Abstract
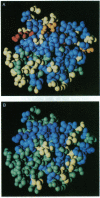
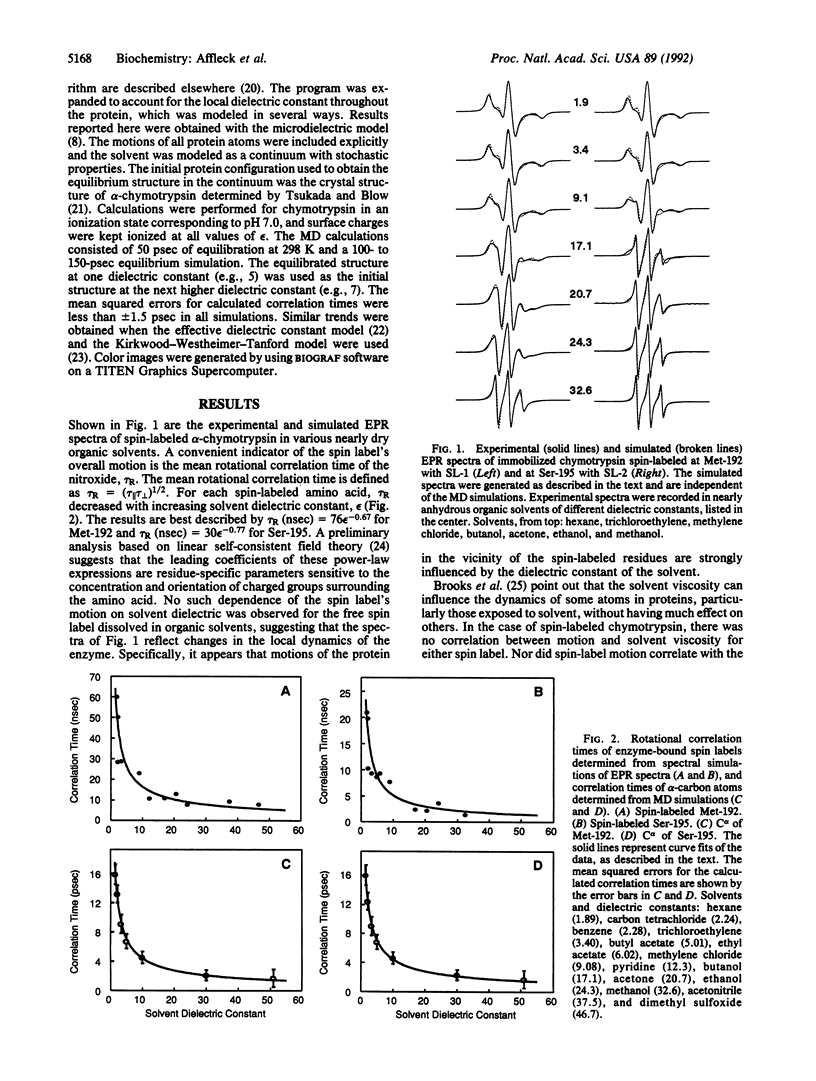
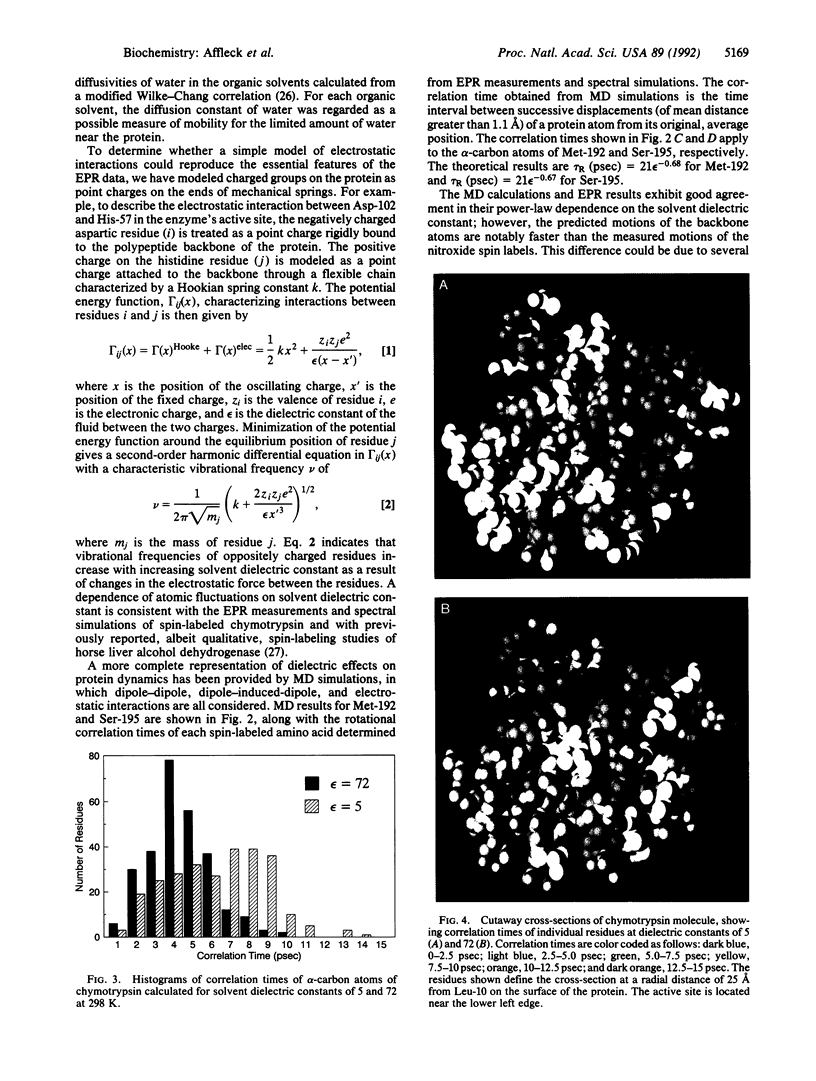
Electron paramagnetic resonance (EPR) spectroscopy and molecular dynamics (MD) simulations were used to investigate the dynamics of alpha-chymotrypsin in solvents ranging in dielectric constant from 72 to 1.9. EPR measurements showed that motions in the vicinity of two spin-labeled amino acids (Met-192 and Ser-195) decreased dramatically with decreasing solvent dielectric constant, a trend consistent with changes in the electrostatic force between charged residues of the protein. EPR results and MD simulations revealed a very similar functional dependence between rates of motion in the protein and the dielectric constant of the bulk solvent; however, predicted motions of protein atoms were markedly faster than measured motions of the spin labels. MD calculations for dielectric constants of 5 and 72 showed the greatest differences near the outer surface of the protein. In general, at the lower dielectric constant many atoms of the protein move more slowly, and many of the slowest residues are near the exterior. These results suggest that altered dynamics may contribute to the unusual properties--e.g., modified stereoselectivities--of enzymes in nearly dry organic solvents.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Affleck R., Xu Z. F., Suzawa V., Focht K., Clark D. S., Dordick J. S. Enzymatic catalysis and dynamics in low-water environments. Proc Natl Acad Sci U S A. 1992 Feb 1;89(3):1100–1104. doi: 10.1073/pnas.89.3.1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey J. E., Clark D. S. Electron paramagnetic resonance studies of immobilized chymotrypsin. Methods Enzymol. 1987;135:502–512. doi: 10.1016/0076-6879(87)35106-7. [DOI] [PubMed] [Google Scholar]
- Berliner L. J., Wong S. S. Spin-labeled sulfonyl fluorides as active site probes of protease structure. I. Comparison of the active site environments in alpha-chymotrypsin and trypsin. J Biol Chem. 1974 Mar 25;249(6):1668–1677. [PubMed] [Google Scholar]
- Bone S. Time-domain reflectrometry studies of water binding and structural flexibility in chymotrypsin. Biochim Biophys Acta. 1987 Nov 5;916(1):128–134. doi: 10.1016/0167-4838(87)90219-6. [DOI] [PubMed] [Google Scholar]
- Cusack S., Smith J., Finney J., Tidor B., Karplus M. Inelastic neutron scattering analysis of picosecond internal protein dynamics. Comparison of harmonic theory with experiment. J Mol Biol. 1988 Aug 20;202(4):903–908. doi: 10.1016/0022-2836(88)90566-9. [DOI] [PubMed] [Google Scholar]
- Karplus M., Petsko G. A. Molecular dynamics simulations in biology. Nature. 1990 Oct 18;347(6294):631–639. doi: 10.1038/347631a0. [DOI] [PubMed] [Google Scholar]
- Kosman D. J., Piette L. H. ESR probing of macromolecules: spin label study of the conformational integrity and transitions in modified alpha-chymotrypsin. Arch Biochem Biophys. 1972 Apr;149(2):452–460. doi: 10.1016/0003-9861(72)90344-x. [DOI] [PubMed] [Google Scholar]
- Perutz M. F. Electrostatic effects in proteins. Science. 1978 Sep 29;201(4362):1187–1191. doi: 10.1126/science.694508. [DOI] [PubMed] [Google Scholar]
- Randolph T. W., Clark D. S., Blanch H. W., Prausnitz J. M. Enzymatic oxidation of cholesterol aggregates in supercritical carbon dioxide. Science. 1988 Jan 22;239(4838):387–390. doi: 10.1126/science.239.4838.387. [DOI] [PubMed] [Google Scholar]
- Ringe D., Petsko G. A. Mapping protein dynamics by X-ray diffraction. Prog Biophys Mol Biol. 1985;45(3):197–235. doi: 10.1016/0079-6107(85)90002-1. [DOI] [PubMed] [Google Scholar]
- Rogers N. K. The modelling of electrostatic interactions in the function of globular proteins. Prog Biophys Mol Biol. 1986;48(1):37–66. doi: 10.1016/0079-6107(86)90009-x. [DOI] [PubMed] [Google Scholar]
- Tsukada H., Blow D. M. Structure of alpha-chymotrypsin refined at 1.68 A resolution. J Mol Biol. 1985 Aug 20;184(4):703–711. doi: 10.1016/0022-2836(85)90314-6. [DOI] [PubMed] [Google Scholar]
- Warshel A., Russell S. T. Calculations of electrostatic interactions in biological systems and in solutions. Q Rev Biophys. 1984 Aug;17(3):283–422. doi: 10.1017/s0033583500005333. [DOI] [PubMed] [Google Scholar]
- Wong S. S., Quiggle K., Triplett C., Berliner L. J. Spin-labeled sulfonyl fluorides as active site probes of protease structure. II. Spin label syntheses and enzyme inhibition. J Biol Chem. 1974 Mar 25;249(6):1678–1682. [PubMed] [Google Scholar]
- Wüthrich K. Protein structure determination in solution by nuclear magnetic resonance spectroscopy. Science. 1989 Jan 6;243(4887):45–50. doi: 10.1126/science.2911719. [DOI] [PubMed] [Google Scholar]
- Zaks A., Klibanov A. M. Enzymatic catalysis in nonaqueous solvents. J Biol Chem. 1988 Mar 5;263(7):3194–3201. [PubMed] [Google Scholar]
- Zaks A., Klibanov A. M. Enzymatic catalysis in organic media at 100 degrees C. Science. 1984 Jun 15;224(4654):1249–1251. doi: 10.1126/science.6729453. [DOI] [PubMed] [Google Scholar]