Abstract
Gli3 is a major regulator of Hedgehog signaling during limb development. In the anterior mesenchyme, GLI3 is proteolytically processed into GLI3R, a truncated repressor form that inhibits Hedgehog signaling. Although numerous studies have identified mechanisms that regulate Gli3 function in vitro, it is not completely understood how Gli3 function is regulated in vivo. In this study, we show a novel mechanism of regulation of GLI3R activities in limb buds by Gata6, a member of the GATA transcription factor family. We show that conditional inactivation of Gata6 prior to limb outgrowth by the Tcre deleter causes preaxial polydactyly, the formation of an anterior extra digit, in hindlimbs. A recent study suggested that Gata6 represses Shh transcription in hindlimb buds. However, we found that ectopic Hedgehog signaling precedes ectopic Shh expression. In conjunction, we observed Gata6 and Gli3 genetically interact, and compound heterozygous mutants develop preaxial polydactyly without ectopic Shh expression, indicating an additional prior mechanism to prevent polydactyly. These results support the idea that Gata6 possesses dual roles during limb development: enhancement of Gli3 repressor function to repress Hedgehog signaling in the anterior limb bud, and negative regulation of Shh expression. Our in vitro and in vivo studies identified that GATA6 physically interacts with GLI3R to facilitate nuclear localization of GLI3R and repressor activities of GLI3R. Both the genetic and biochemical data elucidates a novel mechanism by Gata6 to regulate GLI3R activities in the anterior limb progenitor cells to prevent polydactyly and attain proper development of the mammalian autopod.
Author Summary
Gli3 is a major regulator of Hedgehog signaling in the limb, where Gli3 counteracts Sonic hedgehog (Shh) for patterning and proliferative expansion of limb progenitor cells. In the anterior limb mesenchyme, GLI3 is proteolytically processed into GLI3R, a truncated repressor form that inhibits Hedgehog signaling. In this study, we show a novel mechanism of regulation of GLI3R activities in limb buds by Gata6, a member of GATA transcription factor family. Conditional inactivation of Gata6 in mice caused formation of an extra digit in the anterior hindlimbs, a common congenital limb malformation. This phenotype was associated with ectopic Hedgehog signaling activation, and later ectopic Shh expression, in the anterior of hindlimb buds. We show that Gata6; Gli3 compound heterozygous mutants developed anterior extradigit without ectopic Shh expression, indicating there to be an additional and prior mechanism before ectopic Shh activation that induces extradigit formation. We identified that GATA6 physically interacts with GLI3R and that the interaction facilitates nuclear localization of GLI3R and repressor activities of GLI3R. Therefore, our study identified a novel mechanism by Gata6 to regulate GLI3R activities in the anterior limb mesenchyme to prevent extra digit formation and proper development of the mammalian autopod.
Introduction
Understanding the developmental mechanisms that regulate progenitor cells to generate organs with specific morphology and function is a central topic in developmental biology. The vertebrate limb has been serving as an excellent system for such studies. In particular, mesenchymal progenitor cells in limb buds are specified, patterned and expanded to generate each skeletal element with a distinct morphology at each defined position to create the stereotypical limb skeletal system. The mammalian autopod possesses five digits, termed as d1-d5, in an anterior to posterior order. The number and identity of digits have been used as a readout of specification, patterning, and proliferative expansion of progenitor cells [1].
Sonic Hedgehog (Shh) is expressed in the zone of polarizing activity (ZPA), located at the posterior mesenchyme of the limb bud, and acts as a major regulatory molecule for limb development [1, 2]. Anterior-posterior specification of digit progenitors is regulated by the concentration and duration of progenitor exposure to SHH [3–6]. SHH also regulates the proliferative expansion of mesenchymal progenitor cells to generate a sufficient number of cells to develop into cartilage condensations [7, 8]. Accordingly, ectopic expression of Shh in the anterior portion is associated with preaxial polydactyly, which is characterized by the formation of ectopic digits in the anterior of the limb [9]. By contrast, the most anterior digit (d1) develops in a SHH-independent manner [10, 11]. Recent studies have shown that anterior genetic programs, such as Irx3-Irx5 and Sall4, are required for development of d1, at least in part, by excluding SHH signaling from the anterior mesenchyme [12, 13].
The glioma-associated oncogene family (GLI) proteins are zinc finger DNA binding proteins, which play diverse roles in animal development and diseases [14]. Among the three Gli genes, Gli3 encodes a bi-functional molecule, acting as both an activator (GLI3A) and a repressor (GLI3R), whose balance depends on Hedgehog signaling [14]. In the presence of Hedgehog ligands, its signal transduction at primary cilia causes inhibition of proteolytic processing of GLI3 [15]. This results in the accumulation of a full-length activator form of GLI3 (GLI3A) in the posterior mesenchyme. In contrast, in the absence of Hedgehog signaling, GLI3 is subjected to proteolysis, generating a truncated repressor form (GLI3R), which accumulates in the anterior mesenchyme. Because GLI1 lacks a repressor domain and GLI2 predominantly functions as an activator [16, 17], GLI3R is the major GLI repressor in the limb [18].
Consistent with the important function of Gli3 in limb development, its mutations cause developmental defects in mice and humans [19–21]. In particular, Gli3-/- mice develop polydactyly [21]. Genetic studies in mice demonstrated that a predominant function of Gli3 is to repress Hedgehog signaling target genes [22, 23]. Furthermore, it has been shown that the balance of GLI3A and GLI3R regulates digit number and identity [24–26]. Numerous studies have shown that multiple mechanisms regulate GLI3 functions in vitro, such as posttranslational modifications, degradation, cytoplasmic retention, and primary cilium-mediated processing (reviewed in [14, 27, 28]). In vivo studies in mice demonstrated that Gli3 genetically interacts with Hox genes, Zic3 and Alx4 during limb development [29–31]. Despite these studies, the in vivo control of Gli3 function during proper limb development is still to be elucidated.
The Gata family of zinc finger transcription factors is an important regulator of tissue and organ development. The Gata family is subdivided into the Gata1/2/3 subfamily and the Gata4/5/6 subfamily, which show expression in hematopoietic cell lineages and meso-endoderm lineages, respectively [32, 33]. In particular, Gata6 is essential for endoderm formation and is also involved in the development of various mesoderm- and endoderm-derived organs, such as the cardiovascular system and pancreas [34–37]. Moreover, a recent study suggested that Gata6 functions as a negative regulator of Shh expression in limb buds by binding to its limb bud-specific cis-regulatory element, ZRS [38].
In this study, we found that broad deletion of Gata6 in the limb mesenchymal progenitors caused hindlimb-specific preaxial polydactyly, which is associated with ectopic SHH signaling in the anterior hindlimb bud. We discovered that Gata6 and Gli3 genetically interact to regulate normal patterning of the hindlimb. Furthermore, we show that direct association of GATA6 with GLI3R promoted nuclear localization and transcriptional repressor activity of GLI3R. Our work identified that genetic and biochemical interactions between Gata6 and Gli3 act as essential mechanisms to regulate GLI3R activity for proper autopod patterning.
Results
Inactivation of Gata6 in early mesoderm caused hindlimb specific preaxial polydactyly
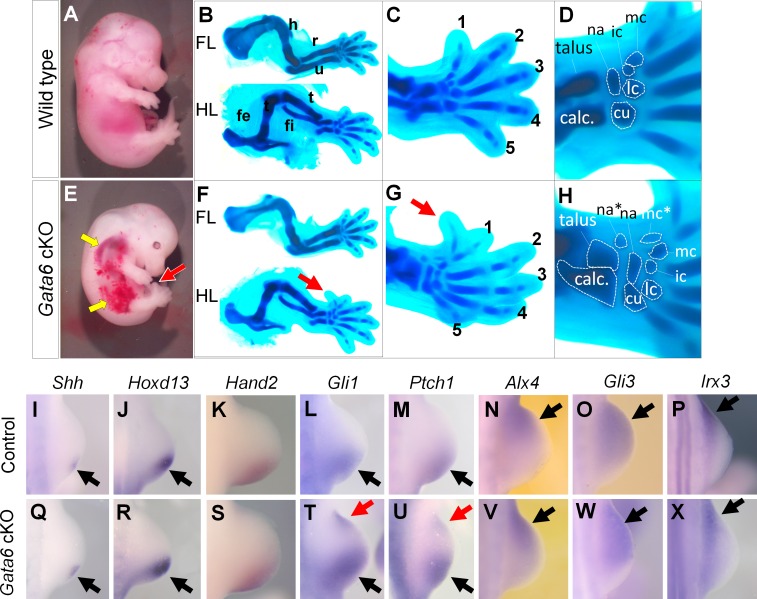
Prior studies have identified expression of Gata6 in developing limb buds [38–40]. Gata6 null embryos die during gastrulation [34, 35]; therefore, we inactivated Gata6 in the meso-endoderm by using the conditional allele of Gata6 (Gata6fl) [41] and the Tcre line, which recombines in the early meso-endoderm [42]. We found that Tcre; Gata6fl/fl mutants (hereafter referred to as Gata6 cKO) die around E12.5–14.5 with broad hemorrhage (Fig 1A and 1E). This result is consistent with a former study, demonstrating a role of proper dosage of Gata4 and Gata6 for vessel integrity [43]. We found that Gata6 cKO embryos exhibited polydactyly in the hindlimb, while forelimbs seem to be unaffected (Fig 1A–1C, 1E and 1G, S1 Table). Alcian blue staining demonstrated that the mutant hindlimbs possess patterned digits, d1-d5, and an extra digit on the anterior edge, which morphologically resembles d1. Based on the position and morphology, tarsal and metatarsal elements were also patterned. Two ectopic tarsal elements, likely the navicular and medial cuneiform, were present proximally to the ectopic 1st metatarsal (Fig 1D and 1H). These observations indicate that the autopod is patterned along the anterior-posterior axis, and the absence of Gata6 induces the formation of an extra anterior digit with the associated tarsal and metatarsal elements.
Fig 1. Loss of Gata6 causes preaxial polydactyly in hindlimbs.
A-H: Lateral views (A, E) of whole E14.5 embryos, and Alcian blue-stained cartilage (B-D, F-H) of wild type (A-D) and Gata6 cKO (F-H) embryos at E14.5. C and G show hindlimb autopod, and D and H show tarsal and metatarsal elements. Red arrows in E-G point to the anterior ectopic digit. Yellow arrows point to hemorrhage in Gata6 cKO embryos. Digits are numbered with 1–5 in C and G. Asterisks in H indicates ectopic elements. calc: calcaneus, cu: cuboid, fe: femur, fi: fibula, ic: intermediate cuneiform, lc: lateral cuneiform, mc: medial cuneiform, na: navicular ti: tibia. I-X: in situ hybridization of wild type (I-P) and Gata6 cKO (Q-X) hindlimb buds at E10.5 with indicated probes. Black and red arrows point to normal and ectopic signals, respectively. See also S1 Table.
Ectopic Hedgehog signaling activation in Gata6 cKO hindlimbs
Preaxial polydactyly is known to be associated with ectopic Sonic Hedgehog (Shh) expression in the anterior margin. At E10.5, we detected posteriorly-localized Shh expression without ectopic anterior expression (n = 4, 39–40 somite stage, Fig 1I and 1Q). Consistent with this normal expression, Hoxd13 (n = 3) and Hand2 (n = 6), upstream regulators of limb bud Shh expression [44], were normally expressed in the posterior mesenchyme (Fig 1J, 1K, 1R and 1S). However, Gli1 (n = 3) and Patch1 (n = 3), targets of Hedgehog signaling, were detected in the anterior margin of Gata6 cKO hindlimb buds (Fig 1L, 1M, 1T and 1U). Expression of anterior marker genes, such as Alx4 (n = 3), Gli3 (n = 4) and Irx3 (n = 3), were not significantly affected in Gata6 cKO hindlimb buds (Fig 1N–1P and 1V–1X).
We also examined gene expression at a later stage. At E11.5, we detected ectopic Shh expression in the anterior border of Gata6 cKO hindlimbs (n = 4, S1 Fig). Consistent with evident ectopic Shh expression, expression of Hoxd13 (n = 3), Gli1 (n = 6), Ptch1 (n = 6) and Gremlin1 (n = 3) was also detected in the anterior margin. This data indicates that ectopic Hedgehog signaling became evident at E10.5 in Gata6 cKO hindlimb buds, although ectopic Shh expression was undetectable. At a later stage (E11.5), ectopic Shh expression became evident and all SHH targets, examined in this study, were detected in the anterior margin.
Shh expression is negatively regulated in the anterior margin by various genes. Thus, we examined expression of negative regulators of Shh expression. In addition to Alx4 and Gli3 (Fig 1) [23, 45], expression of Etv4 (n = 3), Etv5 (n = 5), Tulp3 (n = 3), Twist1 (n = 3), whose loss can cause ectopic Shh expression in the anterior margin [46–52], did not show evident alteration (S2 Fig). Therefore, it is unlikely that these genes account for the preaxial polydactyly phenotype in Gata6 cKO hindlimbs.
Reduction of Shh dosage rescued ectopic SHH signaling but not ectopic anterior digit formation
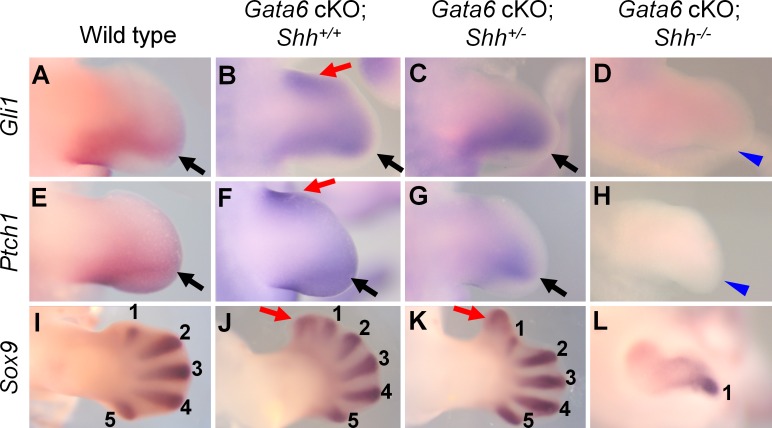
If ectopic Shh expression accounts for the preaxial polydactyly in Gata6 cKO hindlimbs, we would expect that reducing Shh dosage might rescue the phenotype. Therefore, we genetically reduced Shh dosage from the Gata6 cKO background using the Shh null allele [2]. Gata6 cKO; Shh+/- mutants did not survive beyond E12.5, thus, we examined expression of SHH target genes (Gli1 and Ptch1) and expression of Sox9, an early marker of chondrogenic condensation [53].
Removing one allele of Shh from the Gata6 cKO background resulted in posteriorly restricted expression of Gli1 and Ptch1, and the ectopic anterior expression became undetectable (n = 4, Fig 2A–2C and 2E–2G). However, ectopic chondrogenic condensation in the anterior portion was still detected by Sox9 expression at E12.5 (n = 3, Fig 2I–2K). Removing both alleles of Shh from the Gata6 cKO background resulted in the loss of Gli1 and Ptch1 expression and single digit condensation, the same phenotype as Shh-/- limbs (n = 3, Fig 2D, 2H and 2L) [10, 11]. These results indicate that Shh functions downstream of Gata6 during preaxial polydactyly development. However, ectopic chondrogenic condensation in the anterior portion of Gata6 cKO; Shh+/- hindlimbs suggests that additional mechanisms could be involved in the preaxial polydactyly in Gata6 cKO hindlimbs.
Fig 2. Expression pattern of SHH targets and digit condensation in Gata6 cKO; Shh allelic series.
Expression pattern of Gli1 (A-D), Ptch1 (E-H) and Sox9 (I-L) of wild type (A, E, I), Gata6 cKO (B, F, J), Gata6 cKO; Shh+/- (C, G, K) and Gata6 cKO; Shh-/- (D, H, L) hindlimb buds. A-H: E11.5, I-L: E12.5. In A-H, black arrows and red arrows point to normal and ectopic signals, respectively. Blue arrowheads indicate loss of expression in D and H. In I-L, digit condensations are labeled as 1–5, and ectopic condensation is marked with red arrows.
Gli3 genetically interacts with Gata6 in forelimbs and hindlimbs
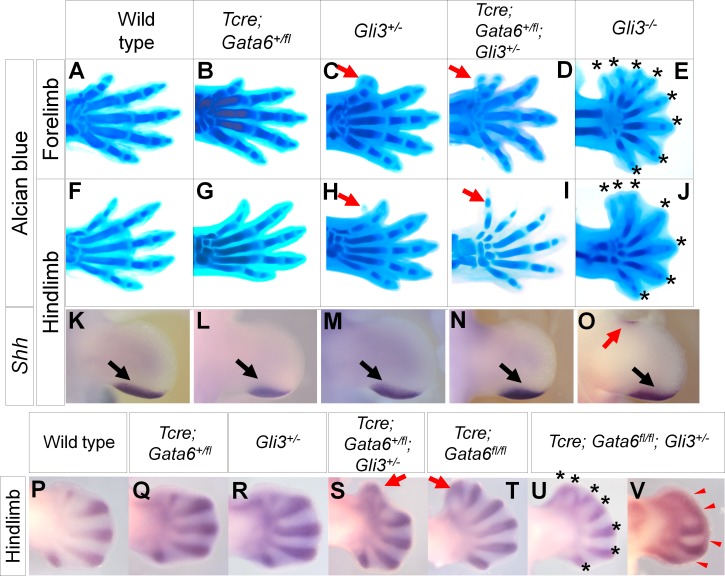
GLI3 is a major regulator of Hedgehog signaling, and thus, Gli3 might be involved in preaxial polydactyly in Gata6 cKO hindlimbs. To test this hypothesis, we genetically removed Gli3 from the Gata6 cKO background. Gli3+/- hindlimbs developed a small spike in the anterior region [21, 54], while most of the Tcre; Gata6+/fl hindlimbs were indistinguishable from the wild-type hindlimbs at E14.5–15.5 (Fig 3F–3H, Table 1). Tcre; Gata6+/fl; Gli3+/- compound heterozygous hindlimbs developed an extra digit in the anterior region (Fig 3I). Unexpectedly, we also found that this interaction operates in forelimbs. Gli3+/- forelimbs developed d1, which was associated with small ectopic cartilage condensation at the distal tip. Contrary to this, Tcre; Gata6+/fl; Gli3+/- compound heterozygous forelimbs developed an evident extra digit with incomplete penetrance (Fig 3A–3D) or an extra digit that partially fused with endogenous d1 with incomplete penetrance (S2 Table). These results demonstrate a genetic interaction between Gli3 and Gata6 in fore- and hind-limbs.
Fig 3. Genetic interaction between Gata6 and Gli3 in preaxial polydactyly development.
A-J: Alcian blue-stained autopod of indicated genotypes at E15.5. A-E: forelimbs, F-J: hindlimbs. Thin red arrows point to bifurcated d1 (C) and small projection (H) in fore- and hind-limbs, respectively in Gli3+/- mutants. Thick red arrows in D and I point to anterior ectopic digits. Asterisks in E and J indicate digit tips of Gli3-/- autopod. K-O: Expression pattern of Shh in hindlimb buds of indicated genotypes at E11.5. Black and red arrows point to normal and ectopic signals, respectively. P-V: Sox9 in situ hybridization in hindlimbs of indicated genotypes at E12.5. Red arrows in S and T point to anterior ectopic digit condensation. Asterisks in U indicate distal tips of digit condensation. Red arrowheads in V point to distally-fused condensation.
Table 1. Number of hindlimbs with indicated phenotypes at E14.5–16.5.
| Genotype | Number of hindlimbs with normal digits | Number of hindlimbs with small projection | Number of hindlimbs with anterior extra digit |
|---|---|---|---|
| Wild type | 140/140 (100%) | 0/140 | 0/140 |
| Gli3+/- | 2/18 (11.1%) | 16/18 (88.9%) | 0/18 (0%) |
| Tcre; Gata6+/fl | 61/66 (92.4%) | 0/66 (0%) | 5/66 (7.6%) |
| Tcre; Gata6+/fl; Gli3+/- | 3/54 (5.6%) | 0/54 (0%) | 51/54 (94.4%) |
Because the Gata6 cKO limb phenotype was evident in hindlimbs, we focused the following analysis on hindlimbs. Ectopic Shh expression can cause preaxial polydactyly, therefore, we examined Shh expression at E11.5. We detected a small domain of anterior ectopic Shh expression in Gli3-/- hindlimbs (n = 3/6, Fig 3O), as previously reported [23]. By contrast, Tcre; Gata6+/fl; Gli3+/- compound heterozygous hindlimbs did not exhibit anterior ectopic Shh expression (n = 6), similar to wild-type, Tcre; Gata6+/fl (n = 6) and Gli3+/- (n = 6) hindlimb buds (Fig 3K–3N). Therefore, preaxial polydactyly in Tcre; Gata6+/fl; Gli3+/- compound heterozygous limbs were unlikely to be caused by ectopic Shh expression. Given that GLI3R prevents ectopic digit formation in the anterior portion [55], these results suggest that an interaction between Gata6 and Gli3 contributes to GLI3R activities.
Gata6 cKO; Gli3+/- embryos do not survive beyond E12.5, therefore, we further examined the interaction between Gata6 and Gli3 by visualizing digit condensation by Sox9 in situ hybridization. Both Gli3+/- and Tcre; Gata6+/fl hindlimbs exhibited similar expression patterns to wild-type hindlimbs at E12.5 (Fig 3P–3R). Correlating with preaxial polydactyly at E15.5, Gata6 cKO and Tcre; Gata6+/fl; Gli3+/- compound heterozygous hindlimbs exhibited ectopic anterior digit condensation (Fig 3S and 3T). Gata6 cKO; Gli3+/- hindlimbs were slightly underdeveloped and exhibited seven digit condensations (n = 2/6, Fig 3U), distally-fused condensation (n = 2/6, Fig 3V) or one extra anterior condensation, similar to Gata6 cKO hindlimbs (n = 2/6). Formation of multiple extra digits and distal fusion of cartilage condensation are characteristics of Gli3-/- limbs [21]. Therefore, we speculate that the Gata6 cKO; Gli3+/- genotype may be in conditions similar to the Gli3-/- genotype in hindlimbs. These results further support the idea that loss of Gata6 leads to reduction of GLI3R activities.
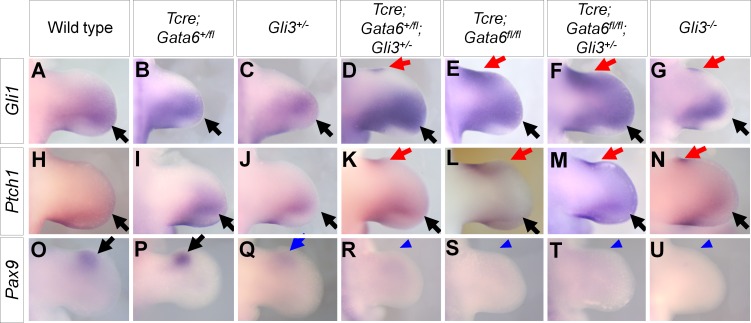
In order to further characterize the Gata6-Gli3 interaction, we examined gene expression at E11.5. Expression of Gli1 and Patch1 was posteriorly restricted in wild-type, Tcre; Gata6+/fl and Gli3+/- hindlimbs (Fig 4A–4C and 4H–4J). Hindlimbs with the Tcre; Gata6+/fl; Gli3+/-, Gata6 cKO, Gata6 cKO; Gli3+/- or Gli3-/- genotypes exhibited anterior ectopic expression of these genes (Fig 4D–4G and 4K–4N). The ectopic expression domain was larger in Gata6 cKO and Gata6 cKO; Gli3+/- hindlimb buds than that in Tcre; Gata6+/fl; Gli3+/- and Gli3-/- hindlimbs, likely due to ectopic Shh expression in the Gata6 cKO background.
Fig 4. Expression pattern of Gli1, Ptch1 and Pax9 in Gata6; Gli3 allelic series.
In situ hybridization of Gli1 (A-G), Ptch1 (H-N) and Pax9 (O-U) of hindlimb buds of indicated genotypes at E11.5. Black and red arrows point to normal and ectopic signals, respectively. Blue arrows and arrowheads indicate reduced and loss of Pax9 signals, respectively.
Pax9, whose expression requires high levels of GLI3R activities [56], was detected in the anterior of wild-type and Tcre; Gata6+/fl hindlimbs, and was reduced in Gli3+/- hindlimb buds (Fig 4O–4Q). In Tcre; Gata6+/fl; Gli3+/-, Gata6 cKO, Gata6 cKO; Gli3+/- hindlimbs, Pax9 expression was undetectable, similar to Gli3-/- hindlimbs (Fig 4R–4U).
These alterations of gene expression at E11.5 are consistent with the idea that GLI3R activities were reduced in hindlimbs with the Tcre; Gata6+/fl; Gli3+/-, Gata6 cKO and Gata6 cKO; Gli3+/- genotypes.
GATA6 and GLI3 functionally and physically interact in vitro
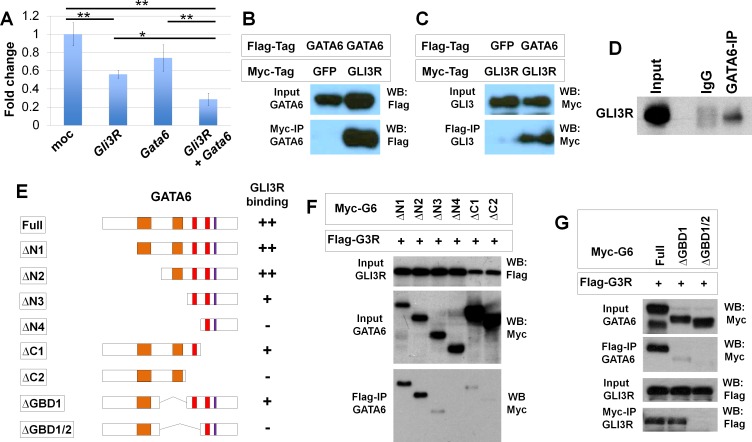
Ectopic Shh expression in the Gata6 cKO background could affect gene expression patterns in hindlimb buds. Therefore, we set up in vitro experiments to further investigate how Gata6 regulates Gli3 function. We first set up luciferase reporter assays using 12xGLI-binding site luciferase [31]. Transfecting a C-terminally truncated form of human GLI3 that could function as GLI3R caused significant reduction of the reporter activities, while transfecting human GATA6 did not affect the reporter activities. Co-transfecting GLI3R and GATA6 caused further reduction of the reporter activities (Fig 5A). These results are consistent with the in vivo data and support the idea that GATA6 functionally interacts with and contributes to GLI3R activities.
Fig 5. Physical and functional interaction between GATA6 and GLI3R.
A: GLI-specific luciferase reporter assay with indicated expression constructs. *: p<0.01, **: p<0.001. B, C: Co-immunoprecipitation assay of Flag-GATA6 and Myc-GLI3R. (B) Pulldown with anti-Myc, detection by anti-Flag. (C) Pulldown with anti-Flag, detection by anti-Myc. D: Co-immunoprecipitation of GATA6 and GLI3R from wild-type hindlimb buds. E: Schematic presentation of deletion mutants of GATA6. Binding with GLI3R in F and G is summarized in the right side of the panel. Orange bars represent transactivation domains. Red and blue bars represent zinc finger DNA binding domains and the nuclear localization signal, respectively. F, G: Co-immunoprecipitation assay of Flag-GLI3R and GATA6 mutants.
Next, we tested whether GATA6 and GLI3R physically interact by co-immunoprecipitation assays. HEK293T cells were transfected with Flag-tagged GATA6, Myc-tagged GLI3R or GFP. Flag-GATA6 and Myc-GLI3R were co-immunoprecipitated, demonstrating that GATA6 and GLI3R can interact (Fig 5B and 5C). We also confirmed that the interaction occurs in vivo. GLI3R was detected in immunoprecipitated complex from E10.25–10.5 wild-type hindlimb buds using ant-GATA6 (Fig 5D). To further characterize their interaction, we mapped the GLI3R interaction domain in GATA6. For this purpose, we generated serial deletion mutants (Fig 5E), and performed co-immunoprecipitation assays with Flag-GLI3R. The ΔN1 and ΔN2 mutants showed a strong interaction with Flag-GLI3R. The ΔN3 and ΔC1 mutants exhibited weak interaction, and we did not detect any interactions of Flag-GLI3R with ΔN4 and ΔC2 (Fig 5F).
We also generated intra-molecular deletion mutants. These mutants lack the GLI3R-binding domain (GBD) 1, which includes the second putative transactivation domain (ΔGBD1), or both GBD1 and GBD2 (ΔGBD1/2). We did not detect any interaction of ΔGBD1/2 with GLI3R, although ΔGBD1 exhibited a weak interaction with GLI3R (Fig 5G). These results suggest that the zinc finger domain 1 (ZFD1) is critical to interact with GLI3R. The weak interaction of ΔN3, ΔC1 and ΔGBD1, which possess the ZFD1, also suggests that both the N- and C-terminal regions around the ZFD1 contribute to the interaction with GLI3R, in collaboration with the ZFD1.
Interaction between GATA6 and GLI3R regulates subcellular localization of GLI3R
Our analyses indicated the presence of genetic and physical interactions between Gata6 and Gli3. Given that both GATA6 and GLI3R act as transcription factors, we next examined subcellular localization of these proteins after co-transfecting HEK293T cells with Flag-GLI3R and either full length or mutant forms of Myc-GATA6.
We observed three patterns of localization (Fig 6A and 6B, S3 Fig). First, co-transfection of either full length GATA6, ΔN1-GATA6 or ΔN2-GATA6, which can interact with GLI3R and possess the nuclear localization signal (NLS), resulted in predominant nuclear localization of both GLI3R and GATA6. Second, we co-transfected ΔN3-GATA6 or ΔN4-GATA6, which possess the NLS, but have either very weak or undetectable interactions with GLI3R. In these transfection assays, GLI3R localization became either predominantly cytoplasmic or localized similarly in both the cytoplasm and nucleus, although GATA6 was predominantly detected in the nucleus. Third, we co-transfected ΔC1-GATA6 or ΔC2-GATA6, which lack the NLS and have very weak or undetectable interactions with GLI3R. We detected GATA6 predominantly in the cytoplasm, consistent with the lack of NLS. GLI3R was also predominantly located in the cytoplasm or located similarly in the nucleus and cytoplasm.
Fig 6. GATA6 regulates subcellular localization of GLI3R.
A: Representative in vitro images of nuclear GATA6+nuclear GLI3R (upper), nuclear GATA6+cytosolic GLI3R (middle) and cytosolic GATA6+cytosolic GLI3R (bottom). B: Quantitation of subcellular localization of GATA6 and GLI3R. N<C: predominantly cytoplasmic, N = C: similarly in cytoplasm and in nucleus, N>C: predominantly nuclear localized. GATA6 mutants, indicated at the bottom, are shown in Fig 5E. The number of cells examined for each set of transfection is indicated in the panel. C-H: Representative images of the anterior-proximal mesenchyme of hindlimb buds at E10.25. C, E, G: wild type, D, F, H: Gata6 cKO. I: Quantitation of subcellular localization of GLI3R in the anterior-proximal mesenchyme of hindlimb buds at E10.25. Gray and black bars represent wild-type and Gata6 cKO samples, respectively. The graph shows percentage of GLI3R localization patterns, such as predominantly nuclear (N>C), similarly in the nucleus and cytoplasm (N = C), or predominantly cytoplasmic (N<C). A total of 597 cells from three wild-type embryos and a total of 528 cells from three Gata6 cKO embryos were examined. * indicates P<0.05. J: Western blot of nuclear fractions from anterior part of wild-type and Gata6 cKO hindlimb buds at E10.25–10.5. Histone H3 (H3) is included as a loading control.
These results indicate a correlation between GLI3R nuclear localization and nuclear GATA6 that possesses a GLI3R-interaction ability. This correlation suggests that physical association between GATA6 and GLI3R contributes to nuclear localization and the repressor activities of GLI3R. We next tested this idea in vivo by examining GLI3R nuclear localization. The earliest molecular alteration in Gata6 cKO hindlimb buds in our study is ectopic Gli1 and Ptch1 expression at E10.5 (Fig 1). Therefore, we re-examined Gata6/GATA6 expression, although their mRNA expression patterns were examined in previous studies [38–40]. Gata6 mRNA was detected in the anterior-proximal part of hindlimb buds at E10.25 (34 somite stage) (S4A Fig), but the strong signals in endoderm -derived tissues seem to mask the limb bud signals. Therefore, we also performed immunofluorescence of GATA6 in combination with limb bud mesenchyme markers, such as Fibroblast growth factor10 (FGF10) [57] or Dual specificity phosphatase6 (DUSP6) [58–60]. Co-staining with these markers on transverse sections indicates that GATA6 is present in the ventral side of the proximal region in anterior hindlimb buds at E10.25 (S4B and S4C Fig). The GATA6 signal was undetectable in limb buds in the middle-posterior region.
In the anterior proximal region of limb buds at E10.25, we detected GLI3R predominantly in the nucleus or similarly in the nucleus and cytoplasm (Fig 6C, 6E, 6G and 6I). By contrast, Gata6 cKO hindlimb buds showed a reduced percentage of cells with predominant nuclear GLI3R signals. Accordingly, we detected an increased percentage of cells with nuclear/cytoplasmic GLI3R (Fig 6D, 6H and 6I). Western blot analysis of nuclear extracts from the anterior part of hindlimb buds showed reduced GLI3R levels in Gata6 cKO, compared to wild-type embryos (Fig 6J). Although the presence of nuclear GLI3R in Gata6 cKO hindlimb buds indicates Gata6-independent GLI3R nuclear localization mechanisms in the anterior mesenchyme, reduced GLI3R levels provide evidence that Gata6 contributes to GLI3R nuclear localization. These results are consistent with the in vitro data, and further support the idea that Gata6 regulation of GLI3R nuclear localization contributes to GLI3R activities during normal limb development.
Discussion
In this study, we found hindlimb-specific preaxial polydactyly in Gata6 mutants. The skeletal phenotype of Gata6 mutants was restricted to hindlimbs, and the forelimbs developed normally. Several possibilities would account for such limb type-specific phenotypes. For instance, a recent study showed that Gata4 is differentially expressed in forelimb buds (high) and hindlimb buds (low) [38]. Gata4 and Gata6 are functionally redundant during heart development and for vascular integrity [36, 43]; therefore, Gata4 might compensate for loss of Gata6 in forelimb buds [38]. Another possibility is that differences in the sensitivity to Hedgehog signaling contribute to different phenotypes in fore- and hind-limbs. It is suggested that levels of Hedgehog signaling are higher in hindlimb mesenchyme than forelimb mesenchyme [12], and that hindlimbs are more sensitive to changes in the levels of Hedgehog signaling. Higher Hedgehog signaling, in combination with reduced GLI3R, might have contributed to hindlimb-specific polydactyly in Gata6 cKO. This idea is consistent with ectopic digit formation in Tcre; Gata6+/fl; Gli3+/- forelimbs, in which GLI3R activities would be lower than and SHH signaling levels would be higher than Gli3+/- forelimbs. These two scenarios are not mutually exclusive, and they might cooperate together to ensure proper Hedgehog signaling and pentadactyly in mammalian limbs.
Our study proposes two mechanisms by which Gata6 regulates proper autopod patterning. One mechanism is by enhancing GLI3R activities to repress Hedgehog signaling in the anterior mesenchyme, and the other is by negative regulation of Shh expression in the anterior mesenchyme.
Genetic studies have shown that preaxial polydactyly is associated with ectopic expression of Shh in the anterior mesenchyme [9]. Expression of Shh is positively and negatively regulated in the posterior and anterior mesenchyme, respectively. Twist1, Alx4, Gli3, Tulp3 and Etv4-Etv5 act as negative regulators, for their loss of function caused ectopic Shh expression [23, 47, 49, 51, 61]. Genetic and biochemical studies have shown that Hand2 and Hoxd13 positively regulate Shh expression through the limb bud-specific cis-regulatory element, ZRS [44, 62]. Anterior Shh expression could be induced by loss of negative regulators or ectopic expression of positive regulators [63]. Given that these regulators did not exhibit significant alteration in Gata6 cKO hindlimb buds, the preaxial polydactyly phenotype in Gata6 cKO limbs is unlikely to be induced through these genes. A recent study suggested that Gata6 represses Shh in the limb through binding to ZRS [38]. Our data is consistent with this report, and demonstrated that Shh and its targets are ectopically expressed in Gata6 cKO hindlimb buds at E11.5. Restoration of normal expression pattern of Gli1 and Ptch1 in Gata6 cKO; Shh+/- hindlimbs also supports the idea that Gata6 is upstream of Shh.
The second role is repressing ectopic Hedgehog signaling by enhancing repressor function of Gli3. Ectopic Shh expression in the Gata6 cKO background affects data interpretations; however, compound heterozygous mutant analyses could enable separate analysis of the two mechanisms and support the second mechanism. Previous studies have shown Gli3 to genetically interact with other genes during limb development. Studies on Hox genes suggested that the Gli3-/- polydactyly phenotype is mediated by Hoxd9 and Hoxd10 [29, 64]. In addition, it has been shown that polydactyly of Gli3-/- limbs becomes milder on the Alx4-/- or Zic3-/- background [30, 31], which suggested that the Gli3-/- polydactyly phenotype requires Alx4 or Zic3. In contrast to these reports, loss of one allele of Gata6 enhanced the polydactyly phenotype of Gli3+/- hindlimbs. Therefore, unlike previous genetic studies, our study identified Gata6 as a negative factor for polydactyly development. Given that GLI3R prevents extra-digit formation in the anterior mesenchyme [55], our results suggest that Gata6 cooperates with GLI3R activities.
It is believed that d1 develops in a Shh-independent manner, while development of d2-d5 requires Shh [5, 6, 10, 11]. Genetic manipulation of Gli3 in mice provided evidence that high levels of GLI3R in the anterior of limb buds is necessary for proper d1 development and ensuring pentadactyly [24, 55, 65]. Expression pattern of Pax9, which requires high levels of GLI3R [56], indicates that Gata6 contributes to GLI3R activities in the anterior of hindlimb buds. In particular, Pax9 was undetectable in Tcre; Gata6+/fl; Gli3+/- hindlimb buds, similar to Gata6 cKO and Gli3-/- hindlimb buds. These altered expression pattern of Pax9 correlates with ectopic digit condensation and preaxial polydactyly, and further supports the idea that Gata6 cooperate with Gli3 for proper GLI3R activities in the anterior of hindlimb buds.
How does Gata6 cooperate with Gli3? Our data support the idea that GATA6 physically interacts with GLI3R, facilitates the nuclear localization of GLI3R, and enhances the repressor activities of GLI3R. Reduced nuclear GLI3R localization in Gata6 cKO hindlimb supports the idea that this interaction-mediated nuclear GLI3R localization would also occur in vivo. A recent study showed that Gata4, 5, and 6 can repress Gli-dependent reporter activation in vitro [66]. This study suggested that GATA inhibits SHH-dependent GLI activator function by protein interaction in the chick presomitic mesoderm. Based on this report and our study, GATA might modulate both GLI3R (this study) and SHH-dependent GLI activator [66] in a context-dependent manner. Since expression of Gata genes is reported in other Gli3-positive developing tissues, such as the branchial arch, somite and central nervous system [16, 67, 68], Gata regulation of GLI3R might be a shared mechanism during the development of other organs.
Materials and Methods
Ethics statement
Animal breeding was performed according to the approval by the Institutional Animal Care and Use Committee of the University of Minnesota. Compressed CO2 gas from a cylinder followed by cervical dislocation was the methods of euthanasia for mice. All efforts were made to minimize suffering.
Mouse lines and embryo
The mouse lines for Gata6fl [41], Gli3- [69] and Tcre [42] were maintained on a mixed genetic background. Skeletal preparation was done as previously published [70]. Whole mount in situ hybridization was done as previously published [13].
Expression constructs
The full-length human GATA6 construct and the human GLI3 construct were published [31, 71]. The GLI3R construct was generated by deleting the 3’ part of full-length cDNA, and cloned into 3xFlag CMV7. GATA6 deletion constructs were generated by PCR-based cloning and cloned in pcDNA3.1 or pCS2.
Immunofluorescence and confocal imaging for GLI3R localization
For in vitro analysis, cells were fixed with 4% PFA for two hours at room temperature, washed with PBS and stained with anti-Flag (Sigma, M2, F3165, dilution 1:500) and anti-Myc tag (Abcam, ab9106, dilution 1:500) antibodies. For in vivo analysis, embryos were fixed for two hours in 4% PFA at 4C, washed with cold PBS, and cryosectioned with the OCT compound at 14 μm thickness. Sections were stained according to a standard procedure [13] without heat-induced epitope retrieval. Anti-GATA6 (R&D Systems, AF1700, dilution 1:400) and anti-GLI3R (Clone 6F5, dilution 1:200) [15, 72] were used. Alexa fluorophore-labelled secondary antibodies were obtained from Invitrogen (1:1000 dilution). Fluorescent confocal images were obtained by using Zeiss LSM 710 laser scanning microscope system (Carl Zeiss Microscopy), and analyzed using ZEN2009 software (Carl Zeiss Microscopy).
For subcellular localization analysis in vitro, images were acquired form six arbitrary areas from two plates. Nuclear/cytoplasmic localization of GLI3R and GATA6 was blindly evaluated in cells that were doubly transfected with GLI3R and GATA6 (or its mutants) except for samples that are transfected with GLI3R alone. For in vivo samples, nuclear/cytoplasmic localization of GLI3R was evaluated similarly in the anterior-proximal domain where GATA6 signals in wild-type hindlimb buds were detected. In Gata6 cKO embryos, the anterior-proximal domain, similar to wild-type embryos, was selected for GLI3R subcellular localization. The quantification was performed similar to in vitro samples.
GATA6 localization in hindlimb buds
In order to clarify GATA6 localization in hindlimb bud mesenchyme, GATA6 was simultaneously detected with limb bud mesenchyme markers, such as FGF10 or DUSP6. Wild-type embryos were fixed, washed and cryosectioned as described above. Sections were simultaneously stained by anti-GATA6 (R&D AF1700 or Cell Signaling #5851, dilution 1:1,600) and anti-FGF10 (Santa Cruz, sc-7917, dilution 1:100) or anti-DUSP6 (Sigma, Clone 3G2, dilution 1:200). Sections were reacted with Alexa fluorophore-labelled secondary antibodies, and fluorescent signals were detected by Zeiss LSM 710 according to a standard procedure [13].
Luciferase reporter assay
NIH3T3 cells in 48-well plates were transfected with the 12xGLI-binding site-TK minimum promoter-luciferase [31] with pRL-TK, GATA6 and/or GLI3R expression constructs by using Fugene6 (Promega). Forty hours after transfection, cells were subjected to analysis using the Dual-Luciferase Reporter Assay System (Promega). Experiments were performed in triplicate, and statistical significance was analyzed by One-way ANOVA followed by the Tukey’s comparison.
Co-immunoprecipitation assay and nuclear GLI3R detection
HEK293T cells were transfected with expression constructs by using the standard calcium phosphate method. Cell lysates, prepared after two days, were passed through 25 gauge syringes to ensure protein extraction from the nucleus, and co-immunoprecipitation assays were performed by using Dynabeads protein G (Invitrogen) and anti-Flag (Sigma, M2, F3165, 2μg) or anti-Myc tag (Abcam, ab9106, 1 μg) antibodies. Proteins were resolved by SDS-PAGE, transferred to PVDF membranes (Millipore, MA, USA), reacted with anti-Myc tag or anti-Flag antibodies, followed by HRP goat anti-mouse or rabbit IgG, and a chemiluminescence detection.
For co-immunoprecipitation assays with in vivo samples, hindlimb buds were collected from wild-type embryos at E10.25–10.5. After pooling, the samples were lysed and subjected to co-immunoprecipitation procedures [73] using anti-GATA6 (Cell Signaling, #5851) and Dynabeads protein G. The protein complex was eluted, and detected by Western using anti-GLI3 (R&D Systems, AF3690, dilution 1:100) and the PicoLUCENT PLUS HRP detection kit (G-Bioscience) according to the manufacturer’s instructions.
For nuclear GLI3R detection by Western, anterior one third of hindlimb buds at E10.25–10.5 were collected, and the nuclear fraction was prepared after dissociating cells by using the NE-PER kit (Thermo Fischer) according to the manufacturer’s instructions. The nuclear extracts were analyzed by Western using anti-GLI3 (R&D Systems, AF3690) and anti-Histone H3 (Abcam, ab-1791).
Supporting Information
In situ hybridization of indicated genes in hindlimb buds of wild type (A-E) and Gata6 cKO (F-J) at E11.5.
(TIFF)
In situ hybridization of indicated genes in hindlimb buds of wild type (A-D) and Gata6 cKO (E-H) at E10.5.
(TIFF)
HEK293 cells were transfected with GLI3R and indicated forms of GATA6 (wild type or deletion mutants). Panels show staining by anti-Myc antibodies (GATA6), anti-Flag antibodies (GLI3) or merged images.
(TIF)
(A) Gata6 mRNA expression. Gata6 is expressed in the anterior proximal region of hindlimb buds (arrowhead). (B, C) Co-immunofluorescence of GATA6 with DUSP6 (B) or FGF10 (C). Transverse sections were stained with antibodies for indicated proteins. Dotted areas indicate hindlimb buds. Shown are sections corresponding to the anterior region. GATA6 is expressed in the ventral side of anterior mesenchyme (white arrows). d: dorsal side, v: ventral side.
(TIF)
Embryos at E13.5–15.5 were collected. The breeding pairs are Gata6fl/fl and TcreTg/Tg; Gata6+/fl.
(DOCX)
Embryos at E14.5–16.5 were collected and scored.
(DOCX)
Acknowledgments
We are grateful to Drs. Christine Iacobuzio-Donahue, Juan Carlos Izpisua Belmonte, Mark Lewandoski, Xin Sun, Stephanie Ware, Rolf Zeller and Yi Zhong for sharing plasmids and/or mouse lines. We are also grateful to Dr. Susan Scales for anti-GLI3R antibodies, to Dr. Michael O’Connor for the use of his LSM710, to Dr. Laura Gammill for suggestions and reagents, to Dr. Naoyuki Wada for critical reading, and to Asha Elgonda, Malina Peterson and Samantha Young for their excellent technical support. We thank Malina Peterson and Austin Johnson for editorial assistance.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was supported by grants from the US National Institute of Arthritis, Musculoskeletal and Skin Diseases (URL: http://www.niams.nih.gov) to YK with the grant numbers R01AR064195 and R21AR063782. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Zeller R, Lopez-Rios J, Zuniga A. Vertebrate limb bud development: moving towards integrative analysis of organogenesis. Nat Rev Genet. 2009;10(12):845–58. 10.1038/nrg2681 [DOI] [PubMed] [Google Scholar]
- 2.Chiang C, Litingtung Y, Lee E, Young KE, Corden JL, Westphal H, et al. Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function. Nature. 1996;383(6599):407–13. . [DOI] [PubMed] [Google Scholar]
- 3.Riddle RD, Johnson RL, Laufer E, Tabin C. Sonic hedgehog mediates the polarizing activity of the ZPA. Cell. 1993;75(7):1401–16. . [DOI] [PubMed] [Google Scholar]
- 4.Yang Y, Drossopoulou G, Chuang PT, Duprez D, Marti E, Bumcrot D, et al. Relationship between dose, distance and time in Sonic Hedgehog-mediated regulation of anteroposterior polarity in the chick limb. Development (Cambridge, England). 1997;124(21):4393–404. . [DOI] [PubMed] [Google Scholar]
- 5.Harfe BD, Scherz PJ, Nissim S, Tian H, McMahon AP, Tabin CJ. Evidence for an expansion-based temporal Shh gradient in specifying vertebrate digit identities. Cell. 2004;118(4):517–28. . [DOI] [PubMed] [Google Scholar]
- 6.Ahn S, Joyner AL. Dynamic changes in the response of cells to positive hedgehog signaling during mouse limb patterning. Cell. 2004;118(4):505–16. . [DOI] [PubMed] [Google Scholar]
- 7.Towers M, Mahood R, Yin Y, Tickle C. Integration of growth and specification in chick wing digit-patterning. Nature. 2008;452(7189):882–6. 10.1038/nature06718 [DOI] [PubMed] [Google Scholar]
- 8.Zhu J, Nakamura E, Nguyen MT, Bao X, Akiyama H, Mackem S. Uncoupling Sonic hedgehog control of pattern and expansion of the developing limb bud. Developmental cell. 2008;14(4):624–32. 10.1016/j.devcel.2008.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Masuya H, Sagai T, Wakana S, Moriwaki K, Shiroishi T. A duplicated zone of polarizing activity in polydactylous mouse mutants. Genes & development. 1995;9(13):1645–53. Epub 1995/07/01. . [DOI] [PubMed] [Google Scholar]
- 10.Chiang C, Litingtung Y, Harris MP, Simandl BK, Li Y, Beachy PA, et al. Manifestation of the limb prepattern: limb development in the absence of sonic hedgehog function. Developmental biology. 2001;236(2):421–35. . [DOI] [PubMed] [Google Scholar]
- 11.Kraus P, Fraidenraich D, Loomis CA. Some distal limb structures develop in mice lacking Sonic hedgehog signaling. Mechanisms of development. 2001;100(1):45–58. . [DOI] [PubMed] [Google Scholar]
- 12.Li D, Sakuma R, Vakili NA, Mo R, Puviindran V, Deimling S, et al. Formation of proximal and anterior limb skeleton requires early function of Irx3 and Irx5 and is negatively regulated by Shh signaling. Developmental cell. 2014;29(2):233–40. 10.1016/j.devcel.2014.03.001 [DOI] [PubMed] [Google Scholar]
- 13.Akiyama R, Kawakami H, Wong J, Oishi I, Nishinakamura R, Kawakami Y. Sall4-Gli3 system in early limb progenitors is essential for the development of limb skeletal elements. Proceedings of the National Academy of Sciences of the United States of America. 2015;112(16):5075–80. 10.1073/pnas.1421949112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hui CC, Angers S. Gli proteins in development and disease. Annu Rev Cell Dev Biol. 2011;27:513–37. Epub 2011/08/02. 10.1146/annurev-cellbio-092910-154048 . [DOI] [PubMed] [Google Scholar]
- 15.Wen X, Lai CK, Evangelista M, Hongo JA, de Sauvage FJ, Scales SJ. Kinetics of hedgehog-dependent full-length Gli3 accumulation in primary cilia and subsequent degradation. Molecular and cellular biology. 2010;30(8):1910–22. 10.1128/MCB.01089-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McDermott A, Gustafsson M, Elsam T, Hui CC, Emerson CP Jr., Borycki AG. Gli2 and Gli3 have redundant and context-dependent function in skeletal muscle formation. Development (Cambridge, England). 2005;132(2):345–57. 10.1242/dev.01537 . [DOI] [PubMed] [Google Scholar]
- 17.Pan Y, Bai CB, Joyner AL, Wang B. Sonic hedgehog signaling regulates Gli2 transcriptional activity by suppressing its processing and degradation. Molecular and cellular biology. 2006;26(9):3365–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang B, Fallon JF, Beachy PA. Hedgehog-regulated processing of Gli3 produces an anterior/posterior repressor gradient in the developing vertebrate limb. Cell. 2000;100(4):423–34. . [DOI] [PubMed] [Google Scholar]
- 19.Winter RM, Huson SM. Greig cephalopolysyndactyly syndrome: a possible mouse homologue (Xt-extra toes). Am J Med Genet. 1988;31(4):793–8. 10.1002/ajmg.1320310411 . [DOI] [PubMed] [Google Scholar]
- 20.Pettigrew AL, Greenberg F, Caskey CT, Ledbetter DH. Greig syndrome associated with an interstitial deletion of 7p: confirmation of the localization of Greig syndrome to 7p13. Hum Genet. 1991;87(4):452–6. . [DOI] [PubMed] [Google Scholar]
- 21.Hui CC, Joyner AL. A mouse model of greig cephalopolysyndactyly syndrome: the extra-toesJ mutation contains an intragenic deletion of the Gli3 gene. Nature genetics. 1993;3(3):241–6. Epub 1993/03/01. 10.1038/ng0393-241 . [DOI] [PubMed] [Google Scholar]
- 22.Hu MC, Mo R, Bhella S, Wilson CW, Chuang PT, Hui CC, et al. GLI3-dependent transcriptional repression of Gli1, Gli2 and kidney patterning genes disrupts renal morphogenesis. Development (Cambridge, England). 2006;133(3):569–78. 10.1242/dev.02220 . [DOI] [PubMed] [Google Scholar]
- 23.Buscher D, Bosse B, Heymer J, Ruther U. Evidence for genetic control of Sonic hedgehog by Gli3 in mouse limb development. Mechanisms of development. 1997;62(2):175–82. Epub 1997/03/01. S0925-4773(97)00656-4 [pii]. . [DOI] [PubMed] [Google Scholar]
- 24.Wang C, Ruther U, Wang B. The Shh-independent activator function of the full-length Gli3 protein and its role in vertebrate limb digit patterning. Developmental biology. 2007;305(2):460–9. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.te Welscher P, Zuniga A, Kuijper S, Drenth T, Goedemans HJ, Meijlink F, et al. Progression of vertebrate limb development through SHH-mediated counteraction of GLI3. Science (New York, NY. 2002;298(5594):827–30. . [DOI] [PubMed] [Google Scholar]
- 26.Litingtung Y, Dahn RD, Li Y, Fallon JF, Chiang C. Shh and Gli3 are dispensable for limb skeleton formation but regulate digit number and identity. Nature. 2002;418(6901):979–83. . [DOI] [PubMed] [Google Scholar]
- 27.Aberger F, Ruiz IAA. Context-dependent signal integration by the GLI code: the oncogenic load, pathways, modifiers and implications for cancer therapy. Seminars in cell & developmental biology. 2014;33:93–104. 10.1016/j.semcdb.2014.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Briscoe J, Therond PP. The mechanisms of Hedgehog signalling and its roles in development and disease. Nature reviews. 2013;14(7):416–29. 10.1038/nrm3598 . [DOI] [PubMed] [Google Scholar]
- 29.Sheth R, Bastida MF, Ros M. Hoxd and Gli3 interactions modulate digit number in the amniote limb. Developmental biology. 2007;310(2):430–41. 10.1016/j.ydbio.2007.07.023 . [DOI] [PubMed] [Google Scholar]
- 30.Panman L, Drenth T, Tewelscher P, Zuniga A, Zeller R. Genetic interaction of Gli3 and Alx4 during limb development. The International journal of developmental biology. 2005;49(4):443–8. Epub 2005/06/22. 051984lp [pii] 10.1387/ijdb.051984lp . [DOI] [PubMed] [Google Scholar]
- 31.Quinn ME, Haaning A, Ware SM. Preaxial polydactyly caused by Gli3 haploinsufficiency is rescued by Zic3 loss of function in mice. Human molecular genetics. 2012;21(8):1888–96. Epub 2012/01/12. 10.1093/hmg/dds002 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chlon TM, Crispino JD. Combinatorial regulation of tissue specification by GATA and FOG factors. Development (Cambridge, England). 2012;139(21):3905–16. 10.1242/dev.080440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Molkentin JD. The zinc finger-containing transcription factors GATA-4, -5, and -6. Ubiquitously expressed regulators of tissue-specific gene expression. The Journal of biological chemistry. 2000;275(50):38949–52. 10.1074/jbc.R000029200 . [DOI] [PubMed] [Google Scholar]
- 34.Morrisey EE, Tang Z, Sigrist K, Lu MM, Jiang F, Ip HS, et al. GATA6 regulates HNF4 and is required for differentiation of visceral endoderm in the mouse embryo. Genes & development. 1998;12(22):3579–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koutsourakis M, Langeveld A, Patient R, Beddington R, Grosveld F. The transcription factor GATA6 is essential for early extraembryonic development. Development (Cambridge, England). 1999;126(9):723–32. . [DOI] [PubMed] [Google Scholar]
- 36.Zhao R, Watt AJ, Battle MA, Li J, Bondow BJ, Duncan SA. Loss of both GATA4 and GATA6 blocks cardiac myocyte differentiation and results in acardia in mice. Developmental biology. 2008;317(2):614–9. 10.1016/j.ydbio.2008.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Decker K, Goldman DC, Grasch CL, Sussel L. Gata6 is an important regulator of mouse pancreas development. Developmental biology. 2006;298(2):415–29. 10.1016/j.ydbio.2006.06.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kozhemyakina E, Ionescu A, Lassar AB. GATA6 is a crucial regulator of Shh in the limb bud. PLoS Genet. 2014;10(1):e1004072 Epub 2014/01/15. 10.1371/journal.pgen.1004072 PGENETICS-D-13-01628 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karamboulas K, Dranse HJ, Underhill TM. Regulation of BMP-dependent chondrogenesis in early limb mesenchyme by TGFbeta signals. Journal of cell science. 2010;123(Pt 12):2068–76. Epub 2010/05/27. 10.1242/jcs.062901 [pii]. . [DOI] [PubMed] [Google Scholar]
- 40.Alexandrovich A, Qureishi A, Coudert AE, Zhang L, Grigoriadis AE, Shah AM, et al. A role for GATA-6 in vertebrate chondrogenesis. Developmental biology. 2008;314(2):457–70. Epub 2008/01/15. 10.1016/j.ydbio.2007.12.001 S0012-1606(07)01570-9 [pii]. . [DOI] [PubMed] [Google Scholar]
- 41.Sodhi CP, Li J, Duncan SA. Generation of mice harbouring a conditional loss-of-function allele of Gata6. BMC developmental biology. 2006;6:19 .16611361 [Google Scholar]
- 42.Perantoni AO, Timofeeva O, Naillat F, Richman C, Pajni-Underwood S, Wilson C, et al. Inactivation of FGF8 in early mesoderm reveals an essential role in kidney development. Development (Cambridge, England). 2005;132(17):3859–71. . [DOI] [PubMed] [Google Scholar]
- 43.Xin M, Davis CA, Molkentin JD, Lien CL, Duncan SA, Richardson JA, et al. A threshold of GATA4 and GATA6 expression is required for cardiovascular development. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(30):11189–94. Epub 2006/07/19. 0604604103 [pii] 10.1073/pnas.0604604103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Galli A, Robay D, Osterwalder M, Bao X, Benazet JD, Tariq M, et al. Distinct roles of Hand2 in initiating polarity and posterior Shh expression during the onset of mouse limb bud development. PLoS Genet. 2010;6(4):e1000901 Epub 2010/04/14. 10.1371/journal.pgen.1000901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qu S, Tucker SC, Ehrlich JS, Levorse JM, Flaherty LA, Wisdom R, et al. Mutations in mouse Aristaless-like4 cause Strong's luxoid polydactyly. Development (Cambridge, England). 1998;125(14):2711–21. . [DOI] [PubMed] [Google Scholar]
- 46.Zhang Z, Verheyden JM, Hassell JA, Sun X. FGF-regulated Etv genes are essential for repressing Shh expression in mouse limb buds. Developmental cell. 2009;16(4):607–13. Epub 2009/04/24. S1534-5807(09)00082-3 [pii] 10.1016/j.devcel.2009.02.008 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mao J, McGlinn E, Huang P, Tabin CJ, McMahon AP. Fgf-dependent Etv4/5 activity is required for posterior restriction of Sonic Hedgehog and promoting outgrowth of the vertebrate limb. Developmental cell. 2009;16(4):600–6. Epub 2009/04/24. S1534-5807(09)00079-3 [pii] 10.1016/j.devcel.2009.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bourgeois P, Bolcato-Bellemin AL, Danse JM, Bloch-Zupan A, Yoshiba K, Stoetzel C, et al. The variable expressivity and incomplete penetrance of the twist-null heterozygous mouse phenotype resemble those of human Saethre-Chotzen syndrome. Human molecular genetics. 1998;7(6):945–57. . [DOI] [PubMed] [Google Scholar]
- 49.Zhang Z, Sui P, Dong A, Hassell J, Cserjesi P, Chen YT, et al. Preaxial polydactyly: interactions among ETV, TWIST1 and HAND2 control anterior-posterior patterning of the limb. Development (Cambridge, England). 2010;137(20):3417–26. Epub 2010/09/10. dev.051789 [pii] 10.1242/dev.051789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Patterson VL, Damrau C, Paudyal A, Reeve B, Grimes DT, Stewart ME, et al. Mouse hitchhiker mutants have spina bifida, dorso-ventral patterning defects and polydactyly: identification of Tulp3 as a novel negative regulator of the Sonic hedgehog pathway. Human molecular genetics. 2009;18(10):1719–39. Epub 2009/02/19. ddp075 [pii] 10.1093/hmg/ddp075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Norman RX, Ko HW, Huang V, Eun CM, Abler LL, Zhang Z, et al. Tubby-like protein 3 (TULP3) regulates patterning in the mouse embryo through inhibition of Hedgehog signaling. Human molecular genetics. 2009;18(10):1740–54. Epub 2009/03/17. ddp113 [pii] 10.1093/hmg/ddp113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cameron DA, Pennimpede T, Petkovich M. Tulp3 is a critical repressor of mouse hedgehog signaling. Dev Dyn. 2009;238(5):1140–9. Epub 2009/04/01. 10.1002/dvdy.21926 . [DOI] [PubMed] [Google Scholar]
- 53.Wright E, Hargrave MR, Christiansen J, Cooper L, Kun J, Evans T, et al. The Sry-related gene Sox9 is expressed during chondrogenesis in mouse embryos. Nature genetics. 1995;9(1):15–20. . [DOI] [PubMed] [Google Scholar]
- 54.Mo R, Freer AM, Zinyk DL, Crackower MA, Michaud J, Heng HH, et al. Specific and redundant functions of Gli2 and Gli3 zinc finger genes in skeletal patterning and development. Development (Cambridge, England). 1997;124(1):113–23. Epub 1997/01/01. . [DOI] [PubMed] [Google Scholar]
- 55.Lopez-Rios J, Speziale D, Robay D, Scotti M, Osterwalder M, Nusspaumer G, et al. GLI3 constrains digit number by controlling both progenitor proliferation and BMP-dependent exit to chondrogenesis. Developmental cell. 2012;22(4):837–48. Epub 2012/04/03. 10.1016/j.devcel.2012.01.006 S1534-5807(12)00040-8 [pii]. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McGlinn E, van Bueren KL, Fiorenza S, Mo R, Poh AM, Forrest A, et al. Pax9 and Jagged1 act downstream of Gli3 in vertebrate limb development. Mechanisms of development. 2005;122(11):1218–33. . [DOI] [PubMed] [Google Scholar]
- 57.Min H, Danilenko DM, Scully SA, Bolon B, Ring BD, Tarpley JE, et al. Fgf-10 is required for both limb and lung development and exhibits striking functional similarity to Drosophila branchless. Genes & development. 1998;12(20):3156–61. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dickinson RJ, Eblaghie MC, Keyse SM, Morriss-Kay GM. Expression of the ERK-specific MAP kinase phosphatase PYST1/MKP3 in mouse embryos during morphogenesis and early organogenesis. Mechanisms of development. 2002;113(2):193–6. [DOI] [PubMed] [Google Scholar]
- 59.Klock A, Herrmann BG. Cloning and expression of the mouse dual-specificity mitogen-activated protein (MAP) kinase phosphatase Mkp3 during mouse embryogenesis. Mechanisms of development. 2002;116(1–2):243–7. [DOI] [PubMed] [Google Scholar]
- 60.Kawakami Y, Rodriguez-Leon J, Koth CM, Buscher D, Itoh T, Raya A, et al. MKP3 mediates the cellular response to FGF8 signalling in the vertebrate limb. Nature cell biology. 2003;5(6):513–9. . [DOI] [PubMed] [Google Scholar]
- 61.Qu S, Niswender KD, Ji Q, van der Meer R, Keeney D, Magnuson MA, et al. Polydactyly and ectopic ZPA formation in Alx-4 mutant mice. Development (Cambridge, England). 1997;124(20):3999–4008. Epub 1997/11/28. . [DOI] [PubMed] [Google Scholar]
- 62.Lettice LA, Horikoshi T, Heaney SJ, van Baren MJ, van der Linde HC, Breedveld GJ, et al. Disruption of a long-range cis-acting regulator for Shh causes preaxial polydactyly. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(11):7548–53. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Charite J, McFadden DG, Olson EN. The bHLH transcription factor dHAND controls Sonic hedgehog expression and establishment of the zone of polarizing activity during limb development. Development (Cambridge, England). 2000;127(11):2461–70. Epub 2000/05/11. . [DOI] [PubMed] [Google Scholar]
- 64.Zakany J, Zacchetti G, Duboule D. Interactions between HOXD and Gli3 genes control the limb apical ectodermal ridge via Fgf10. Developmental biology. 2007;306(2):883–93. . [DOI] [PubMed] [Google Scholar]
- 65.Hill P, Gotz K, Ruther U. A SHH-independent regulation of Gli3 is a significant determinant of anteroposterior patterning of the limb bud. Developmental biology. 2009;328(2):506–16. 10.1016/j.ydbio.2009.02.017 . [DOI] [PubMed] [Google Scholar]
- 66.Daoud G, Kempf H, Kumar D, Kozhemyakina E, Holowacz T, Kim DW, et al. BMP-mediated induction of GATA4/5/6 blocks somitic responsiveness to SHH. Development (Cambridge, England). 2014;141(20):3978–87. 10.1242/dev.111906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nardelli J, Thiesson D, Fujiwara Y, Tsai FY, Orkin SH. Expression and genetic interaction of transcription factors GATA-2 and GATA-3 during development of the mouse central nervous system. Developmental biology. 1999;210(2):305–21. 10.1006/dbio.1999.9278 . [DOI] [PubMed] [Google Scholar]
- 68.Ruest LB, Xiang X, Lim KC, Levi G, Clouthier DE. Endothelin-A receptor-dependent and -independent signaling pathways in establishing mandibular identity. Development (Cambridge, England). 2004;131(18):4413–23. 10.1242/dev.01291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Buscher D, Grotewold L, Ruther U. The XtJ allele generates a Gli3 fusion transcript. Mamm Genome. 1998;9(8):676–8. . [DOI] [PubMed] [Google Scholar]
- 70.Kawakami Y, Tsuda M, Takahashi S, Taniguchi N, Esteban CR, Zemmyo M, et al. Transcriptional coactivator PGC-1alpha regulates chondrogenesis via association with Sox9. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(7):2414–9. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhong Y, Wang Z, Fu B, Pan F, Yachida S, Dhara M, et al. GATA6 activates Wnt signaling in pancreatic cancer by negatively regulating the Wnt antagonist Dickkopf-1. PloS one. 2011;6(7):e22129 10.1371/journal.pone.0022129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Osterwalder M, Speziale D, Shoukry M, Mohan R, Ivanek R, Kohler M, et al. HAND2 targets define a network of transcriptional regulators that compartmentalize the early limb bud mesenchyme. Developmental cell. 2014;31(3):345–57. Epub 2014/12/03. 10.1016/j.devcel.2014.09.018 S1534-5807(14)00624-8 [pii]. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Suzuki A, Raya A, Kawakami Y, Morita M, Matsui T, Nakashima K, et al. Nanog binds to Smad1 and blocks bone morphogenetic protein-induced differentiation of embryonic stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(27):10294–9. . [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
In situ hybridization of indicated genes in hindlimb buds of wild type (A-E) and Gata6 cKO (F-J) at E11.5.
(TIFF)
In situ hybridization of indicated genes in hindlimb buds of wild type (A-D) and Gata6 cKO (E-H) at E10.5.
(TIFF)
HEK293 cells were transfected with GLI3R and indicated forms of GATA6 (wild type or deletion mutants). Panels show staining by anti-Myc antibodies (GATA6), anti-Flag antibodies (GLI3) or merged images.
(TIF)
(A) Gata6 mRNA expression. Gata6 is expressed in the anterior proximal region of hindlimb buds (arrowhead). (B, C) Co-immunofluorescence of GATA6 with DUSP6 (B) or FGF10 (C). Transverse sections were stained with antibodies for indicated proteins. Dotted areas indicate hindlimb buds. Shown are sections corresponding to the anterior region. GATA6 is expressed in the ventral side of anterior mesenchyme (white arrows). d: dorsal side, v: ventral side.
(TIF)
Embryos at E13.5–15.5 were collected. The breeding pairs are Gata6fl/fl and TcreTg/Tg; Gata6+/fl.
(DOCX)
Embryos at E14.5–16.5 were collected and scored.
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.