Figure 6. Nuclear localization of RON activates cFLIP transcriptionally.
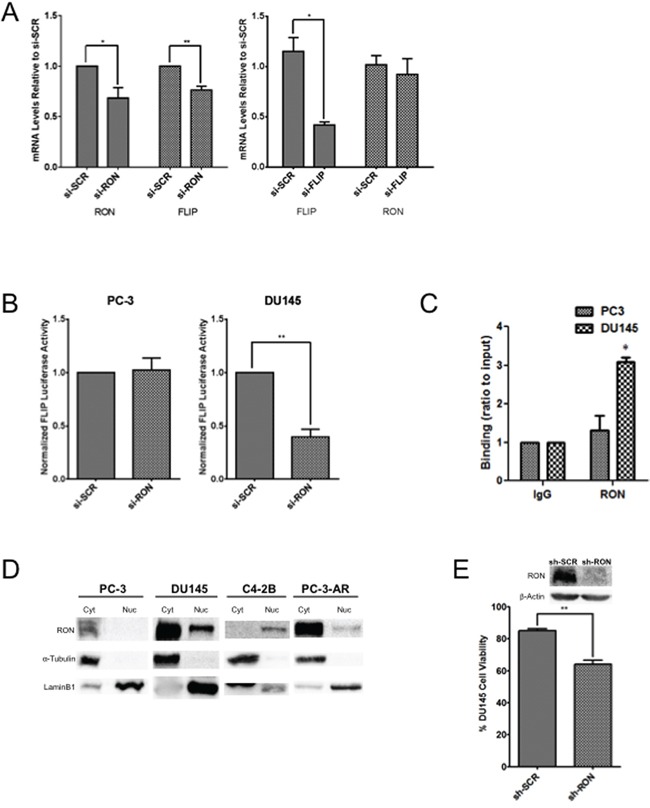
A. Total RNA extracted from DU145 cells (n= 2 biological replicates with triplicate replicates) transiently transfected with si-RON or si-c-FLIP and scrambled control was used in real-time PCR using RON and c-FLIP-specific primers. B. Logarithmically growing PC-3 or DU145 cells (n= 3 biological replicates with triplicate replicates) were transfected with pGL3-c-FLIP reporter along with Renilla luciferase. 48h after transfection, luciferase activity was measured. Normalized luciferase/renilla activity was calculated with respect to scrambled siRNA. The data shown are average + sd of three independent experiments conducted in triplicate. C. DNA from IgG or RON-immunoprecipitated lysates from PC-3 (n=2 biological replicates each with triplicate technical replicates) or DU145 cells (n=2 biological replicates each with triplicate technical replicates) was amplified by real-time PCR using primers for the RON binding site on the c-FLIP promoter. DNA binding was calculated (in arbitrary units) by normalizing to input DNA. IgG was used as a negative control. Fold enrichment was calculated as 100*2−(Ct[Target]-Ct[Input]) and the amplification value from immunoprecipitated DNA was normalized to 10% input (p≤0.05). The data presented is mean+s.e.m from two indepdent experiments each with three technical replicates. Statistical significance was determined using two-sided t-test with no adjustment for multiple comparisons. D. Nuclear and cytosolic extracts prepared from PC-3 (n=3), PC-3AR (n=3), C4-2B (n=3) and DU145 (n=2) cells (all biological replicates) were probed for RON. α-Tubulin and Lamin B1 were used as loading controls for cytosolic and nuclear proteins, respectively. E. Percent cell viability (average+SD) of DU145 stable Scramble or RON silenced cells (n= 3 biological replicates) growing under androgen-depleted conditions for 120h from three independent experiments is presented. (* = p≤ 0.05; ** = p ≤ 0.01; *** = p ≤ 0.001).
