fragment 367 disappeared during the first 100 days, while fragment 206 appeared after 127 days and was present until the end of the sampling period. In addition to fragment 206, fragments 60, 66, 95, 376, and 567 were present most of time during days 127 to 811. CA revealed that the majority of the changes occurred during days 0 to 104 (Fig. 6B), while for days 127 to 811 the points tended to cluster together near the origin, indicating that the community structures were similar and relatively stable during this lengthy period. The TCE transformation rates for these clusters were significantly different, and the values were 0.19 and 0.044 liter mg−1 day−1 for the day 0 to day 104 cluster and the day 127 to day 811 cluster, respectively (Fig. 6B and Table 2).
To investigate changes in community structure at the functional level, phenol hydroxylase genes were amplified by PCR, digested with two restriction enzymes, and separated by electrophoresis (Fig. 7). Cluster analysis of the phenol hydroxylase fingerprints revealed that populations with the gene in the phenol-fed reactor exhibited periods that corresponded approximately to the periods found by T-RFLP analysis (Fig. 7). The fingerprints for days 0 to 198 of the phenol-fed reactor were different from the fingerprints for days 226 to 525, when the intensity of a ∼135-bp band increased and a band between 89 and 104 bp appeared. Furthermore, the fingerprints from days 630 to 811 were similar to profiles for days 0 to 198, which is consistent with the T-RFLP data (Fig. 5 and 6A). The phenol hydroxylase-bearing populations in the phenol-plus-TCE-fed reactor showed two periods of change (Fig. 7). During the first 50 days the community was stable, and then it changed, with the emergence of a band at ∼135 bp and the disappearance of two bands at ∼210 and ∼230 bp. This profile remained stable until the end of the experiment (day 811). Cluster analysis of the phenol hydroxylase fingerprints from both reactors confirmed that the two communities were similar during the initial days and the later period of the phenol-fed reactor, when the highest TCE transformation rates were observed. The communities were also similar during days 226 to 525 and days 127 to 811 of the phenol-fed reactor and the phenol-plus-TCE-fed reactor, respectively (Fig. 7), which were periods with low or intermediate TCE transformation rates.
FIG. 7.
Community restriction patterns of PCR-amplified phenol hydroxylase genes digested with RsaI and MspI. The number above each lane indicates the time (in days). The bars at the top indicate the samples with similar fingerprint patterns within and between reactors as determined by unweighted pair group with mathematical average cluster analysis. Lanes M contained markers. The numbers next to the arrows indicate the sizes of the fragments (in base pairs) in the marker lanes.
DISCUSSION
Shih et al. previously showed that pulse feeding of phenol selected for a more efficient TCE-degrading community than continuous feeding of phenol selected for (36). However, the efficiency of TCE degradation was determined by assays of reactor samples, and the effects of TCE application on the community were not measured. In the present study, we employed the phenol feeding strategy of Shih et al., modified by addition of a TCE feeding step to determine the long-term effects of TCE application. Long-term TCE application resulted in a microbial community with reduced TCE transformation rates (0.093 and 0.049 liter mg−1 day−1 for the phenol- and phenol-plus-TCE-fed reactors, respectively), but the rates were relatively stable over 2 years. Toxicity from the TCE epoxide is well documented in P. putida F1 and B. cepacia G4 (32, 41, 43). Although the effects of TCE toxicity can be seen within 1 h of exposure, long-term effects have not been studied. Mars et al. (26) suggested that TCE epoxide toxicity resulted in an increase in maintenance energy cost and the appearance of variants of B. cepacia G4 in batch culture which had lost the catabolic plasmid for TCE cooxidation. After 100 days of reactor operation we observed low but relatively stable phenol degradation rates in the phenol-plus-TCE-fed reactor. In contrast, in the phenol-fed community there were frequent changes in phenol degradation rates that were linked to changes in TCE degradation rates (Fig. 1). Our findings indicate that long-term TCE application in a sequential feeding mode resulted in a community that degraded TCE more slowly but was more stable. It has been proposed that one of the adaptations to TCE toxicity is a reduced rate of TCE cometabolism (23). Apparently, TCE application selected against the organisms that degraded TCE quickly but allowed organisms that degraded TCE more slowly to succeed.
Community analysis by T-RFLP of 16S rDNAs revealed that there was correspondence between community structure and function in the reactors. Changes in TCE transformation rates for the phenol-fed reactor occurred periodically and corresponded to changes in community structure. CA of all the fragments obtained with two enzymes (HaeIII and CfoI) showed that different periods had particular community structures associated with them, and when TCE transformation rates were high, the community structures were similar (Fig. 5 and 6A). Fragment 367 was present during days 0 to 198 and 605 to 729 in the phenol-fed reactor, when the highest TCE transformation rates were observed. In contrast, fragments 206 and 95 were present during days 226 to 567, when the TCE transformation rates were intermediate or lower. Interestingly, a similar pattern was observed in the phenol-plus-TCE-fed reactor, in which fragment 367 was present when the transformation rates were high (days 0 to 104) and was replaced by fragments 206 and 95 when the transformation rates were low (127 to 811 days). Changes in the distribution of phenol-degrading populations can result from adaptations to phenol degradation or interactions between community members (e.g., predation by protozoans, which were present in both reactors). Mutations in the promoter region of the phenol degradation genes (11), repression by organic acids (4, 31), and activities of unknown gene repressors (6) are events that are possible in a mixed community and could affect the fitness of phenol-degrading populations, causing changes in community structure.
While the T-RFLP data suggest that there was a community structure-function relationship, it is possible that the changes observed at the 16S rDNA level were not related to changes in the functional genes. Analysis of the genes encoding phenol hydroxylase (an enzyme often involved in TCE cooxidation) revealed changes in community structure that corresponded to changes in the phenol hydroxylase fingerprints. Different phenol hydroxylase genotypes were present when the reactors had higher transformation rates (days 0 to 198 and 630 to 811 in the phenol-fed reactor and days 0 to 50 in the phenol-plus-TCE-fed reactor), suggesting that some phenol hydroxylases were more efficient in TCE cooxidation (Fig. 7). Furthermore, some phenol hydroxylase genotypes disappeared shortly after TCE was injected into one of the reactors, suggesting that genotypes that were efficient in TCE cometabolism were also sensitive to TCE toxicity.
Understanding community succession has been one of the most challenging topics in contemporary ecology. Current ecological theories of succession predict contrasting outcomes of the process. Succession, as defined by Clements (13), is an ordered unidirectional process of species replacement that culminates in a stable community (i.e., a climax community). More recent theories, however, propose that during succession, disturbances are frequent and communities never reach a climax stage. Our findings suggest that microbial community succession was continuous in the phenol-fed reactor, while a relatively stable community was formed in the phenol-plus-TCE-fed reactor. Furthermore, CA of the phenol-plus-TCE-fed reactor community revealed that most of the changes in the community occurred during the first 100 days (Fig. 6B) and then the community stabilized for 700 days, suggesting that the community reached a climax stage, as proposed by Clements (13). In contrast, the community in the phenol-fed reactor changed periodically over the 2-year period and never reached a stable phase. Interestingly, the community structure of the phenol-fed reactor during the initial days (days 0 to 198) was similar to the community structure at the end of the experiment (days 605 to 811). The similarity between community structures was determined by observing terminal restriction fragments present in the periods that were apparently displaced during the intermediate days (days 226 to 567). This is unusual in ecology since lost species usually do not return to a community. In our case, species were probably never entirely displaced from the community because they could remain as a refugia and their levels were below the detection limit of our methodologies (17, 27). Nevertheless, it can be hypothesized from the changes in community structure that some microbial communities are dynamic and never reach a climax stage, but rather are shifting mosaics in which changes are unpredictable, as observed in some plant communities (29), and only limited temporal stability occurs.
A remaining question is why the phenol-plus-TCE-fed community was more stable than the phenol-fed community. Cook (14) reviewed modern successional theory and concluded that one of the points of commonality in the new theories is that disturbances are frequent and they likely affect the outcome of succession. It is possible that the changes in community structure seen in the phenol-fed reactor were a response to random disturbances that occurred as part of the normal reactor operation. However, because of the way that the reactors were designed and operated, random disturbance should have affected both communities. Then why was one of our communities more stable? A possible answer to this question lies in the effects of TCE on the community. Toxicity studies with pure cultures have suggested that the epoxide generated from TCE cometabolism damages cellular components, rendering cells inactive (41). Furthermore, recent studies on B. cepacia G4 suggest that this strain secretes toxic soluble intermediates from the degradation of TCE, which affect cells that do not degrade TCE (43). Therefore, it is reasonable to hypothesize that TCE application selected for a community that was more resistant to TCE toxicity, and depending on the protection mechanism, the community could have been more resistant to environmental disturbances. For instance, an increase in extracellular polymeric substance accumulation was observed in the phenol-plus-TCE-fed reactor (7), a phenomenon that resulted in the formation of star-like flocs, which to some extent resembled biofilms. Biofilm communities are very stable and resilient to perturbations, suggesting that an analogous mechanism could have conferred increased stability to the TCE-fed community.
The results of this study suggest that the stability of microbial communities can be dependent upon random disturbances and the selective pressure applied to the ecosystem. More than 2 years of phenol and TCE feeding resulted in a community with reduced but stable TCE degradation rates and a relatively stable community structure. Conversely, long-term feeding of phenol resulted in a different community with higher TCE transformation rates but with continuous and unpredictable changes in community structure and function. These findings suggest that microbial communities in nature in similar niches could have contrasting dynamics depending upon particular physical, chemical, or biological conditions imposed.
Acknowledgments
We thank Carl Ramm for advice on CA, Robert SanInocencio for assistance with the reactors, and Wang-kuan Chang and Shannon J. Flynn for help in early stages of this project.
This work was funded under grant R-817873-02-0 from the U.S. Environmental Protection Agency and under grant R-819605 from the Office of Research and Development, U.S. Environmental Protection Agency, to the Great Lakes and Mid-Atlantic Hazardous Substance Research Center and by NSF grant DEB 9120006 to the Center for Microbial Ecology.
REFERENCES
- 1.Agency for Toxic Substances and Disease Registry. 1994. National exposure registry. Trichloroethylene (TCE) subregistry baseline technical report (revised). NTIS publication PB95-154589. Agency for Toxic Substances and Disease Registry, U. S. Public Health Service, Department of Health and Human Services, Atlanta, Ga.
- 2.Alexander, M. 1994. Biodegradation and bioremediation. Academic Press, San Diego, Calif.
- 3.Amann, R. I., W. Ludwig, and K. H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ampe, F., D. Léonard, and N. Lindley. 1998. Repression of phenol catabolism by organic acids in Ralstonia eutropha. Appl. Environ. Microbiol. 64:1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anderson, J. E., and P. L. McCarthy. 1997. Transformation yields of chlorinated ethenes by a methanotrophic mixed culture expressing particulate methane monooxygenase. Appl. Environ. Microbiol. 63:687-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arai, H., S. Akahira, T. Ohishi, M. Maeda, and T. Kudo. 1998. Adaptation of Comamonas testosteroni TA441 to utilize phenol: organization and regulation of the genes involved in phenol degradation. Microbiology 144:2895-2903. [DOI] [PubMed] [Google Scholar]
- 7.Ayala-del-Río, H. L. 2002. Long-term effects of phenol and phenol plus trichloroethene application on microbial communities in aerobic sequencing batch reactors. Ph.D. dissertation. Michigan State University, East Lansing.
- 8.Blackwood, C. B., T. Marsh, S.-H. Kim, and E. A. Paul. 2003. Terminal restriction fragment length polymorphism data analysis for quantitative comparison of microbial communities. Appl. Environ. Microbiol. 69:926-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Braker, G., H. L. Ayala-del-Rio, A. H. Devol, A. Fesefeldt, and J. M. Tiedje. 2001. Community structure of denitrifiers, Bacteria, and Archaea along redox gradients in Pacific Northwest marine sediments by terminal restriction fragment length polymorphism analysis of amplified nitrite reductase (nirS) and 16 rRNA genes. Appl. Environ. Microbiol. 67:1893-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bruckner, J. V., B. D. Davis, and J. N. Blancato. 1989. Metabolism, toxicity, and carcinogenicity of trichloroethylene. Crit. Rev. Toxicol. 20:31-50. [DOI] [PubMed] [Google Scholar]
- 11.Burchhardt, G., I. Schmidt, H. Cuypers, L. Petruschka, A. Völker, and H. Herrmann. 1997. Studies on spontaneous promoter-up mutations in the transcriptional activator-encoding gene phlR and their effects on the degradation of phenol in Escherichia coli and Pseudomonas putida. Mol. Gen. Genet. 254:539-547. [DOI] [PubMed] [Google Scholar]
- 12.Chang, W. 1996. Kinetics characterization of cometabolizing communities and adaptation to nongrowth substrate. Ph.D. dissertation. Michigan State University, East Lansing.
- 13.Clements, F. E. 1916. Plant succession: an analysis of the development of vegetation. Carnegie Inst. Washington Publ., Washington, D.C.
- 14.Cook, J. E. 1996. Implications of modern successional theory for habitat typing: a review. For. Sci. 42:67-75. [Google Scholar]
- 15.Dolan, M. E., and P. L. McCarty. 1995. Small column microcosm for assessing methane-stimulated vinyl-chloride transformation in aquifer samples. Environ. Sci. Technol. 29:1892-1897. [DOI] [PubMed] [Google Scholar]
- 16.Dunbar, J., O. Ticknor Lawrence, and C. Kuske. 2000. Assessment of microbial diversity in four southwestern United States soils by 16S rRNA gene terminal restriction fragment analysis. Appl. Environ. Microbiol. 66:2943-2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fernández, A., S. Huang, S. Seston, J. Xing, R. Hickey, C. Criddle, and J. Tiedje. 1999. How stable is stable? Function versus community composition. Appl. Environ. Microbiol. 65:3697-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fries, M. R., L. J. Forney, and J. M. Tiedje. 1997. Phenol- and toluene-degrading microbial populations from an aquifer in which successful trichloroethene cometabolism occurred. Appl. Environ. Microbiol. 63:1523-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fries, M. R., G. D. Hopkins, P. L. McCarty, L. J. Forney, and J. M. Tiedje. 1997. Microbial succession during a field evaluation of phenol and toluene as the primary substrates for trichloroethene cometabolism. Appl. Environ. Microbiol. 63:5-1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giovannoni, S. J. 1991. The polymerase chain reaction, p. 177-201. In E. Stackebrandt (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, New York, N.Y.
- 21.Hopkins, G. D., and P. L. McCarty. 1995. Field-evaluation of in-situ aerobic cometabolism of trichloroethylene and three dichloroethylene isomers using phenol and toluene as the primary substrates. Environ. Sci. Technol. 29:1628-1637. [DOI] [PubMed] [Google Scholar]
- 22.Infante, P. F., and T. A. Tsongas. 1987. Mutagenic and oncogenic effects of chloromethanes, chloroethenes, and halogenated analogs of vinyl chloride. Environ. Sci. Res. 25:301-327. [Google Scholar]
- 23.Ishida, H., and K. Nakamura. 2000. Trichloroethylene degradation by Ralstonia sp. KN1-10A constitutively expressing phenol hydroxylase: transformation products, NADH limitation and product toxicity. J. Biosci. Bioeng. 5:438-445. [DOI] [PubMed] [Google Scholar]
- 24.Leahy, J. G., A. M. Byrne, and R. H. Olsen. 1996. Comparison of factors influencing trichloroethylene degradation by toluene-oxidizing bacteria. Appl. Environ. Microbiol. 62:825-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Legendre, P., and L. Legendre. 1998. Numerical ecology, 2nd English ed. Elsevier Science B. V., Amsterdam, The Netherlands.
- 26.Mars, A. E., J. Houwing, J. Dolfing, and D. B. Janssen. 1996. Degradation of toluene and trichloroethylene by Burkholderia cepacia G4 in growth-limited fed-batch culture. Appl. Environ. Microbiol. 62:886-891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Massol-Deyá, A., R. Weller, L. Ríos-Hernández, J. Z. Zhous, R. F. Hickey, and J. M. Tiedje. 1997. Succession and convergence of biofilm communities in fixed-film reactors treating aromatic hydrocarbons in groundwater. Appl. Environ. Microbiol. 63:270-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCarty, P. L., M. N. Goltz, G. D. Hopkins, M. E. Dolan, J. P. Allan, B. T. Kawakami, and T. J. Carrothers. 1998. Full scale evaluation of in situ cometabolic degradation of trichloroethylene in groundwater through toluene injection. Environ. Sci. Technol. 32:88-100. [Google Scholar]
- 29.McCune, B., and G. Cottam. 1985. The successional status of a southern Wisconsin oak woods. Ecology 66:1270-1278. [Google Scholar]
- 30.Mohn, W. W., and J. M. Tiedje. 1992. Microbial reductive dehalogenation. Microbiol. Rev. 56:482-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Müller, C., L. Petruschka, H. Cuypers, G. Burchhardt, and H. Herrmann. 1996. Carbon catabolite repression of phenol degradation in Pseudomonas putida is mediated by the inhibition of the activator protein PhlR. J. Bacteriol. 178:2030-2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Newman, L. M., and L. P. Wackett. 1997. Trichloroethylene oxidation by purified toluene 2-monooxygenase: products, kinetics, and turnover-dependent inactivation. J. Bacteriol. 179:90-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Osborn, A. M., E. R. B. Moore, and K. N. Timmis. 2000. An evaluation of terminal-restriction fragment length polymorphism (T-RFLP) analysis for the study of microbial community structure and dynamics. Environ. Microbiol. 2:39-50. [DOI] [PubMed] [Google Scholar]
- 34.Scala, D. J., and L. J. Kerkhof. 2000. Horizontal heterogeneity of denitrifying bacterial communities in marine in marine sediments by terminal-restriction fragment length polymorphism analysis. Appl. Environ. Microbiol. 66:1980-1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Segar, R. L., S. L. Dewys, and G. E. Speitel. 1995. Sustained trichloroethylene cometabolism by phenol-degrading bacteria in sequencing biofilm reactors. Water Environ. Res. 67:764-774. [Google Scholar]
- 36.Shih, C. C., M. E. Davey, J. Zhou, J. M. Tiedje, and C. S. Criddle. 1996. Effects of phenol feeding pattern on microbial community structure and cometabolism of trichloroethylene. Appl. Environ. Microbiol. 62:2953-2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.ter Braak, C. J. F. 1995. Ordination, p. 91-105. In R. H. G. Jongman, C. J. F. ter Braak, and V. Torgeren (ed.), Data analysis in community and landscape ecology. Cambridge University Press, Cambridge, United Kingdom.
- 38.Thioulouse, J., D. Chessel, S. Dolédec, and J. M. Olivier. 1997. ADE-4: a multivariate analysis and graphical display software. Stat. Comp. 7:75-83. [Google Scholar]
- 39.Vogel, T. M., C. S. Criddle, and P. L. McCarty. 1987. Transformations of halogenated aliphatic compounds. Environ. Sci. Technol. 21:722-736. [DOI] [PubMed] [Google Scholar]
- 40.Wackett, L. P. 1995. Bacterial cometabolism of halogenated organic compounds, p. 217-241. In L. Y. Young and C. E. Cerniglia (ed.), Microbial transformations and degradation of toxic organic chemicals. Wiley-Liss, New York, N.Y.
- 41.Wackett, L. P., and S. R. Householder. 1989. Toxicity of trichloroethylene to Pseudomonas putida F1 is mediated by toluene dioxygenase. Appl. Environ. Microbiol. 55:2723-2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Watanabe, K., M. Teramoto, H. Futamata, and S. Harayama. 1998. Molecular detection, isolation, and physiological characterization of functionally dominant phenol-degrading bacteria in activated sludge. Appl. Environ. Microbiol. 64:4396-4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yeager, C. M., P. J. Bottomley, and D. J. Arp. 2001. Cytoxicity associated with trichloroethylene oxidation in Burkholderia cepacia G4. Appl. Environ. Microbiol. 67:2107-2115. [DOI] [PMC free article] [PubMed] [Google Scholar]