Abstract
Mass spectrometry imaging (MSI) and in-situ single cell mass spectrometry (SCMS) analysis under ambient conditions are two emerging fields with great potential for the detailed mass spectrometry (MS) analysis of biomolecules from biological samples. The single-probe, a miniaturized device with integrated sampling and ionization capabilities, is capable of performing both ambient MSI and in-situ SCMS analysis. For ambient MSI, the single-probe uses surface micro-extraction to continually conduct MS analysis of the sample, and this technique allows the creation of MS images with high spatial resolution (8.5 µm) from biological samples such as mouse brain and kidney sections. Ambient MSI has the advantage that little to no sample preparation is needed before the analysis, which reduces the amount of potential artifacts present in data acquisition and allows a more representative analysis of the sample to be acquired. For in-situ SCMS, the single-probe tip can be directly inserted into live eukaryotic cells such as HeLa cells, due to the small sampling tip size (< 10 µm), and this technique is capable of detecting a wide range of metabolites inside individual cells at near real-time. SCMS enables a greater sensitivity and accuracy of chemical information to be acquired at the single cell level, which could improve our understanding of biological processes at a more fundamental level than previously possible. The single-probe device can be potentially coupled with a variety of mass spectrometers for broad ranges of MSI and SCMS studies.
Keywords: Biochemistry, Issue 112, Mass spectrometry imaging, high spatial resolution, single cell mass spectrometry, microscale sampling, ambient ionization, single-probe device.
Introduction
Mass spectrometry imaging (MSI) is a relatively new molecular imaging technique to provide the spatial distribution of the compounds of interest on surfaces. During the MSI analysis, mass spectrometry (MS) measurements are recorded across the surface on an individual pixel basis to create a 2D image of the species of interest 1. MSI techniques have the ability to provide a spatially resolved feature distribution for a large range of metabolites, allowing a much greater amount of information to be obtained from a sample than from using traditional molecular imaging techniques, and they have the potential to greatly improve the analysis of biological samples for biological and pharmacology studies 2. MSI can be broadly separated into non-ambient and ambient approaches. The non-ambient MSI analysis techniques, such as matrix assisted laser desorption ionization (MALDI) MS 3 and time of flight secondary ion MS (ToF SIMS) 4, are capable of high spatial resolution (around 5 µm and 100 nm, respectively) and high sensitivity. However, these methods require extensive sample preparation, such as the application of matrix molecules to the sample surface, and a vacuum sampling environment, which could introduce artifacts to the data obtained. Ambient techniques such as desorption electrospray ionization (DESI) MS 5, laser ablation electrospray ionization (LAESI) MS 6, and nano-DESI MS 7 are capable of MSI of samples with little to no prior preparation under the ambient environment, which is able to produce MS images that potentially reflect the sample in its most native state. However, most of these techniques generally lack the high spatial resolution and detection sensitivity compared with the non-ambient techniques, with experiments typically conducted at around 150 µm per pixel 8.
Single cell analysis (SCA) is a growing field that has the ability to characterize the chemical composition of biological samples at the cellular level. SCA enables the analysis of biological systems at a more fundamental level than traditional cell analysis techniques, which produce an averaged result of a population of cells, potentially providing insights that are previously intractable 9. MS techniques have recently been applied to SCA (termed single cell mass spectrometry or SCMS) using non-ambient techniques such as MALDI MS 10 and ToF SIMS 11 in which cells are pretreated before analysis, and with ambient techniques such as LAESI MS 12 and direct extraction methods, such as live single-cell video-MS 13, 14, to analyze a wide variety of cell types such as egg, plant, and cancer. Ambient techniques have the advantage of being applied to live cells, which again minimizes the artifacts, leading to a better representation of the metabolites in the live cells. The direct extraction based methods described above, however, perform the sample extraction and analysis process at two different steps, which result in a time gap during the analysis that could potentially alter the metabolites present within the sample.
The single-probe, a miniaturized multifunctional device that is capable of conducting high spatial resolution ambient MSI on biological tissue sections 15 and near real-time in-situ SCMS on live single cells 16. The single-probe has an integrated construction that is made up of a pulled dual-bore quartz capillary coupled with a solvent providing inlet and a nano-ESI emitter made from fused silica capillaries, enabling solvent delivery and analyte extraction to be performed from a single device. In the ambient MSI mode, the single-probe is placed over the sample tissue and surface micro-extraction occurs, allowing a rastered MS image to be made at high spatial resolution. Particularly, the tapered tip of the single-probe is small enough to be inserted into live eukaryotic cells for in-situ SCMS analysis, where the metabolite detection takes less than two seconds between probe insertion and MS detection, allowing chemical information to be taken in near real-time. Here are the protocols to fabricate the single-probe device and to conduct both the ambient MSI and SCMS modes using the single-probe MS techniques.
Protocol
Animal use and welfare should adhere to the NIH Guide for the Care and Use of Laboratory Animals following protocols reviewed and approved by the Institutional Animal Care and Use Committee (IACUC). Mouse tissue samples were provided by collaborator Dr. Chuanbin Mao.
1. Mouse Tissue Section Preparation
Place a whole mouse organ of interest (brain, kidney, liver, etc.) into the center of a small plastic well (e.g., 12-well cell culture plate), and submerge in tissue embedding compound up to about 10 mm height. Make sure there is no bubble formed in the tissue embedding compound and that the organ is placed in the desired orientation (i.e., sagittal, coronal, etc.).
Immediately place tissues into liquid nitrogen for flash freezing. For long-term storage, store the frozen samples in a -80 °C freezer.
Take the frozen mouse organ and thaw to -15 °C in a temperature controlled cryomicrotome.
Secure tissue onto a steel base with approximately 500 µl of tissue embedding compound and place onto a cryomicrotome sectioning mount so that the desired sectioning orientation is presented to the knife.
Section the tissue to a 12 µm thickness. Place sectioned tissue slices onto polycarbonate microscope slides and leave to dry for 30 min at room temperature. For long-term storage, store the frozen slide in a -80 °C freezer.
2. Cell Culture
Note: Cell culture was performed in biological safety cabinet (Biosafety Level II) under sterile conditions. HeLa cell line was used as a model system, and cells were cultured in complete culture medium with the following conventional protocols:
Warm reagents (i.e., trypsin, phosphate buffered saline (PBS), and cell culture medium) to 37 °C. Note: The cell culture medium contains inorganic salts, amino acids, vitamins, and others. For a complete list of constituents, refer to the formulation from the manufacturer.
Obtain cell sample (e.g., 1 ml HeLa cell suspension) and add it into 9 ml of complete cell culture medium in a standard 10-cm cell culture plate. The initial cell number is around 0.5 x 106 cells/ml. Keep cells in culture at 37 °C with 5% CO2 for 2-3 days until the growing surface is covered at 70-80% on the cell culture plate. Record cell passage number for each successive round.
- Perform cell passaging (i.e., cell splitting) in the cell culture plate.
- Aspirate growth medium, and use 5 ml of 1x PBS to rinse the cells. Remove PBS using a sterile aspiration tip, and incubate the cells with 2.5 ml of trypsin (0.25%) for ~5 min at 37 °C to detach the cells from the culture plate. Note: The actual trypsin treatment time need to be optimized according to the specific trypsin product purchased from the manufacturer. Inadequate treatment time leaves the cells attached to the plate, whereas excessive treatment leads to cell death.
- Stop trypsin activity by adding 7.5 ml complete cell culture medium, and then uniformly resuspend the cells (total volume 10 ml). Use the cell suspension for culture (step 2.2) or preparation of SCMS samples (step 2.4).
- Prepare the cell samples for SCMS experiments.
- Place individual micro cover slides in a 12-well plate, and add 1.8 ml cell culture medium and 0.2 ml of cell suspension into the well.
- Gently mix cells with mild agitation of the plate, and incubate in a 5% CO2 environment at 37 °C for ~24 hr. To perform drug treatment to the cultured cells, add a drug compound solution (e.g., in DMSO (dimethyl sulfoxide)) into 12-well cell culture plate. Note: The final drug concentration (e.g., 10 nM, 100 nM, 1 µM, and 10 µM) and treatment time (e.g., 4 hr) are varied according to the specific purpose of studies. Cells are attached to the micro cover slides and ready for the CSMS experiments (step 6).
3. Single-probe Fabrication
Place the dual-bore quartz tubing (inner diameter (I.D.) 127 µm, outer diameter (O.D.) 500 µm) into a laser micropipette puller and pull a dual-bore quartz needle. Use the following parameters as starting points: Heat = 400, Fil = 3, Vel = 80, Del =150, and Pul = 250 (all units are manufacturer's units). Ensure the pulled dual-bore quartz needle has a tapered tip for optimal probe properties. Cut the pulled tip so that there is ~5 mm long of unpulled dual-bore quartz capillary left at the other end. Note: The actual parameters of the laser puller should be optimized according to the specific conditions of the instrument.
Cut a ~80 mm section of fused silica capillary (I.D. 40 µm, O.D. 105 µm) as the solvent providing capillary, and insert it into one bore at the flat end of the dual-bore quartz needle.
Cut a ~40 mm section of fused silica capillary (I.D. 40 µm, O.D. 105 µm) and use a razor to shave ~5 mm of the polyimide coating from the mid-point. Use a propane flame to quickly heat and pull the fused capillary into a nano-electrospray ionization (ESI) emitter with a fine taper. Cut a nano-ESI emitter (~7-10 mm long), and insert it into the other bore at the flat end of the dual-bore quartz needle. Alternatively, use the laser puller to produce a fine taper.
Apply a minimal amount of (~1-2 µl) UV curing resin onto the flat end of the dual-bore quartz needle, and solidify the resin using a LED UV lamp for ~20 sec to secure the solvent providing capillary and the nano-ESI emitter. The procedures to assemble the individual parts into a single-probe are shown in Figure 1a.
Cut a standard microscope glass slide (1" x 3") into half lengthways. Place the single-probe onto one end of the glass slide so that the nano-ESI emitter is pointed outwards. Apply the regular epoxy to the body of the single-probe so that it becomes secured onto the glass slide (Figure 1b). Leave overnight for hardening. Couple the fabricated single-probe with the integrated single-probe setup (Figure 1c), which is attached to a mass spectrometer as illustrated in Figure 1d.
4. Build the Integrated Single-probe MS Setup
- Modify the ion source interface flange of the mass spectrometer and fabricate the stand (with adjustable position and height) of the digital stereoscope (Figures 1e and 1f).
- Drill an ion source interface flange with two holes allowing for the attachment of an aluminum optical board. Make a slide rail device and a height adjustment rod (attached to a XY stage for fine position tuning), such that the digital stereoscope system can be attached to the aluminum optical board (Figure 1e).
Attach the modified digital stereo microscope, a USB digital microscope, a miniature manual XYZ translation stage with a flexible clamp holder, the motorized XYZ translation stage system to the aluminum optical board, which is mounted on the customized ion source interface flange of the mass spectrometer (Figures 1c and 1f). Use the flexible clamp holder to fix the glass slide attached with a single-probe.
Attach the single-probe setup to the mass spectrometer (Figure 1f). Adjust the flexible clamp holder and the miniature XYZ stage to place the emitter of the single-probe in front of the inlet of the mass spectrometer. Use the USB digital microscope (with adjustable view angle) on the side of single-probe to provide a zoomed-in image of the single-probe tip or the nano-ESI emitter, and the digital stereoscope (with adjustable height) above the single-probe to view the cells and the probe tip. Note: Using the corresponding ion source flange, this integrated single-probe system can be coupled to any other types of mass spectrometers equipped with ambient ionization sources.
5. Ambient MSI
Thaw the sample section at room temperature and place it onto the motorized XYZ translation stage system underneath the single-probe. Adjust the sample position by changing the coordinates in the control software.
Using the syringe to pump the sampling solvent at an appropriate rate (e.g., 0.2 µl/min), and apply the ionization voltage (e.g., 5 kV). The selection of the sampling solvent is flexible, and the common ones include MeOH:water (9:1) and acetonitrile. The dead volume of the nano-ESI emitter was estimated to be ~3 nl, and the time between the probe-surface contact and ion signal observation is usually less than 1 sec 15. Note: The customized ion source interface flange allows the ionization voltage to be delivered from the mass spectrometer to a conductive union through a crocodile clip. The ionization voltage is then transmitted through a conductive union to the solvent inside the capillary and the single-probe channels, and applied onto the nano-ESI emitter to ionize the sampled analytes. Ensure that the ionization voltage is turned off when connecting the crocodile clip with the conductive union.
Adjust the height of the single-probe so that it is resting just above the surface of the sample and able to perform surface-extraction of metabolites. Carefully lift the Z-stage, and then use the USB digital microscope (on the side of the single-probe) to monitor the distance change between the single-probe tip and tissue surface. Monitor changes in the mass spectrum during this height adjustment, and stop lifting the Z-stage when a change of the ion signal from solvent background to tissue metabolites is observed.
Repeat step 5.3 three times to set three different points within the stage control program for automated surface flattening adjustment. Place the tip of the single-probe at three points on the sample surface at a distance of about 10 mm apart from each other. Perform height adjustment by pressing the up and down icons, and lock the three points into position under the "Plan method".
Set other parameters for rastering across the section of interest within the sample using this program. For the mouse kidney sections presented here, use a 10.0 µm/sec rastering speed and 20 µm distance between lines. The motorized stage system has a 0.1 µm minimum increment motion. The distance between the single-probe tip and tissue is obtained from step 5.3.
Set up a method for the automated acquisition of MS spectra from the mass spectrometer. For high mass resolution MSI on mouse kidney sample, use the following parameters: mass resolution 60,000 (m/Δm), ~5 kV positive mode, 1 microscan, 150 msec max injection time, and AGC on. All acquired MS spectra representing individual lines of the MS image had the same number of scans with uniform time spacing between each scan, indicating that the pixel sizes for the images produced were uniformly distributed.
- Initiate the MSI data acquisition. Initiate the MS acquisition sequence for the mass spectrometer, and then initiate the rastering sequence for the XYZ control program.
- For example, in the MS data acquisition program utilized here, go to "Sequence setup", select "New sequence", generate a set of files for a new sequence numbered from 01 to X, where X is the number of lines used for the desired MS image to be taken, and then press "Run sequence".
- Use a homemade electronic device to allow the software to produce a contact closure signal for the mass spectrometer to collect the data. The circuit diagram is shown in the supplementary figure (Figure S1) as a reference.
- Construct MS images from raw MS files using appropriate MSI visualization software. For example, when using the software package developed by Laskin's group at PNNL 17, perform the following steps.
- Click "Brows File." Select the first file obtained from the MSI experiment. Specify where the file starts and finishes under "Number of Lines." Select a range of m/z values for the MS image range under "Enter MZ Range".
- Press the "Start" button to initiate the image creation process. Once the MS image is made, click "Save Image" under "Toolbar" to store images in the computer.
6. In-situ Live SCMS
Setup the single-probe system as per instructions for MSI. Adjust the solvent (e.g., MeOH/H2O or acetonitrile) flow rate (e.g., ~25 nl/min).
Wash the cultured cells, which are attached on the micro cover glass slides, with PBS to remove cultural media and extracellular drug components. Place cell containing glass slide onto the motorized XYZ translation stage system for experiment. Note: Alternatively, use the fresh cell culture medium (without containing fetal bovine serum) to rinse the cultured cells. Less ion suppression has been observed. In addition, cell can survive for longer time during the experiment where the ambient temperature (~20 °C) is significantly lower than the culture temperature (37 °C). The drug type, solution concentration, and treatment time vary in different studies.
Focus the digital stereo microscope (above the sample) onto the tip of the single-probe to monitor cell penetration during the analysis. Use the USB digital microscope (on the side of the single-probe) to monitor the working conditions of the nano-ESI emitter on the single-probe.
- Use the motorized XYZ stage control program and digital stereo microscope (above cells) to locate a cell of interest, and precisely position the single-probe tip above the sample. Start MS data acquisition before the single-probe tip is inserted into the cell.
- Use the following parameters as references for MS analysis using a high resolution mass spectrometer: mass resolution 100,000 (m/Δm), ~3 kV positive and negative mode, 1 microscan, 150 msec max injection time, AGC mode on. Automated acquisition of MS spectra is done by clicking "Start" in the MS data acquisition program.
- Lift the motorized Z-stage by clicking the icon to penetrate the cell membrane and keep recording the MS signal generated from the cell. A time delay of 1-2 sec is usually observed between the probe insertion and the MS signal detection. As the other confirmation of cell penetration, a dramatic change of the MS signals can be observed upon the penetration of the cell membrane. The MS signals of intracellular compounds usually can last for ~15-20 sec before a significantly decrease.
- Lower down the cell containing plate to pull the single-probe tip out from the cell. It usually takes <15 sec for the ion signals of the cellular compounds to approach the noise level. Let the solvent flow for ~3 min to completely flush the single-probe. Meanwhile, position the motorized XYZ stage system to locate the next cell to be analyzed. Each cell experiment requires ~3 min to be accomplished.
Representative Results
The single-probe was successfully used for the ambient MSI analysis of sectioned mouse kidney tissue 15. The device uses the mechanism of surface liquid micro-extraction (Figure 1a), which provides highly efficient analyte extraction from a small area, leading to abundant ion signals intensities in the MSI results. For example, the signal intensities of more than 107 have been achieved for some abundant metabolites (Figure 2a). A large number of metabolites were detected in this manner, including a number of sphingomyelin (SM) and phosphatidylcholine (PC) species such as [SM (34:1) + Na]+ (725.5575 m/z), [PC (32:0) + H]+ (734.5700 m/z), [PC (34:1) + Na]+ (782.5696 m/z), and [PC (38:5 + Na)]+ (814.5726 m/z). These compounds were identified with high mass resolution and mass accuracy when coupled to a high resolution mass spectrometer. For example, the identification was achieved with less than 4 ppm m/z mass accuracy (i.e., the difference between the observed and theoretical values) for each metabolite (Figure 2b) in results presented here. In addition, tandem MS analyses (i.e., MS/MS) were also conducted for more confident identification of species of interest.15
Due to the capability of performing efficient liquid micro-extraction on a small area, the single-probe device can be used to perform high spatial resolution MSI experiments under ambient conditions15. For example, detailed MS images of mouse kidney sections have been obtained illustrating the spatial distribution of selected metabolites (Figure 2c). The spatial resolution of the MS image was determined to be 8.5 µm, following the widely used metric of having the transition point of a sharp feature determined within a 20-80% intensity change of the MS signal. 18 In the case of phospholipid [PC (38:5 + Na)]+ on the mouse kidney section, the feature transition between the inner medulla and the outer medulla takes place across one scan cycle in the chronogram, showing an intensity change greater than 20-80% range. Based on the sample moving speed (10.0 µm/sec) and MS data acquisition rate (0.85 sec/spectrum), the sample moving distance in one MS scan cycle (8.5 µm), i.e., the MSI spatial resolution, can be calculated (Figure 2d). This spatial resolution is amongst the highest yet achieved for ambient MSI techniques conducted on biological samples.
For SCMS the single-probe was able to achieve the analysis of individual live HeLa cells 16. The tip size of the single-probe is typically less than 10 µm (Figure 3a), which is small enough to be directly inserted into many types of eukaryotic cells, of which the diameter is ~10 µm, for extraction and MS analysis. The insertion process of the single-probe tip into a cell can be visually monitored using a digital stereo microscope (Figure 3b), and the cell membrane penetration can be confirmed through the rapid and significant change of mass spectra from PBS (or fresh cell culture medium) to intracellular compounds (Figures 3c and 3d). The experiments can be conducted in both positive and negative ion modes to detect broader types of molecular species. For example, 18 different lipid species were identified in the positive mode, including sphingomyelins (SM) and phosphatidylcholines (PC), whereas adenosine phosphates (AMP, ADP, and ATP) were detected in the negative ion mode (Figures 3c and d). The time delay between the single-probe insertion into a cell and the signal detection was typically less than two seconds, allowing a near real-time detection of cellular metabolites. SCMS was also applied to experiments where cells were treated with anticancer drugs (e.g., OSW-1, paclitaxel, and doxorubicin) 19]. The corresponding drugs can be detected within HeLa cells after 4-hour treatment at a series of concentrations (i.e., 10 nM, 100 nM, 1 µM, and 10 µM) in DMSO (dimethyl sulfoxide), using the untreated cells (add DMSO only) as the controls. The MS signals of drugs were not present within the extracellular PBS or the control (Figure 3e), but were detected within the single cells using the single-probe MS technique (only 100 nM treatment results are shown in Figure 3f). Because cells were rinsed with PBS (or fresh cell culture medium) to remove extracellular compounds and contaminations, the detection of endogenous metabolites (e.g., cell lipids and adenosine phosphates) and exogenous compounds (e.g., anticancer drugs) indicates that the Single-probe MS technique can be used to analyze intracellular compounds.
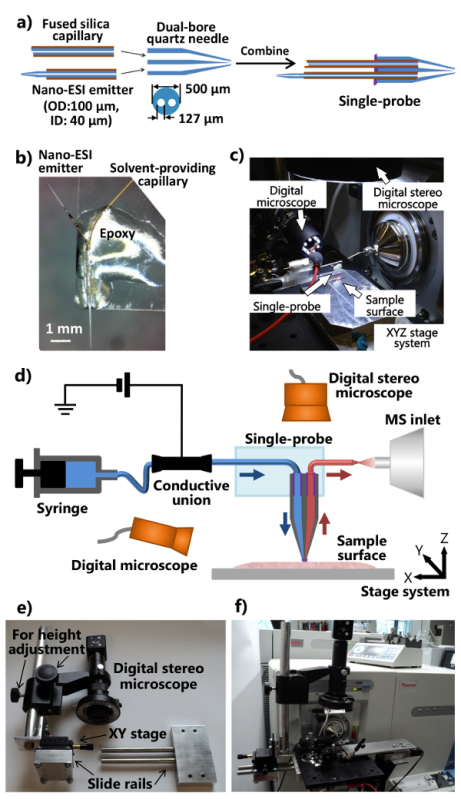 Figure 1. Fabrication and setup of the single-probe for ambient MSI and SCMS analysis.a) Fabrication procedures of the single-probe. b) Photograph of a fabricated single-probe attached to a glass slide. c) Photograph of the single-probe setup attached to a mass spectrometer. d) Diagram of the single-probe setup coupled with a mass spectrometer. During an experiment, the sampling solvent is continuously provided from the syringe, the ionization voltage is applied to the conductive union from the mass spectrometer, two digital microscopes are used to monitor sample placement, the motorized XYZ stage system is used to control sample motion, and a mass spectrometer is used for analysis. e) Photograph of the customized digital stereoscope system. f) Photograph showing the digital stereoscope attached to the ion source interface flange through an optical board. Please click here to view a larger version of this figure.
Figure 1. Fabrication and setup of the single-probe for ambient MSI and SCMS analysis.a) Fabrication procedures of the single-probe. b) Photograph of a fabricated single-probe attached to a glass slide. c) Photograph of the single-probe setup attached to a mass spectrometer. d) Diagram of the single-probe setup coupled with a mass spectrometer. During an experiment, the sampling solvent is continuously provided from the syringe, the ionization voltage is applied to the conductive union from the mass spectrometer, two digital microscopes are used to monitor sample placement, the motorized XYZ stage system is used to control sample motion, and a mass spectrometer is used for analysis. e) Photograph of the customized digital stereoscope system. f) Photograph showing the digital stereoscope attached to the ion source interface flange through an optical board. Please click here to view a larger version of this figure.
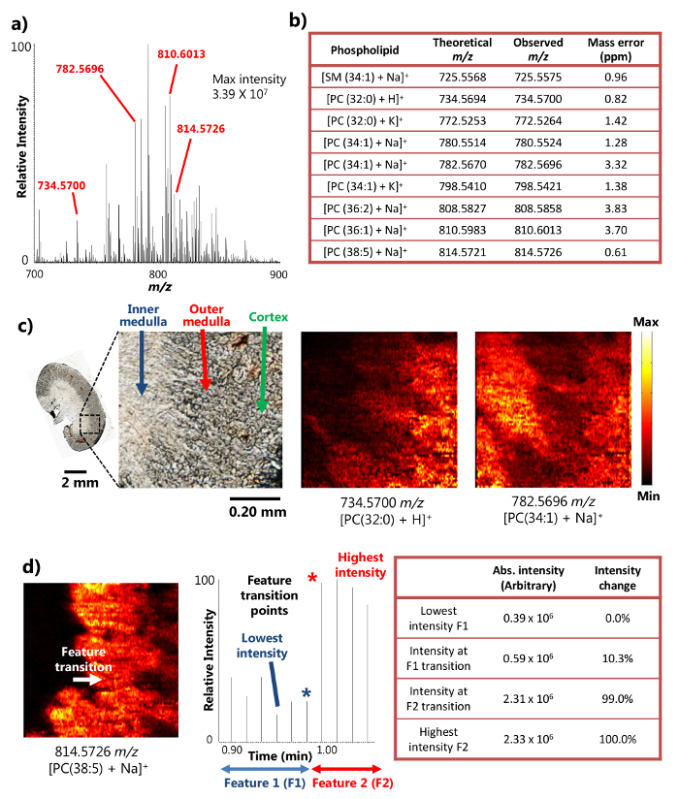 Figure 2. Results from an ambient MSI study of a mouse kidney section with high spatial and mass resolution.a) A representative mass spectrum from the single-probe MSI. The maximum intensity of detected metabolites can reach 3.39 x 107 (arbitrary units). b) A selection of the detected metabolites presented with their mass accuracy. c) MS images of [PC(32:0) + H]+ and [PC(34:1) + Na]+ taken from a mouse kidney section at 8.5 µm spatial resolution. PC: phosphatidylcholine. Scale bar: 2 mm; 0.20 mm (inset). d) Determination of spatial resolution of the MS image for [PC(38:5) + Na]+ (adapted with permission from reference 15). Please click here to view a larger version of this figure.
Figure 2. Results from an ambient MSI study of a mouse kidney section with high spatial and mass resolution.a) A representative mass spectrum from the single-probe MSI. The maximum intensity of detected metabolites can reach 3.39 x 107 (arbitrary units). b) A selection of the detected metabolites presented with their mass accuracy. c) MS images of [PC(32:0) + H]+ and [PC(34:1) + Na]+ taken from a mouse kidney section at 8.5 µm spatial resolution. PC: phosphatidylcholine. Scale bar: 2 mm; 0.20 mm (inset). d) Determination of spatial resolution of the MS image for [PC(38:5) + Na]+ (adapted with permission from reference 15). Please click here to view a larger version of this figure.
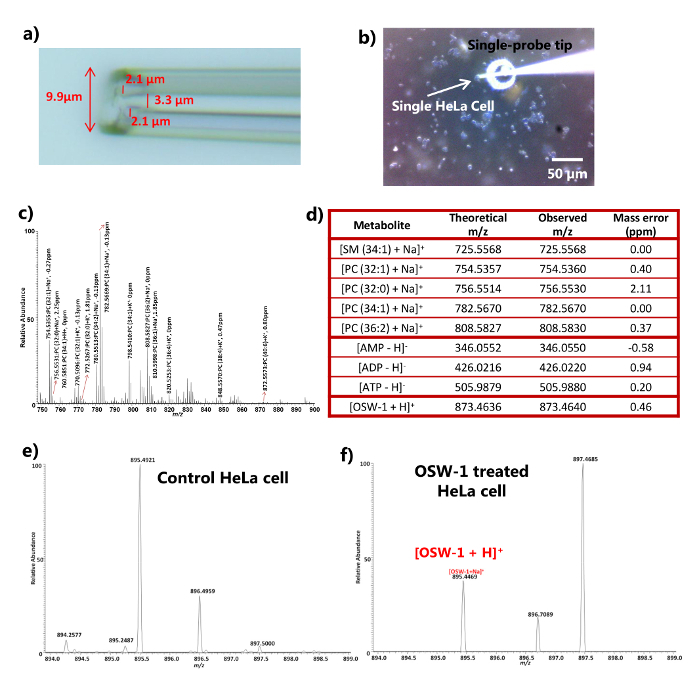 Figure 3. Results from an ambient SCMS analysis of drug treated HeLa cells with high mass resolution.a) Zoomed-in photograph of the single-probe tip showing a typical size of < 10 µm in diameter. b) Photograph taken at the point of single-probe insertion into a HeLa cell. Scale bar: 50 µm. c) A typical positive ion mode mass spectrum with the identifications of a number of PC (phosphatidylcholine) species. d) A representative list of metabolites detected from SCMS analysis of HeLa cells both in the positive and negative ion modes. e-f) Mass spectra for the control and treated (100 nM OSW-1) cells (adapted with permission from reference 16). Please click here to view a larger version of this figure.
Figure 3. Results from an ambient SCMS analysis of drug treated HeLa cells with high mass resolution.a) Zoomed-in photograph of the single-probe tip showing a typical size of < 10 µm in diameter. b) Photograph taken at the point of single-probe insertion into a HeLa cell. Scale bar: 50 µm. c) A typical positive ion mode mass spectrum with the identifications of a number of PC (phosphatidylcholine) species. d) A representative list of metabolites detected from SCMS analysis of HeLa cells both in the positive and negative ion modes. e-f) Mass spectra for the control and treated (100 nM OSW-1) cells (adapted with permission from reference 16). Please click here to view a larger version of this figure.
Figure S1. Circuit diagram of the electronic device used to produce contact closure signal for mass spectrometer to collect data. Please click here to view or download this figure.
Discussion
The single-probe is a multifunctional device that can be used for both MSI and SCMS experiments. The single-probe setup (including translation stage systems, microscopes, ion source interface flange, etc.) is designed as an add-on component that can be flexibly adapted to the existing mass spectrometer. A rapid exchange between the single-probe setup and the conventional ESI ion source can be accomplished within one minute. In principle, using the appropriate ion source interface flange, the single-probe setup can be adapted to the any other mass spectrometers. Additionally, the sampling solvent containing a variety of reagents can be used with the single-probe setup for reactive MSI and SCMS experiments, which greatly enhances the detection of broader ranges of biomolecules. In addition to animal tissues and cell lines, the single-probe is also capable of analyzing other biological systems such as plants. Therefore, with the same experimental setup and similar user training, a variety of studies can be performed using a single instrument and by the same users, allowing for efficient and versatile experiments to be accomplished with the minimum training time and instrumentation cost.
The key component of the single-probe MS technique is the probe itself. The quality of the single-probe has a significant influence on its performance, which largely determines the quality of both MSI and SCMS experiments. When fabricating single-probes, make sure that the capillaries inside of the dual-bore tubing are securely glued to eliminate the chance of solvent leak during the experiments. It is critical to use a minimum amount of UV curable epoxy, such that the orifices and capillaries are not clogged during the probe fabrication.
The single-probe has been used to conduct high spatial and mass resolution ambient MSI on biological samples 15. The major advantage of ambient MSI over non-ambient methods is that sample preparation is kept at a minimum with no need for a vacuum sampling environment, which allows the sample to be analyzed in a near native state 8. One of the major hurdles for most other ambient MSI technique has been a lack of spatial resolution 1. Compared with the desorption based MSI techniques (such as DESI and LAESI), the small tip size of the single-probe allows a more robust and efficient surface liquid micro-extraction to be performed over a small area, leading to a high spatial resolution of 8.5 µm, which is amongst the highest ones achieved using ambient MSI techniques 15. In addition, adjusting the components of the sampling solvent provides extra flexibility to conduct the experiments. For example, sampling solvents containing reagents (e.g., dicationic compounds) have been used to perform reactive MSI experiments, allowing for a significant increase in the number of metabolites identified per experiment 20. The other advantage of the single-probe is the integrated design, which provides the ease of operation during the entire data acquisition process. Because the distance between the tip and tissue surface is very sensitive for ion signal intensity and stability, obtaining a flat tissue section and conducting surface flattening adjustment to minimize the distance variance is a key for high quality MSI experiments. It follows that the single-probe MSI techniques are not suitable to obtain high spatial MS images of uneven surfaces.
In addition to fabricating a high quality probe, carefully tuning the instrument is essential for a successful MSI experiment. Among all tuning steps, adjusting the height of the single-probe tip above the tissue section surface is the most critical one. When adjusting the probe height, pump the sampling solvent and turn on the ionization voltage, so that only the solvent background ion signals can be observed. Then monitor the change of the mass spectrum while carefully reducing the probe-surface distance by lifting the motorized Z-stage until strong and stable ion signals from tissue section can be observed; this probe height will be used for MSI data collection during the experiment. In addition, an optimized solvent flow rate is essential for MSI experiments. Adjust the flow rate with the optimized probe height. Ensure that there is no solvent spread on the tissue surface (i.e., flow rate is too high) or bubble formation inside the nano-ESI emitter (i.e., flow rate is too low).
The single-probe is a multifunctional device for bioanalysis. In addition to the MSI experiments, it is capable of conducting near real-time in-situ SCMS to elucidate detailed chemical information from live eukaryotic cells16, which is a major advantage compared with other vacuum based SCMS techniques (such as MALDI10 and SIMS21). The small size of the probe tip provides the ability to be inserted into a live eukaryotic cell and to extract and ionize the intracellular compounds for immediate MS analysis. Similarly, the sampling solvents containing reagents (e.g., dicationic compounds) can be used in the SCMS experiments, and a broader range of cellular constituents can be detected in a live single cell than ever before (ongoing research, data are not shown). Although the real-time analysis will provide the chemical profiles of live single cells, due to the cell penetration of membrane and extraction of cellular contents, the cell under investigation will be killed after the experiment, implying that the single-probe SCMS technique is still a destructive method. In addition, the probe tip and nano-ESI emitter in the single-probe can be easily clogged for inexperienced users. To reduce the chance of device clogging, ensure to avoid touching the nucleus when inserting the single-probe tip into a cell. If clogging occurs, the device can be regenerated by heating up the clogged probe tip or the nano-ESI emitter using a homebuilt heating coil16. Another limitation of the single-probe SCMS technique is that only the adhesive cells (i.e., cells are attached to surfaces) can be analyzed using current setup. However, by incorporating the cell manipulation system into the single-probe MS apparatus, broader types of cell can be studied in future.
Similar to the MSI experiment, obtaining a high quality probe and an optimized solvent flow rate is critical for SCMS studies. When tuning the solvent flow rate, the single-probe tip is placed above the sample (i.e., no contact with the cell or culture medium), and ensure that there is no solvent dripping from the probe tip or bubble formation inside the nano-ESI emitter.
Disclosures
We have no conflict of interest to declare with the work presented here.
Acknowledgments
The authors would like to thank Dr. Laskin (the Pacific Northwest National Laboratory) for sharing the motorized stage control software and MSI visualization program. We also thank Dr. Mao (the University of Oklahoma) for providing mouse organ samples and Mr. Chad E. Cunningham (the University of Oklahoma) for the assistance in machining and electronics work. This research was supported by grants from the Research Council of the University of Oklahoma Norman Campus, the American Society for Mass Spectrometry Research Award (sponsored by Waters Corporation), Oklahoma Center for the Advancement of Science and Technology (Grant HR 14-152), and National Institutes of Health (R01GM116116).
References
- Vickerman JC. Molecular imaging and depth profiling by mass spectrometry-SIMS, MALDI or DESI? Analyst. 2011;136(11):2199–2217. doi: 10.1039/c1an00008j. [DOI] [PubMed] [Google Scholar]
- Schwamborn K. Imaging mass spectrometry in biomarker discovery and validation. J. Proteomics. 2012;75(16):4990–4998. doi: 10.1016/j.jprot.2012.06.015. [DOI] [PubMed] [Google Scholar]
- Schwamborn K, Caprioli RM. MALDI Imaging Mass Spectrometry - Painting Molecular Pictures. Mol Oncol. 2010;4(6):529–538. doi: 10.1016/j.molonc.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft ML, Klitzing HA. Imaging lipids with secondary ion mass spectrometry. Biochim. Biophys. Acta. 2014;1841(8):1108–1119. doi: 10.1016/j.bbalip.2014.03.003. [DOI] [PubMed] [Google Scholar]
- Wiseman JM, et al. Desorption electrospray ionization mass spectrometry: Imaging drugs and metabolites in tissues. Proc Natl Acad Sci U S A. 2008;105(47):18120–18125. doi: 10.1073/pnas.0801066105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemes P, Woods AS, Vertes A. Simultaneous imaging of small metabolites and lipids in rat brain tissues at atmospheric pressure by laser ablation electrospray ionization mass spectrometry. Anal. Chem. 2010;82(3):982–988. doi: 10.1021/ac902245p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskin J, Heath BS, Roach PJ, Cazares L, Semmes OJ. Tissue Imaging Using Nanospray Desorption Electrospray Ionization Mass Spectrometry. Anal. Chem. 2012;84(1):141–148. doi: 10.1021/ac2021322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Dill AL, Eberlin LS, Cooks RG, Ifa DR. Mass spectrometry imaging under ambient conditions. Mass. Spectrom. Rev. 2013;32(3):218–243. doi: 10.1002/mas.21360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschuler SJ, Wu LF. Cellular heterogeneity: do differences make a difference? Cell. 2010;141(4):559–563. doi: 10.1016/j.cell.2010.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amantonico A, Urban PL, Fagerer SR, Balabin RM, Zenobi R. Single-Cell MALDI-MS as an Analytical Tool for Studying Intrapopulation Metabolic Heterogeneity of Unicellular Organisms. Anal. Chem. 2010;82(17):7394–7400. doi: 10.1021/ac1015326. [DOI] [PubMed] [Google Scholar]
- Passarelli MK, Ewing AG, Winograd N. Single-cell lipidomics: characterizing and imaging lipids on the surface of individual Aplysia californica neurons with cluster secondary ion mass spectrometry. Anal. Chem. 2013;85(4):2231–2238. doi: 10.1021/ac303038j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha B, et al. Subcellular metabolite and lipid analysis of Xenopus laevis eggs by LAESI mass spectrometry. PLoS One. 2014;9(12):e115173. doi: 10.1371/journal.pone.0115173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno H, Tsuyama N, Harada T, Masujima T. Live single-cell video-mass spectrometry for cellular and subcellular molecular detection and cell classification. J. Mass. Spectrom. 2008;43(12):1692–1700. doi: 10.1002/jms.1460. [DOI] [PubMed] [Google Scholar]
- Zhang L, et al. In Situ metabolic analysis of single plant cells by capillary microsampling and electrospray ionization mass spectrometry with ion mobility separation. Analyst. 2014;139(20):5079–5085. doi: 10.1039/c4an01018c. [DOI] [PubMed] [Google Scholar]
- Rao W, Pan N, Yang Z. High Resolution Tissue Imaging Using the Single-probe Mass Spectrometry under Ambient Conditions. J. Am. Soc. Mass. Spectrom. 2015;26(6):986–993. doi: 10.1007/s13361-015-1091-4. [DOI] [PubMed] [Google Scholar]
- Pan N, et al. The single-probe: a miniaturized multifunctional device for single cell mass spectrometry analysis. Anal. Chem. 2014;86(19):9376–9380. doi: 10.1021/ac5029038. [DOI] [PubMed] [Google Scholar]
- Thomas M, et al. Visualization of High Resolution Spatial Mass Spectrometric Data during Acquisition. 2012 Annual International Conference of the Ieee Engineering in Medicine and Biology Society (Embc) 2012. pp. 5545–5548. [DOI] [PubMed]
- Luxembourg SL, Mize TH, McDonnell LA, Heeren RM. High-spatial resolution mass spectrometric imaging of peptide and protein distributions on a surface. Anal. Chem. 2004;76(18):5339–5344. doi: 10.1021/ac049692q. [DOI] [PubMed] [Google Scholar]
- Zhou Y, et al. OSW-1: a natural compound with potent anticancer activity and a novel mechanism of action. J Natl Cancer Inst. 2005;97(23):1781–1785. doi: 10.1093/jnci/dji404. [DOI] [PubMed] [Google Scholar]
- Rao W, Pan N, Tian X, Yang Z. High-Resolution Ambient MS Imaging of Negative Ions in Positive Ion Mode: Using Dicationic Reagents with the Single-Probe. J. Am. Soc. Mass. Spectrom. 2016;27(1):124–134. doi: 10.1007/s13361-015-1287-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrowski SG, Kurczy ME, Roddy TP, Winograd N, Ewing AG. Secondary ion MS imaging to relatively quantify cholesterol in the membranes of individual cells from differentially treated populations. Anal. Chem. 2007;79(10):3554–3560. doi: 10.1021/ac061825f. [DOI] [PMC free article] [PubMed] [Google Scholar]


