Healthy donors exposed to Japanese encephalitis (JE) virus show a CD8+ T cell response that cross reacts with other flaviviruses. Patients that recovered from JE show a CD4+ T cell response that targets structural proteins of JE virus.
Abstract
Japanese encephalitis (JE) virus (JEV) is an important cause of encephalitis in children of South and Southeast Asia. However, the majority of individuals exposed to JEV only develop mild symptoms associated with long-lasting adaptive immunity. The related flavivirus dengue virus (DENV) cocirculates in many JEV-endemic areas, and clinical data suggest cross-protection between DENV and JEV. To address the role of T cell responses in protection against JEV, we conducted the first full-breadth analysis of the human memory T cell response using a synthetic peptide library. Ex vivo interferon-γ (IFN-γ) responses to JEV in healthy JEV-exposed donors were mostly CD8+ and targeted nonstructural (NS) proteins, whereas IFN-γ responses in recovered JE patients were mostly CD4+ and targeted structural proteins and the secreted protein NS1. Among patients, a high quality, polyfunctional CD4+ T cell response was associated with complete recovery from JE. T cell responses from healthy donors showed a high degree of cross-reactivity to DENV that was less apparent in recovered JE patients despite equal exposure. These data reveal divergent functional CD4+ and CD8+ T cell responses linked to different clinical outcomes of JEV infection, associated with distinct targeting and broad flavivirus cross-reactivity including epitopes from DENV, West Nile, and Zika virus.
Japanese encephalitis (JE) virus (JEV) is a member of the family Flavivirus, genus Flaviviridae. JEV is an arthropod-borne virus (arbovirus) endemic to rural parts of South and Southeast Asia. JE is the most commonly diagnosed encephalitis in Asia and is an important cause of disability and death in children in this region. Only ~0.1–1% of JEV infections in humans results in encephalitis (Halstead and Grosz, 1962). The remainder is clinically silent or results in a mild febrile illness (Watt and Jongsakul, 2003). The overwhelming majority of JEV-exposed individuals thus develop long-lasting naturally induced adaptive immunity, although this wanes in later life (Solomon, 2004).
JEV is a single-stranded, positive sense RNA virus; the single 10-kb open reading frame encodes three structural proteins, envelope (E), premembrane (prM), and core (C), and seven nonstructural (NS) proteins designated NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5 (Sumiyoshi et al., 1987). There are an estimated 68,000 cases of JE annually and probably around 17,000 deaths (Campbell et al., 2011). Clinical outcomes vary from complete recovery to recovery with neuropsychiatric sequelae to death (Solomon et al., 2002). The role of the immune response to JEV in determining whether infection is asymptomatic or results in disease is incompletely understood.
Protection by neutralizing antibodies (NAbs) to JEV in animal models is well established, whether passively administered, preformed by immunization, or rapidly developed after challenge by a memory response (Konishi et al., 1999; Gao et al., 2010; Larena et al., 2011; Van Gessel et al., 2011). Higher titers of JEV NAbs correlate with better protection (Lubiniecki et al., 1973). Development of JEV NAbs after vaccination correlates with protection in humans (Hoke et al., 1988), and an anamnestic antibody response is associated with a good outcome from disease (Libraty et al., 2002; Winter et al., 2004). Conversely, low JEV antibody levels in serum and cerebrospinal fluid (CSF) of patients with JE are associated with death (Libraty et al., 2002; Solomon et al., 2002).
Cell-mediated immunity to JEV is less well studied. Observations from animal models suggest that T cells play a subsidiary role in protection from JE but could also contribute to immunopathology (Larena et al., 2011). Adoptive transfer of primed T cells can protect animals from intracerebral JEV challenge (Murali-Krishna et al., 1996), even in the absence of antibodies in some cases (Ashok and Rangarajan, 1999).
Lymphocyte responses to JEV have been detected in humans for many years (Chaturvedi et al., 1979; Konishi et al., 1995). Studies of healthy JEV-exposed residents of endemic areas demonstrated CD4+ and CD8+ T cell responses to several JEV proteins dominated by NS3, especially amino acids 193–324 (Kumar et al., 2004c). In recovered JE patients, IFN-γ responses to NS3 amino acids 193–324 were infrequent and smaller than in healthy JEV-exposed controls, despite proliferative responses being preserved (Kumar et al., 2004b). Low IFN-γ production was associated with poor outcome, indicating a role for IFN-γ in recovery from JE. However, other cytokine responses and responses to other JEV proteins have not been examined in JE patients.
JEV cocirculates with other flaviviruses in much of South and Southeast Asia, notably the four serotypes of dengue virus (DENV). Prior DENV exposure reduces the severity of JE (Edelman et al., 1975; Libraty et al., 2002); however, the effect of JEV immunity on subsequent DENV infection is less clear. A large study of an inactivated JE vaccine in Thailand showed those patients who developed dengue hemorrhagic fever (DHF) had lower illness severity scores if they had received the JE vaccine, suggesting a beneficial effect (Hoke et al., 1988). On the other hand, a later careful prospective study in the same location showed that the presence of JEV NAbs at the start of the study (in the absence of DENV NAbs) gave a 2.75-fold increase in the risk of dengue fever (DF; Anderson et al., 2011).
DENV infection primes broadly cross-reactive T cell responses; such responses between DENV serotypes have been studied extensively, particularly with reference to DHF (Rothman, 2010). Limited data suggest that cross-reactive T cell responses between JEV and DENV exist in inactivated JE vaccine recipients (Aihara et al., 1998). However, the potential for cross-reactive T cell responses between JEV and other flaviviruses after natural infection and their potential for protection is not known.
Collectively, the available data suggest that T cell responses could be protective in human JE; however, no study to date has compared responses across the entire length of the JEV polyprotein between encephalitis and asymptomatic infection. Moreover, many of the published DENV epitopes show marked similarity to the corresponding sequences in JEV, suggesting that immunological cross-recognition by T cells is theoretically possible. Given that prior DENV infection is protective in JE and DENV infection primes broadly cross-reactive T cells, we hypothesized that in regions endemic to both viruses, patients who had suffered from JE would have memory T cell responses that had less cross-reactivity with DENV than healthy JEV-exposed subjects. Therefore, we undertook a study to systematically map JEV epitopes using a full-length synthetic peptide library for JEV. We assessed the specificity, function, and magnitude of the T cell response and measured the degree of T cell receptor cross-reactivity in healthy JEV-exposed donors and recovered JE patients.
RESULTS
Subjects
The study took place in Karnataka State, South India, a JE-endemic area. 103 participants were recruited into the study (Table 1). 51 were healthy Indian residents of JEV-endemic areas, and 39 were recovered JE patients. Healthy donors were recruited in Bangalore and at the Vijayanagar Institute of Medical Sciences (VIMS), Bellary. Recovered JE patients were recruited at VIMS. 35 healthy donors (69%) were defined as JEV exposed on the basis of residence in a JEV-endemic area during childhood and a positive NAb assay. A training cohort of 13 participants was recruited in the UK, comprising a mixture of flavivirus-exposed and unexposed subjects. Two subjects recruited in the UK reported classical dengue illness (one DF and one DHF) with positive DENV IgM, and one more without diagnostic testing; all three had acquired infection in dengue-endemic areas and were anti-DENV NAb positive.
Table 1. Characteristics of the study participants.
| Characteristics | Recovered JE patients | Healthy, JEV NAb-positive Indian controls | Healthy, JEV NAb-negative Indian controls | UK cohort | |
|---|---|---|---|---|---|
| n | 39 | 35 | 16 | 13 | |
| Median age (range) | 13 (1–25) | 26 (7–55) | 25 (22–55) | 26 (23–60) | |
| Sex | 22 M, 17 F | 24 M, 11 F | 10 M, 6 F | 7 M, 6 F | |
| Years since JE | 6 (0.5–12) | NA | NA | NA | |
| Number tested for DENV NAb | 14 | 14 | 3 | 8 | |
| DENV NAb positive (any serotype) | 14 | 14 | 3 | 3 | |
| Dengue illness | 0 | 3 | 0 | 4 | |
| Residence in a JE-endemic area | All | All | All | 1 | |
| Travel to a JE-endemic area | NA | NA | NA | 8 | |
| JE vaccine | 0 | 0 | 0 | 6a | |
| Liverpool outcome score | 5 | 24 | NA | NA | NA |
| 4 | 5 | ||||
| 3 | 7 | ||||
| 2 | 3 |
The Liverpool outcome score is a five-point score grading recovery from encephalitis in children and adolescents (Lewthwaite et al., 2010a). 5, full recovery; 4, minor disability; 3, moderate disability; 2, severe disability; 1, death (not applicable here).
Six subjects in the UK cohort received the JE vaccine, four before and two during the study period.
T cell responses in healthy JEV-exposed subjects are dominated by diverse, highly cross-reactive CD8+ cells
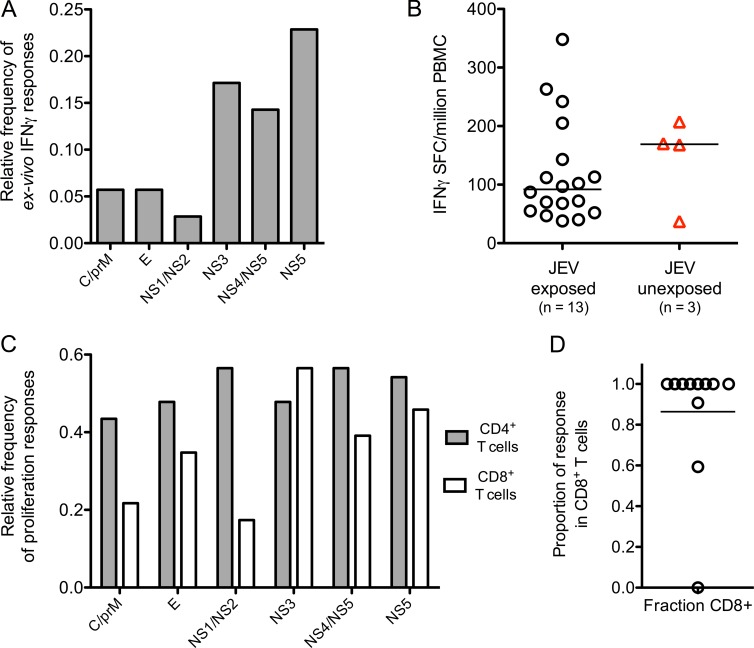
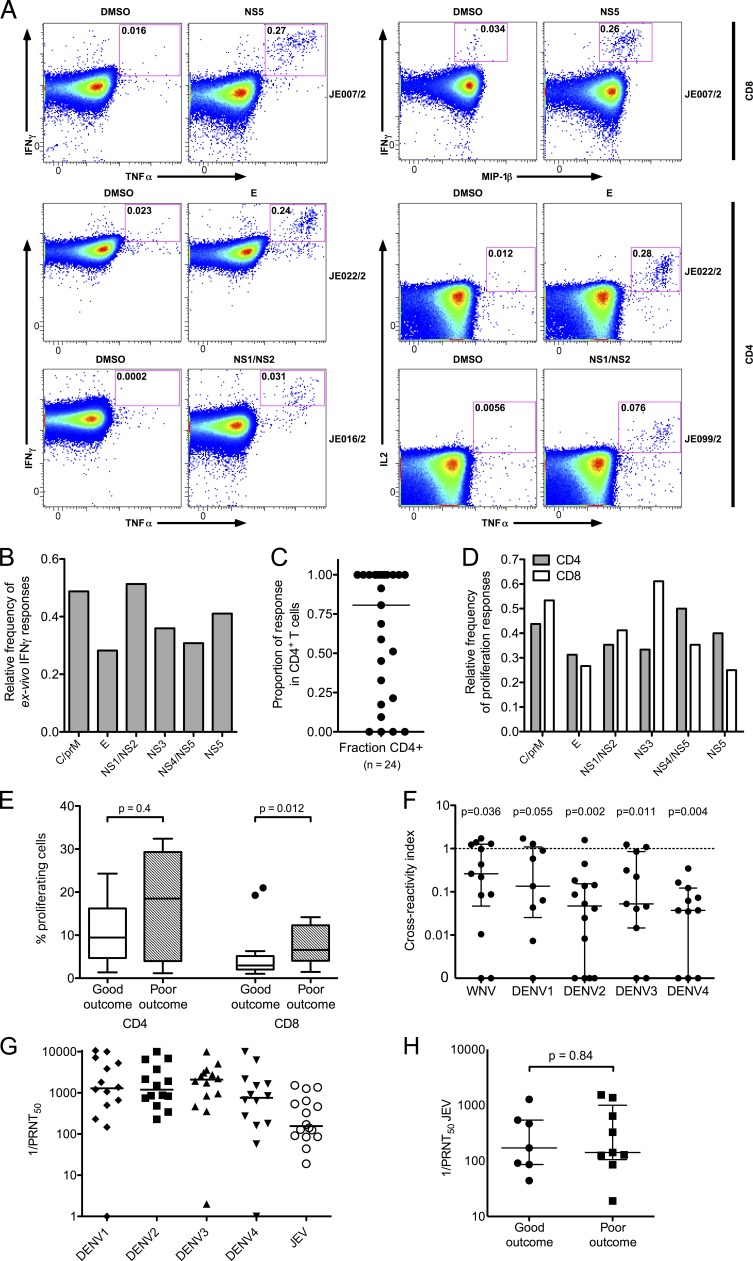
35 JEV NAb (Fig. 1 B)+ healthy South Indian donors were screened by ELISPOT assay or whole blood (WB) intracellular cytokine staining (ICS) for JEV T cell responses. 17 (49%) showed positive ex vivo IFN-γ assays (Fig. 1, A and B). In a subset of 24 subjects tested, 22 (92%) showed proliferative responses (Fig. 1 C). IFN-γ responses were predominantly directed against NS3, NS4, and NS5 (Fig. 1 A) and were modest in size (Fig. 1, B and D), whereas proliferative responses recognized all viral proteins (Fig. 1 C). Flow cytometric analysis showed that 80% of ex vivo IFN-γ responses were CD8+ (Fig. 1 D).
Figure 1.
Breadth of the T cell response to peptide pools of JEV in JEV-exposed healthy donors. (A) Relative frequency of ex vivo IFN-γ responses were measured by ELISPOT or ICS in JEV-exposed healthy donors (n = 35, 29 for ELISPOT, and 6 for ICS). Peptide pools are shown grouped by viral proteins. For a subset of five subjects, ICS and ELISPOT were performed at least three times with consistent results. C, core. E, envelope. (B) Spot-forming cells (SFCs) per million PBMCs were measured by ELISPOT in 13 healthy JEV-exposed donors (18 responses, black circles) and three DENV-exposed subjects (four responses, red triangles). (C) Proliferative responses were measured by CFSE dilution and flow cytometry in healthy JEV-exposed donors once per subject. Data are relative frequency (n = 24) for CD4+ and CD8+ T cells. (D) Based on data from ICS assays, the proportion of the total IFN-γ response produced by CD8+ T cells in each healthy JEV-exposed donor was calculated. The bar depicts the median. n = 11.
Clinical data suggest cross-protection between DENV and JEV. Two subjects with documented dengue illness (but who were unlikely to have been JEV exposed) and one JEV NAb-negative volunteer showed IFN-γ ELISPOT responses to the JEV peptide library (Fig. 1 B, red); no responses were detected in healthy DENV- and JEV-unexposed controls (unpublished data). The two subjects reporting dengue were also positive for JEV NAbs, though anti-DENV titers were higher, consistent with prior DENV infection (JEV 50% plaque reduction neutralization titer [PRNT50] 1 in 266 and 1 in 85 and DENV PRNT50 1 in 4,515 [DENV1] and 1 in 12,413 [DENV3], respectively). Therefore, we set out to determine whether JEV and DENV responses cross react. First, responses were mapped by ELISPOT or by expanding short-term T cell lines from donors showing ex vivo responses followed by deconvolution of pools in ICS assays. Next, cross-reactivity was tested using variant peptides from DENV (and other flaviviruses) corresponding to the mapped peptides of JEV.
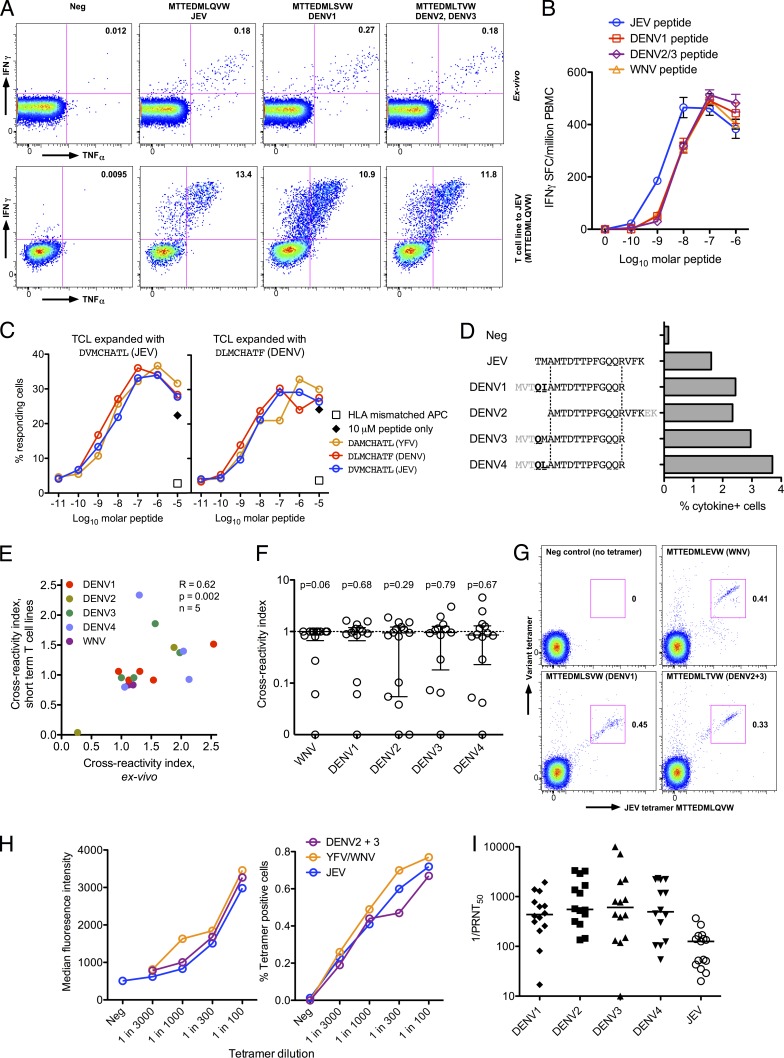
Using this approach, we first studied two naturally JEV-exposed subjects (H001/1 and H008/4) and one reporting DF (H001/4) in detail. CD8+ T cell responses were identical in size and functional characteristics to peptide sequence variants from other flaviviruses (Fig. 2 A [top] and B). T cell lines showed similar responses in functional assays for whichever peptide was tested (Fig. 2 A, bottom), irrespective of which peptide was used to expand the line (Fig. 2 C). Titrations of variant peptides showed responses detectable in the nanomolar range and that cross-reactivity was not limited to high peptide concentration (Fig. 2, B and C), although there was some variation in the efficiency of individual peptides.
Figure 2.
CD8+ T cell responses are highly flavivirus cross-reactive in healthy JEV-exposed donors. (A) ICS assays were used to detect IFN-γ+/TNF-α+ cells from healthy JEV-exposed donor H008/4. Example flow cytometry data from an ex vivo assay (top) and a short-term T cell line (bottom) show responses to variant peptides of JEV NS5 MTTEDMLQVW, gated on live, CD3+, and CD8+ cells, representative of three experiments. Similar results were obtained with DENV4 and WNV peptides (not depicted). Axes are log10 fluorescence units. (B) IFN-γ responses to peptide titrations of the same NS5 peptides as in A and WNV peptide MTTEDMLEVW were measured by ex vivo ELISPOT. The results are representative of two independent experiments. SFC, spot-forming cell. (C) Cytokine (IFN-γ+, TNF-α+, or MIP-1β+ in any combination) responses to NS3 peptide titrations of JEV, DENV1–4, and yellow fever virus (YFV) presented on a B cell line matched for HLA B*08:01 were measured by ICS. Responding cells were CD8+ T cell lines (TCL) from a subject reporting dengue illness and yellow fever vaccination but not JEV exposure (H001/4), expanded with JEV (left) or DENV (right) peptides, each assayed against all three peptides. The black diamonds indicate peptides with no B cell line. Open squares indicate a peptide-pulsed HLA-mismatched B cell line. Peptide titrations by ex vivo ELISPOT or ICS gave similar results, and expansion of T cell lines from two further independent samples showed equal cross-reactivity. (D) IFN-γ+/TNF-α+ cells of a CD8+ T cell line from healthy JEV-exposed donor H007/3 were measured by ICS to a JEV NS5 peptide and the DENV variant peptides indicated, showing epitope conservation in variant peptides. The same experiment was performed twice ex vivo with similar results. (E) Cross-reactivity of ex vivo responses and short-term T cell lines were measured by ICS. Cross-reactivity index (variant response/JEV response from the same assay) between JEV and DENV1–4/WNV in five subjects of ex vivo measurements correlated with T cell lines (Spearman’s R = 0.62; P = 0.002). Ex vivo assays were performed at least twice in all donors except one with similar results. (F) Cross-reactivity of short-term T cell lines expanded with JEV peptides with WNV, and DENV1–4 measured by ICS. Data are cross-reactivity indices as in E of all T cell responses identified in healthy JEV-exposed donors. Wilcoxon signed rank tests showed no significant differences from a hypothetical value of 1 (indicating an equal response to JEV and variant peptides). In two subjects, these assays were repeated three times with good agreement. (G) CD8+ T cells from donor H008/4 were stained with JEV peptide–HLA class I tetramers (x axis) and variant peptide tetramers (y axis) and analyzed by flow cytometry. Data are representative of two independent experiments. (H) CD8+ T cells were tetramer stained and analyzed as in G. Titrations of three different tetramers are shown: JEV (MTTEDMLQVW), WNV (MTTEDMLEVW), and DENV2+3 (MTTEDMLTVW; common epitope). Data are median fluorescence intensity (left) and percentage of CD8+ T cells stained (right). Repeating this experiment using a short-term T cell line expanded with JEV peptides gave similar results. (I) Neutralizing antibodies against JEV and all four DENV serotypes were measured by PRNT50. The bars depict the median. Four subjects had some assays repeated with similar results.
We then extended this analysis across the cohort using peptides of West Nile virus (WNV; a flavivirus closely related to JEV), DENV2, and E, NS1, NS3, and NS5 proteins from DENV1, 3, and 4 (see Materials and methods). Once library peptides were mapped, the same T cell lines were then tested against the equivalent peptides from DENV1–4 and WNV based on a ClustalW alignment of the library polyprotein sequence (an example is shown in Fig. 2 D). Responses to the variant peptides were normalized across different assays by dividing the result by the value for JEV peptides in the same assay, with a cross-reactivity index of 1 indicating an equal response to JEV and variant peptides. In five subjects, cross-reactive responses tested both ex vivo and on T cell lines showed good agreement (Fig. 2 E). Next, we studied 10 healthy JEV-exposed donors in whom 15 epitopes were mapped. In all but three, responses were highly cross-reactive (Fig. 2 F) and were not significantly different from the hypothetical value of 1 (indicating equivalent responses) by a Wilcoxon signed rank test. 8 out of the 10 donors showed responses that recognized peptides from at least two other flaviviruses (often more) as efficiently as JEV.
Cross-reactivity was confirmed by dual tetramer staining between the JEV epitope and three variant epitopes from WNV, DENV1, and DENV2/3 (Fig. 2 G), at least in one individual. Cross-reactivity occurred at the single cell level with apparently equal affinity (Fig. 2 H). Finally, to determine past exposure to DENV, PRNT50 to DENV1–4 was measured in those subjects who had cross-reactivity assays performed; the cohort was extensively DENV exposed (Fig. 2 I).
Overall, these experiments confirmed clear JEV-specific T cell memory responses in JEV NAb-positive subjects from a JE-endemic area. These responses were predominantly CD8+, highly cross-reactive among flaviviruses, and mostly targeted NS proteins, including in donors with clear DENV exposure.
CD8+ T cell responses in healthy JEV-exposed subjects are polyfunctional and show a cytotoxic phenotype
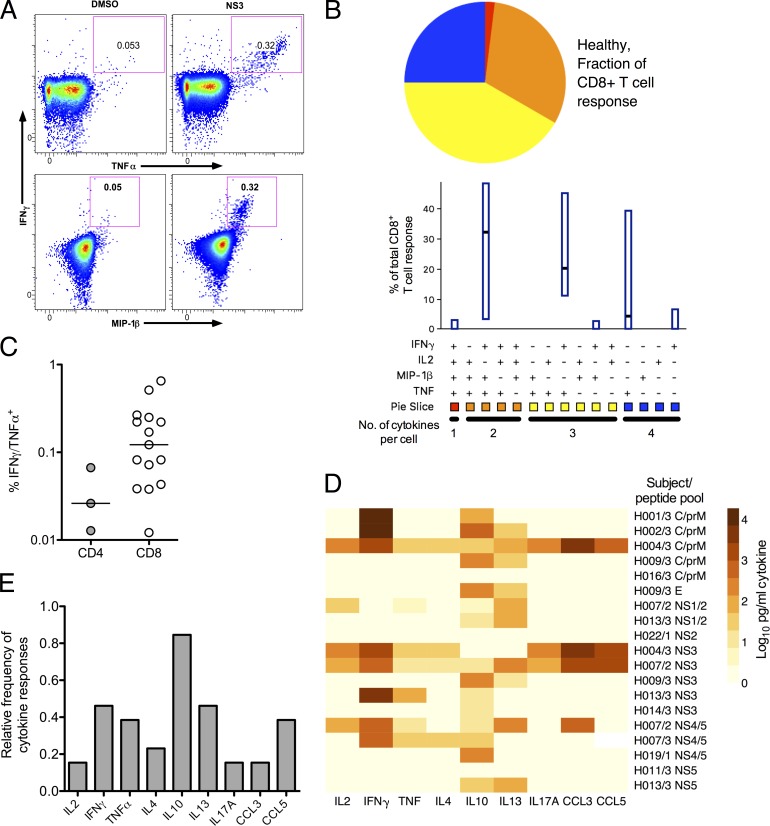
We next addressed the functionality of these responses. We asked whether cytokines were cosecreted by JEV-specific T cells using WB ICS assays; example flow cytometry data are shown in Fig. 3 A. The CD8+ T cell response was dominated by IFN-γ+/TNF-α+/MIP-1β+ and IFN-γ+/TNF-α+ cells (Fig. 3 B). Overall, 75% of the responding cells made more than one cytokine. The small number of CD4+ T cell responses precluded this analysis for this population; CD4+ T cell responses were smaller as well as less frequently detected (Fig. 3 C).
Figure 3.
Function of T cell responses to peptide pools of JEV in healthy JEV-exposed donors. (A) WB ICS assays were used to measure cytokine+ cells from 11 healthy JEV-exposed donors. Example data from a WB ICS experiment (donor H014/3) are shown. Axes are log10 fluorescence units. (Top) IFN-γ versus TNF-α. (Bottom) IFN-γ versus MIP-1β. (B) WB ICS data were analyzed using SPICE software for 10 donors; 1 donor did not have the full flow cytometry panel performed. The bars indicate median and interquartile range (IQR). Pie slices indicate the fraction of response of the group that expresses cytokines four, three, two, and one in any combination. (C) Magnitude of CD4+ and CD8+ WB ICS IFN-γ+/TNF-α+ responses in healthy JEV-exposed donors measured by ICS (n = 11, 15 CD8+ responses and 3 CD4+ responses). (D) Secreted cytokine concentrations were measured by bead array from 12 healthy JEV-exposed donors after 5 d of stimulation with JEV peptide pools. The heat map depicts log10-transformed picograms/milliliter. Some subjects had more than one peptide pool assayed. Data are depicted after subtraction of values obtained from unstimulated cells. This experiment was performed once. (E) Relative frequency of secreted cytokine responses from the same data as in D after 2 and 5 d of stimulation with JEV peptide pools. Subjects were considered positive if any pool was positive at either time point.
To address the production of other cytokines, bead array assays were performed in a subset of 13 donors on day 2 and day 5 supernatants from proliferation assays showing the largest responses, which were often different pools from the ex vivo IFN-γ assays (Fig. 3 D, data for day 5). This showed some evidence of more diverse cellular responses with release of IL-10 (the cytokine most frequently detected in these assays) in 11 (85%) of the donors (Fig. 3, D and E); IL-4 and IL-13 were also detected, but in much smaller amounts and less frequently. Other T cell–derived cytokines and chemokines were detected infrequently; five subjects (39%) made either CCL3 or CCL5 responses, and only two subjects (15%) made IL-17A (Fig. 3 E).
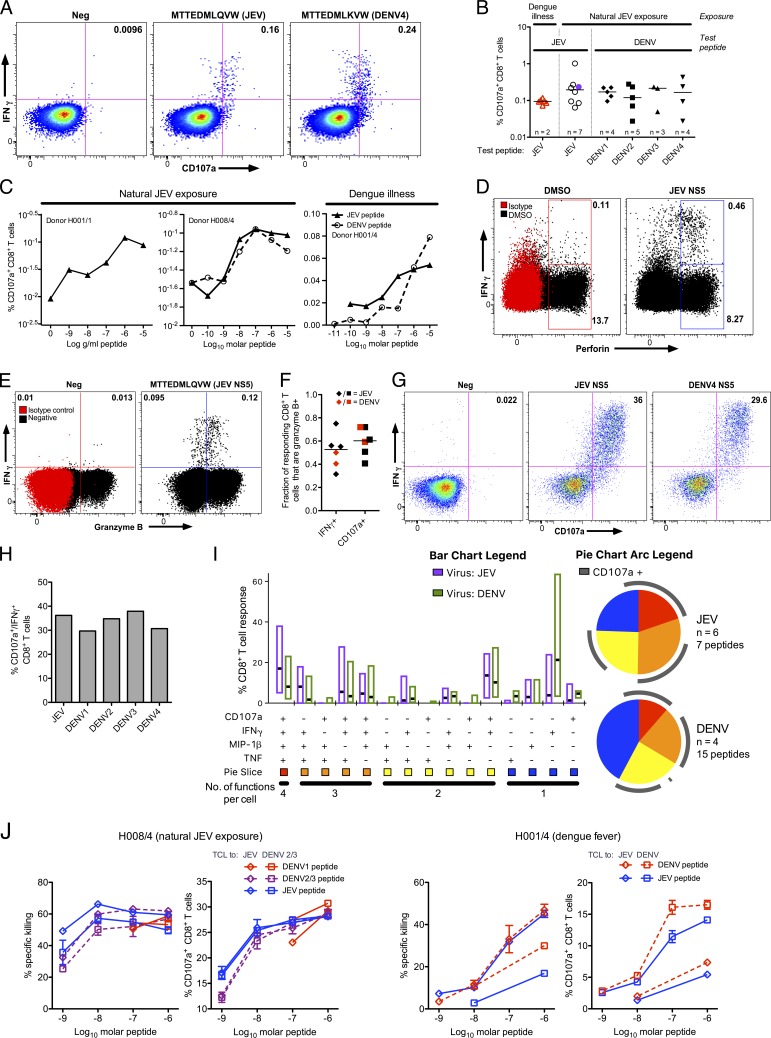
In selected individuals, where resampling was possible after CD8+ T cell responses were mapped, the cytotoxic capacity of these cells was studied in more detail. JEV-specific CD8+ T cell responses degranulated in response to peptide stimulation (Fig. 4 A), and degranulation responses were cross-reactive (Fig. 4, A–C). Responses were detectable to at least 10 nM of peptide ex vivo, indicating that they are likely to be functional in vivo (Fig. 4 C). Responding cells expressed perforin and granzyme B (Fig. 4, D–F) consistent with killing capacity. To confirm these results, short-term T cell lines were expanded to responding peptides; these, too, degranulated (Fig. 4 G) and were cross-reactive between JEV and DENV (Fig. 4 H). Cross-reactive degranulation responses were equivalent in overall functionality between JEV and DENV, with 55–60% of responding cells degranulating in both cases (Fig. 4 I). Finally, cross-reactive CD8+ T cell lines were able to kill peptide-loaded cells, whether expanded to JEV or DENV peptides (Fig. 4 J).
Figure 4.
CD8+ T cell responses from JEV-exposed donors show a cytotoxic phenotype. (A) Degranulation of CD8+ T cells in response to JEV and DENV variant peptides was measured by CD107a staining/flow cytometry. Representative data are from one of two experiments in a healthy JEV-exposed donor (H008/4). (B) Degranulation responses to variant flavivirus peptides were measured as in A in naturally JEV-exposed subjects (n = 7; six healthy and one recovered JE patient) and subjects reporting dengue illness (n = 2). Dengue illness and recovered JE are indicated by open, red triangles and purple circles, respectively. (C) Degranulation responses were measured as in A to titrations of JEV and DENV variant peptides in two healthy JEV-exposed donors (left, H001/1; middle, H008/4) and a subject reporting dengue illness (right, H001/4). (D and E) JEV-specific CD8+ T cells were identified by IFN-γ ICS and costained for perforin (D) and granzyme B (E). Representative data from one (perforin, JE054/2; granzyme B, H008/4) of four subjects are shown. (F) IFN-γ/granzyme B+ and CD107a/granzyme B+ double-positive CD8+ T cells were detected in the same experiments as in D and E in one healthy JEV-exposed and two dengue-exposed donors in response to all peptides tested: four JEV and two DENV peptides. (G) Degranulation of a short-term T cell line from a healthy JEV-exposed donor (H007/2) in response to JEV and DENV variant peptides measured as in A and representative of five donors tested. (H) Data from the same experiment as in G showing cross-reactivity for DENV peptides. Cross-reactivity was observed in two independent experiments, and one included CD107a staining. (I) CD8+ T cell IFN-γ, TNF-α, MIP-1β, and CD107a responses were measured by ICS and analyzed using SPICE. Six healthy JEV-exposed donors (seven JEV responses), four of whom had variant DENV peptides tested (15 DENV peptides), were included. In two subjects, repeat experiments showed similar results. The bars indicate median and IQR, and pie slices indicate the fraction of response showing four, three, two, and one function in any combination. (J) Peptide-pulsed, CFSE-labeled, HLA-matched targets were incubated with CD8+ T cell line effector cells, and the percent specific killing was measured by flow cytometry in response to JEV and DENV peptides. The peptides used were: donor H001/4 (DVMCHATL [JEV] and DLMCHATF [DENV]) and donor H008/4 (MTTEDMLQVW [JEV], MTTEDMLSVW [DENV1], and MTTEDMLTVW [DENV2/3]). Diamonds indicate lines expanded with JEV peptide, and squares indicate DENV peptide. Assays were performed in duplicate for each T cell line/peptide pair. Error bars represent standard error of the mean.
Screening for T cell responses in convalescent JE patients
We next addressed T cell responses in recovered JE patients. 39 recovered JE patients were screened by ELISPOT or WB ICS; 28 (72%) showed ex vivo IFN-γ responses. Sample flow cytometry data are shown in Fig. 5 A. Ex vivo IFN-γ responses targeted all JEV proteins (Fig. 5 B), with slightly more responses against the C/prM and NS1/NS2 pools. Around 80% of the IFN-γ response came from CD4+ T cells (Fig. 5 C), and several donors showed exclusively CD4+ responses. Proliferative responses were similarly distributed among JEV proteins (Fig. 5, D and E). Responding peptides were mapped using short-term T cell lines using the same strategy as the healthy JEV-exposed donors, and 14 peptides from seven recovered patients were tested in cross-reactivity assays (Fig. 5 F). Mapped responses in recovered JE patients were mostly JEV specific and showed a cross-reactivity index significantly different from the hypothetical value of 1 (indicating equivalent responses) by Wilcoxon signed rank test in all cases except DENV1, which approached significance despite fewer variant peptides tested. Importantly, NAbs to DENV were present in all the recovered JE patients with peptides mapped and cross-reactivity assays performed (Fig. 5 G and Table 1), suggesting the lack of cross-reactivity in recovered JE patients is not a result of lack of DENV exposure. In the same subset, JEV NAb titers were also tested and showed no difference between good outcome and poor groups (Fig. 5 H).
Figure 5.
Breadth of the T cell response to JEV peptide pools in recovered JE patients. (A) WB ICS assays were used to measure cytokine+ cells in recovered JE patients. Example flow cytometry data from four patients are shown. Axes are log10 fluorescence units. (Top) CD8+ responses, IFN-γ versus TNF-α (left) and IFN-γ versus MIP-1β (right). (Bottom) CD4+ responses, IFN-γ versus TNF-α (left) and IL2 versus TNF-α (right). (B) Ex vivo IFN-γ responses from 39 recovered JE patients were measured by ELISPOT (n = 17) or ICS (n = 22), and relative frequency of responses to each peptide pool was calculated. (C) The relative proportion of the cytokine response produced by CD4+ T cells in individual patients was measured by ICS in the same experiments as in A and B. The bar depicts the median. (D) Proliferation responses were measured by CFSE dilution and flow cytometry in 18 recovered JE patients of whom 15 showed responses. The relative frequency for CD4+ T cells and CD8+ T cells is shown. (E) Proliferative responses from the same experiments as in D were equal in size between good outcome and poor outcome for CD4+ cells (25 responses in eight patients with good outcome and 13 responses in four patients with poor outcome; Mann Whitney U test); CD8+ responses were larger in poor outcome (23 responses in eight patients with good outcome and 17 responses in six patients with poor outcome). The bars depict the median and IQR, whiskers are 1.5 × IQR, and outliers are shown as points. (F) Cross-reactivity of short-term T cell lines expanded with JEV peptides with DENV1–4 and WNV were measured by ICS. Data are cross-reactivity indices (as in Fig. 2 E) of 14 T cell responses identified in seven recovered JE patients. Wilcoxon signed rank tests showed significant differences from a hypothetical value of 1 (i.e., the response to JEV and variant peptides was not equivalent) for all viruses except DENV1. Number of peptides tested: WNV = 13; DENV1 = 9; DENV2 = 14; DENV3 = 11; DENV4 = 11. (G) NAbs against JEV and all four DENV serotypes were measured by PRNT50. The bars depict the median. Assays were generally performed once per person, and nine subjects had some assays repeated with similar results. (H) JEV PRNT50 were the same in JE patients with good (n = 7) and poor (n = 9) outcome (Mann Whitney U test). The bars depict the median and IQR. C, core protein. E, envelope protein.
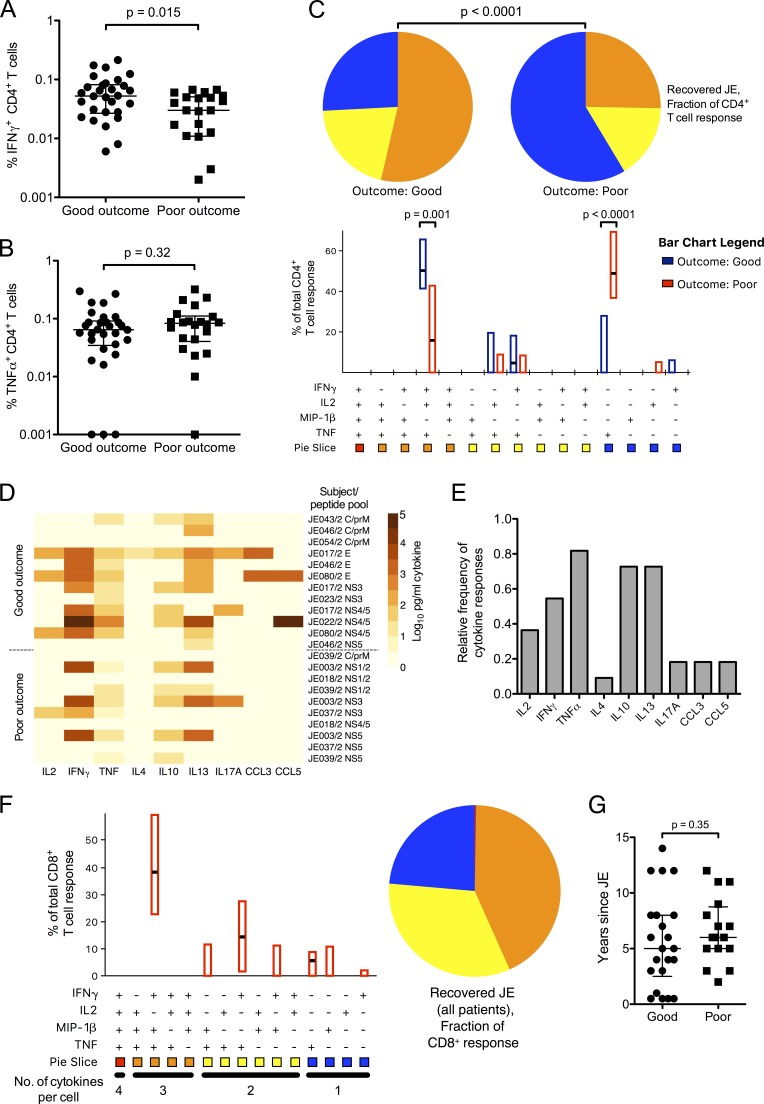
A polyfunctional, IFN-γ–dominated CD4+ T cell response correlates with better clinical outcome in convalescent JE patients
JE patients were categorized into good outcome (complete recovery with no neurological deficit; Liverpool outcome score of 5) or poor outcome (incomplete recovery; Liverpool outcome score of 2–4 or abnormal neurological examination; see Materials and methods). There was no difference in targeting of viral proteins or the proportion of the response expressed by CD4+ T cells when analyzed by outcome (unpublished data). IFN-γ ICS responses, however, were larger in patients with a good outcome (Fig. 6 A). TNF-α production was equivalent between the two groups (Fig. 6 B) but was derived mostly from TNF-α single positive cells in those with poor outcome (Fig. 6 C). Complete recovery was strongly associated with a polyfunctional IFN-γ+/IL2+/TNF-α+ CD4+ T cell response, indicating that a poor quality memory response, which is proinflammatory but lacking in antiviral/helper capacity, is associated with recovery from JE with long-term neurological damage.
Figure 6.
T cell cytokine responses to JEV peptide pools in recovered JE patients. (A–C) Recovered JE patients showing CD4+ T cell responses were categorized into good outcome (complete recovery; n = 16) and poor outcome (recovery with residual neurological deficit/disability >6 mo after disease; n = 11). Cytokine responses were measured by WB ICS once per subject. 30 responses were detected in the good outcome group and 22 responses in the poor outcome group. (A) IFN-γ responses were smaller in those with poor outcome (Mann Whitey U test). (B) TNF-α responses showed no difference with outcome (Mann Whitey U test). (C) CD4+ T cell responses were analyzed using SPICE according to the outcome from JE. Patients with poor outcome had fewer polyfunctional cells (SPICE pie comparison, 10,000 replicates), with the difference largely accounted for by fewer IFN-γ+/TNF-α+/IL2+ cells (P = 0.001 for a Mann Whitney U test) and more TNF-α+ cells (P < 0.0001). The data were analyzed for individual responses; summing by subjects retained the strong significance of the result. (D) Secreted cytokine concentrations were measured by bead array from 11 recovered JE patients after 5 d of stimulation with JEV peptide pools. The heat map depicts log10-transformed picograms/milliliter. Some subjects had more than one peptide pool assayed. Data are depicted after subtraction of values from unstimulated cells and are displayed according to outcome from JE. (Top) Good outcome, n = 7. (Bottom) Poor outcome, n = 4. This experiment was performed once. C, core protein. prM, pre-membrane protein. E, envelope protein. NS, non-structural protein. (E) Relative frequency of secreted cytokine responses from the same experiments as in D after 2 and 5 d of stimulation with JEV peptide pools. Subjects were considered positive if any pool was positive at either time point. (F) CD8+ T cell responses from recovered JE patients were assayed once per subject by WB ICS (n = 14; 18 responses) and analyzed using SPICE. (G) There was no difference in years since admission with clinical JE and sampling for this study between the good and poor outcome groups (Mann Whitney U test). Good outcome, n = 22. Poor outcome, n = 16.
To explore cytokine production further, in a subgroup of 11 patients, secreted cytokine responses were assayed from the day 2 and day 5 supernatants of proliferation assays as for the healthy JEV-exposed group (Fig. 6 D, data for day 5). In these assays, across both time points, IL-10 and IL-13 were detected in several patients from both groups (Fig. 6, D and E), and proinflammatory TNF-α responses were frequent. Similar to the healthy group, other T cell–derived cytokine and chemokine responses were rare. Three subjects made CCL3 or CCL5 responses, and three made IL-17A. Looking specifically at CD8+ T cell responses in recovered JE patients by WB ICS, these were mostly polyfunctional and characterized by IFN-γ+/TNF-α+/MIP-1β+ and IFN-γ+/TNF-α+ cells (Fig. 6 F).
Importantly, all the patients were sampled at least 6 mo after acute illness, by which time most recovery is expected to have occurred. Time from illness to sampling was the same in both groups (Fig. 6 G), indicating that this variable did not confound the results. Interestingly, the observed defect in CD4+ T cell cytokine production was not apparent in proliferative responses, which were equal in magnitude between the two groups; CD8+ proliferative responses were slightly larger in those with poor outcome (Fig. 5 E). Overall, these results indicated that T cell responses in patients who have recovered from JE were predominantly CD4+ and JEV specific. Smaller IFN-γ responses in conjunction with poor quality, proinflammatory (TNF-α+) CD4+ responses were associated with a poor outcome.
T cell responses are differentially targeted in JE patients and healthy donors
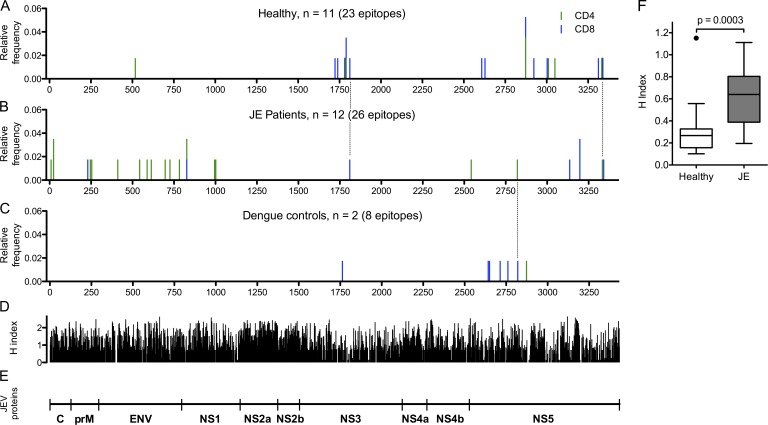
During screening, ex vivo IFN-γ responses in healthy JEV-exposed donors were directed mostly at NS3, NS4, and NS5 (Fig. 1 A), whereas recovered JE patients responded to all JEV proteins (Fig. 5 B). Mapped peptides were plotted according to their location in the JEV polyprotein for healthy, JEV-exposed donors (Fig. 7 A), recovered JE patients (Fig. 7 B), and two UK participants reporting dengue illness (Fig. 7 C). Sequences of all the JEV peptides identified are in Table 2, and the variant peptides showing responses in cross-reactivity assays are shown in Table 3. Almost all the mapped responses in structural proteins were identified in JE patients, and one healthy JEV-exposed donor showed a CD4+ response to the E protein of JEV (Fig. 7, A and B; and Table 2). In contrast, the responses in healthy JEV-exposed donors were focused on NS3 and NS5, and only one peptide was successfully mapped in NS3 in a JE patient. This was despite multiple attempts at expanding T cell lines to NS3 in vitro, which failed to produce responses in all except one individual. The two subjects reporting dengue illness showed similar findings to the healthy JEV-exposed donors (Fig. 7 C). The mean Shannon entropy (H index) of Indian isolates of JEV and DENV corresponding to the position of JEV peptides eliciting T cell responses in recovered JE patients was significantly greater than in healthy JEV-exposed donors (P = 0.0003; Fig. 7 F), indicating that heathy donors target conserved regions of the flavivirus polyprotein.
Figure 7.
Peptide epitopes of JEV identified in healthy JEV-exposed donors, recovered JE patients, and DENV-exposed donors. (A–C) Peptides were identified in ICS assays of short-term T cell lines expanded with peptide pools showing ex vivo responses or by ex vivo ELISPOT. Both peptides that have been mapped down to the minimal epitope (eight peptides) and library peptides of 15–18 amino acids are shown. CD4+ responses are green, and CD8+ responses are blue. Location of peptide responses in healthy JEV-exposed donors (A), recovered JE patients (B), and two subjects reporting dengue illness (C) are shown. Three peptides in common between JE patients and either healthy group are marked by dotted lines. (D) Shannon entropy (H index) was calculated using all the available DENV and JEV complete polyprotein sequences from India (13 DENV1 sequences, 10 DENV2 sequences, 8 DENV3 sequences, 5 DENV4 sequences, and 8 JEV sequences) and the sequences of the DENV1–4 and JEV peptide libraries used in this study. The H index varies from 0, corresponding to a single conserved amino acid residue at that position, to 4.322, where all 20 amino acids are represented equally. (E) Schematic representation of JEV proteins corresponding to their size. C, core protein. prM, pre-membrane protein. ENV, envelope protein. NS, non-structural protein. (F) The mean Shannon entropy of the flavivirus regions corresponding to 15 unique peptides identified in the healthy JEV-exposed group and 25 unique peptides identified in the recovered JE group (using the same virus polyprotein sequences as in D) was significantly lower in the epitopes identified from recovered JE patients (Mann Whitney U test). The bars depict the median and IQR, whiskers are 1.5 × IQR, and outliers are shown as points.
Table 2. JEV peptides eliciting T cell responses identified in this study.
| Subject | Peptide | Subset | JEV protein | Amino acid location | Type | HLA restriction | HLA class I | HLA class II | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | B | C | DR | |||||||||
| JE082/2 | GKNRAINMLKRGLPRVF | CD4 | C | 9–25 | JE | 11:01, 33:01 | 13:01, 44:02 | DR12, DR14, DR52 | ||||
| VFPLVGVKRVVMSLLDGR | CD4 | C | 24–41 | |||||||||
| JE093/2 | MLKRGLPRVFPLVGVKRVVMSLLDGR | CD4 | C | 16–41 | JE | 24:02, 24:02 | 13:01, 27:02 | DR15, DR51 | ||||
| JE043/2 | SLVNKKEAWLDSTKATRY | CD8 | M | 230–247 | JE | 01:01, 11:01 | 52:01, 57:01 | DR3, DR15, DR52, DR51 | ||||
| WIIRNPGYAFLAAVLGWM | CD4 | M | 254–271 | |||||||||
| JE101/2 | RYLMKTENWIIRNPGYAF | CD4 | M | 246–263 | JE | 30:01, 33:01 | 13:01, 44:02 | DR7, DR53 | ||||
| JE017/2 | AKFSCTSKAIGRTIQ | CD4 | E | 411–425 | JE | 03:01, 11:02 | 07:02, 35:01 | DR4, DR11, DR52, DR53 | ||||
| KQSVVALGSQEGGLHQAL | CD4 | E | 543–560 | |||||||||
| CRLKMDKLALKGTTYGMCTEKFSF | CD4 | E | 581–604 | |||||||||
| HGTVVIELSYSGSDGPCK | CD4 | E | 613–630 | |||||||||
| LGKAFSTTLKGAQRLAAL | CD4 | E | 697–714 | |||||||||
| WDFGSIGGVFNSIGKAVHQVFGGAF | CD4 | E | 719–735 | |||||||||
| GVLVFLATNVHADTGCAI | CD4 | E | 783–801 | |||||||||
| H021/1 | PWTSPSSTAWRNRELLM | CD4 | E | 518–534 | H | 02:01, 11:01 | 07:02, 52:01 | DR15, DR51 | ||||
| JE003/2 | KYLPETPRSLAKIVHK | CD4 + CD8 | NS1 | 827–842 | JE | 03:01, 33:01 | 15:01, 37:01 | DR10, DR15, DR51 | ||||
| AVHSDLSYWIESRYNDTWKLERAVF | CD4 | NS1 | 987–1011 | |||||||||
| WIESRYNDTWKLERAVFGEVKSCTW | CD4 | NS1 | 995–1019 | |||||||||
| JE001/2 | EAWVDRYKYLPETPRSLAKIVHK | CD4 | NS1 | 820–842 | JE | 11:01, 31:01 | 35:01, 52:21 | DR4, DR15, DR53, DR51 | ||||
| TYALNTFTNIAVQLVRLM | CD8 | NS5 | 3136–3153 | |||||||||
| H014/3 | TAVLAPTRVVAAEMAEAL | CD8 | NS3 | 1723–1740 | H | 03:01, 30:01 | 07:02, 13:01 | 06:02, 07:01 | ||||
| H001/1 | ALRGLPVRY | CD8 | NS3 | 1739–1747 | H | 11:01, 32:01 | 15:01, 55:01 | 01:02, 04:01 | ||||
| H001/4 | DVMCHATL | CD8 | NS3 | 1763–1770 | D | B*08:01 | 01:01, 03:01 | 08:01, 44:02 | 05:01, 07:01 | DR8, DR17, DR52 | ||
| H019/1 | NLFVMDEAHFTDPASI | CD8 | NS3 | 1784–1799 | H | 02:01 | 38:01, 40:06 | 07:01, 15:02 | ||||
| EAHFTDPASIAARGYI | CD8 | NS3 | 1790–1805 | |||||||||
| H013/3 | NLFVMDEAHFTDPASI | CD4 | NS3 | 1784–1799 | H | 02:01, 24:02 | 35:03, 40:06 | 02:21, 15:02 | ||||
| EAHFTDPASIAARGYI | CD4 + CD8 | NS3 | 1790–1805 | |||||||||
| GEAAAIFMT | CD8 | NS3 | 1812–1820 | |||||||||
| JE098/2 | ELGEAAAIFMTATPP | CD8 | NS3 | 1810–1824 | JE | 02:01 | 35:01, 40:06 | 04:01, 07:01 | ||||
| H005/1 | GCGRGGWSYYAATLKKV | CD8 | NS5 | 2608–2624 | H | 02:01, 30:01 | 13:01, 38:01 | 06:02, 12:02 | ||||
| RGYTKGGAGHEEPMLM | CD8 | NS5 | 2628–2643 | |||||||||
| H010/4 | LMQSYGWNL | CD8 | NS5 | 2642–2650 | D | 01:01, 02:01 | 08:01, 35:01 | 04:01, 07:01 | ||||
| LVSLKSGVDVFYKPS | CD8 | NS5 | 2650–2664 | |||||||||
| YMPKVIEKMEV | CD8 | NS5 | 2714–2724 | |||||||||
| NMTSQVLLGRMDRTVWR | CD8 | NS5 | 2761–2777 | |||||||||
| HPYRTWTY | CD8 | NS5 | 2827–2834 | B*35:01 | ||||||||
| TPFGQQRVF | CD4 + CD8 | NS5 | 2873–2884 | |||||||||
| JE080/2 | KLNAMSREEFFKYRREAIIEVDR | CD4 | NS5 | 2542–2564 | JE | 02:01, 33:01 | 15:05, 44:02 | DR7, DR53 | ||||
| TWHKDPEHPYRTWTYHGSYEVK | CD4 | NS5 | 2820–2841 | |||||||||
| H007/2 | TMAMTDTTPFGQQRVFK | CD4 + CD8 | NS5 | 2869–2885 | H | 01:01, 68:01 | 57:01 | DR53, DR7 | ||||
| H011/3 | TMAMTDTTPFGQQRVFK | CD8 | NS5 | 2869–2885 | H | 02:01, 68:01 | 37:01, 51:01 | 06:02, 15:02 | ||||
| H007/3 | TMAMTDTTPFGQQRVFK | CD4 + CD8 | NS5 | 2869–2885 | H | 11:01, 68:01 | 07:02, 52:02 | 07:01, 12:02 | ||||
| CTKEEFIKKVNSNAAL | CD8 | NS5 | 2923–2938 | |||||||||
| DIAGKQGGKMYADDTAGW | CD4 | NS5 | 3050–3067 | |||||||||
| H002/3 | IWFMWLGARY | CD8 | NS5 | 3003–3012 | H | 01:01, 29:01 | 07:05, 57:01 | 06:02, 15:04 | ||||
| RYLEFEALGFL | CD4 | NS5 | 3011–3021 | |||||||||
| JE054/2 | AISGDDCVVKPLDDRFATALHFL | CD8 | NS5 | 3191–3213 | JE | 11:01, 24:02 | 15:25, 51:01 | 07:01, 16:01 | ||||
| JE074/2 | CVVKPLDDRFATALHFL | CD8 | NS5 | 3197–3213 | JE | |||||||
| H008/4 | MTTEDMLQVW | CD4 + CD8 | NS5 | 3336–3345 | H | B*58:01 | 01:01, 33:03 | 58:01, 15:17 | 07:01, 03:04 | DR13, DR15, DR51, DR52 | ||
| CSAVPVDW | CD8 | NS5 | 3312–3319 | B*58:01 | ||||||||
| JE446/2 | GEWMTTEDMLQVWNRVWI | CD4 | NS5 | 3333–3350 | JE | 01:01, 33:01 | 52:01, 58:01 | DR14, DR15, DR52, DR51 | ||||
| MLQVWNRVWIEENEWMM | CD8 | NS5 | 3341–3357 |
JE, recovered JE patient; H, healthy JEV-exposed donor; D, dengue exposed. In total, 57 responses to 40 peptides (or adjacent pairs of peptides) were identified.
Table 3. Variant peptides eliciting T cell responses.
| JEV peptide sequence | Subject type | Viruses with identical sequence | Subset | Variant | Virus/BEI peptide location |
|---|---|---|---|---|---|
| CSAVPVDW | H | DENV1 | CD8 | CSSVPVDW | MVEV |
| MTTEDMLQVW | H | CD8 | MTTEDMLRVW | JEV | |
| MTTEDMLSVW | DENV1 | ||||
| MTTEDMLTVW | DENV2,3 | ||||
| MTTEDMLKVW | DENV4 | ||||
| MTTEDMLEVW | WNV, YFV | ||||
| MTTEDMLDVW | TBEV | ||||
| ALRGLPVRY | H | ZIKV | CD8 | ALRGLPIRY | WNV, DENV2,4 |
| NLFVMDEAHFTDPASIEAHFTDPASIAARGYI | H | DENV2,3 | CD8 | NYNMIIMDEAHFTDPA | DENV1 NS3 p49 |
| IMDEAHFTDPASIARRG | DENV1 NS3 p50 | ||||
| NYNLIIMDEAHFTDPASI | DENV2 NS3 p38 | ||||
| NYNLIIMDEAHFTDPA | DENV3 NS3 p48 | ||||
| PNYNLIVMDEAHFTD | DENV4 NS3 p49 | ||||
| LIVMDEAHFTDPSSVAA | DENV4 NS3 p50 | ||||
| AHFTDPSSVAARGYIST | DENV4 NS3 p51 | ||||
| TMAMTDTTPFGQQRVFK | H | WNV | CD4 + CD8 | MVTQIAMTDTTPFGQQRa | DENV1 NS5 p60 |
| AMTDTTPFGQQRVFKEKa | DENV2 NS5 p61 | ||||
| MVTQMAMTDTTPFGQQRa | DENV3 NS5 p60 | ||||
| MVTQLAMTDTTPFGQQRa | DENV4 NS5 p60 | ||||
| GEAAAIFMT | H | WNV, DENV1,3,4, ZIKV | CD8 | MGEAAAIFMTATPPGSVb | DENV1 NS3 p54, DENV3 NS3 p52 |
| VEMGEAAAIFMTATPPGb | DENV4 NS3 p54 | ||||
| TAVLAPTRVVAAEMAEAL | H | WNV | CD8 | NVRTLILAPTRVVASEM | DENV1 NS3 p38 |
| LRTLILAPTRVVAAEM | DENV2 NS3 p30 | ||||
| TLILAPTRVVAAEMEEA | DENV3 NS3 p38, DENV4 NS3 p39 | ||||
| DVMCHATL | D | WNV, MVEV, SLEV | CD8 | DLMCHATF | DENV1–4, ZIKV |
| DAMCHATL | YFV | ||||
| AVHSDLSYWIESRYNDTW | JE | CD4 | NLAIHSDLSYWIESRL | WNV NS1 p26 | |
| WIESRYNDTWKLERAVF | JE | CD4 | RLNDTWKLERAVLGEVK | WNV NS1 p28 | |
| IESEKNETWKLARASFI | DENV NS1 p36 | ||||
| KQSVVALGSQEGGLHQAL | JE | CD4 | HATKQSVIALGSQEGALH | WNV E p34 | |
| ALGSQEGALHQALAGAI | WNV E p35 | ||||
| VVVLGSQEGAMHTALTG | DENV1 E p44 | ||||
| HAKKQDVVVLGSQEGAMH | DENV2 E p34 | ||||
| VLGSQEGAMHTALTGA | DENV2 E p35 | ||||
| HAKKQEVVVLGSQEGAMH | DENV3 E p35, DENV1b | ||||
| VLGSQEGAMHTALTGA | DENV3 E p36, | ||||
| HAKRQDVTVLGSQEGAMH | DENV4 E p36 | ||||
| VLGSQEGAMHSALAGA | DENV4 E p37 | ||||
| HGTVVIELSYSGSDGPCK | JE | CD4 | HGTVVLELQYTGTDGPCK | WNV E p44 | |
| LGKAFSTTLKGAQRLAAL | JE | CD4 | HKSGSSIGKAFTTTLKGA | WNV E p55 | |
| KAFTTTLKGAQRLAAL | WNV E p56 | ||||
| WDFGSIGGVFNSIGKAV | JE | CD4 | WDFGSVGGVFTSVGKAVH | WNV E p59 | |
| GVFNSIGKAVHQVFGGAF | VFTSVGKAVHQVFGGAFR | WNV E p60 | |||
| AISGDDCVVKPLDDRFCVVKPLDDRFATALHFL | JE | CD4 + CD8 | AVSGDDCVVKPLDDRFA | WNV NS5 p91+92c | |
| VVKPLDDRFATSLHFLNA | |||||
| ISGDDCVVKPIDDRFAT | DENV1 NS5 p115+116c | ||||
| VVKPIDDRFATALTALN | |||||
| ISGDDCVVKPLDDRFA | DENV2 NS5 p116+117c | ||||
| CVVKPLDDRFASALTAL | |||||
| MAISGDDCVVKPIDDRF | DENV3 NS5 p115+116c | ||||
| DCVVKPIDDRFANALLA | |||||
| MAISGDDCVVKPLDERF | DENV4 NS5 p116+117c | ||||
| DCVVKPLDERFSTSLLF |
JE, recovered JE patient; H, healthy JEV exposed; D, dengue exposed; MVEV, Murray Valley encephalitis virus; YFV, yellow fever virus; TBEV, tick-borne encephalitis virus; SLEV, St. Louis encephalitis virus; ZIKV, Zika virus. Variant peptides were synthesized or obtained from BEI resources; and their location within each peptide set (see Materials and methods) is given in the last column. Bold/underlined amino acids represent differences from the JEV sequence.
Likely to represent conserved epitope DTTPFGQQR (see Fig. 2 D).
Identical viral sequences, but corresponding peptides vary slightly.
Pairs of peptides tested together in the same assay.
The absence of T cell responses in NS1 in the healthy JEV-exposed donors could not be explained by a lack of potential HLA-binding motifs. The HLArestrictor server (Erup Larsen et al., 2011) identified 122 HLA class I–binding motifs for the alleles ranked strong binder, with a predicted accuracy of >80%. A more limited analysis of the HLA class II DRB1 alleles 03:01, 04:01, 07:01, 11:01, and 15:01 using SYFPETHI (Rammensee et al., 1999) showed 197 15-mer peptides with scores ≥20.
Few peptides were identified in more than one individual despite overlap of HLA alleles across the cohort (Table 2). One exception was peptide TMAMTDTTPFGQQRVFK at position 2,869–2,885 of the viral polyprotein (NS5 amino acids 342–358), which showed CD8+ T cell responses in three healthy JEV-exposed donors and one DENV-exposed donor. This corresponds to a highly conserved 20–amino acid segment that is nearly identical across all the medically important mosquito-borne flaviviruses, visible as a trough in Shannon entropy (H index) in NS5 (Fig. 7 D). In our study, three healthy HLA A*68:01–positive JEV-exposed donors who recognized JEV NS5 342–358 also recognized DENV peptides that were truncated for the C-terminal three amino acids (Fig. 2 D), suggesting they were recognizing DTTPFGQQR, previously described as one of the most immunogenic DENV CD8+ T cell epitopes out of 408 identified (Weiskopf et al., 2013). In a fourth DENV-exposed, HLA B*35:01–positive subject (H010/4), the JEV NS5 342–358 response was mapped to TPFGQQRVF (Table 2), also previously shown to be presented by this allele (Rivino et al., 2013).
These data suggest that T cell responses from recovered JE patients target JEV-specific regions that are less conserved among flaviviruses (high H index). Healthy JEV-exposed donors made responses against conserved regions that were highly cross-reactive among different flaviviruses (Fig. 2 F).
DISCUSSION
In this study, we have presented data showing that, across the entire JEV polyprotein, memory T cell responses in healthy JEV-exposed donors are (1) predominantly directed against NS3, NS4, and NS5, (2) are CD8+, and (3) cross react extensively between flaviviruses. Recovered JE patients, on the other hand, mount mostly JEV-specific CD4+ T cell responses and target epitopes predominantly within the JEV structural proteins and the secreted protein NS1. Among the recovered JE patients, the quality of the ex vivo CD4+ T cell response was associated with outcome; in individuals with poor outcome, the presence of a significant TNF-α+-only CD4+ T cell population suggests a potential mechanism whereby T cells might contribute to damaging inflammation in JE. These data are compatible with the notion, supported by animal studies (Larena et al., 2011, 2013), that T cells may play a role both in protection and immunopathology in JE. In addition, this study describes the first polyfunctional T cell responses to JEV, the first minimal epitopes of JEV in humans, and the first demonstration of cytotoxic CD8+ T cell responses to JEV in naturally exposed individuals.
Antiviral T cells exhibiting multiple functions are well described after many viral infections and have been observed after infection or vaccination with several flaviviruses (Akondy et al., 2009; Piazza et al., 2010; Friberg et al., 2011b). In this study, we have described polyfunctional T cell responses to JEV in both CD4+ and CD8+ subsets and found that the quality of the CD4+ T cell response was the factor most strongly associated with complete recovery from JE. Evidence for the protective capacity of polyfunctional T cell responses in humans is largely correlative (Betts et al., 2006), as is the case here. On average, >50% of the responding cells expressed two or more cytokines, and the subjects in this study were sampled a median ≥6 yr after the viral encounter. Therefore, these responses likely reflect a long-lived stable memory population rather than newly responding or terminally differentiated single cytokine-positive cells (Seder et al., 2008).
In a subgroup of subjects, CD8+ T cell degranulation responses were studied. Sampling limitations meant we could not test these responses in the whole cohort, but in all the subjects tested, CD8+ T cells degranulated in response to JEV peptides in the 10-nM range. Degranulation and perforin and granzyme B expression correlate closely with the ability to kill (Betts et al., 2003; Pardo et al., 2004), which we have confirmed using peptide-loaded cells. These cytotoxic responses also cross react between JEV and DENV; in other words, a DENV-primed cell could in principle kill a JEV-infected target or vice versa. Killing of cells expressing pox virus–vectored JEV prM/E/NS1 proteins has previously been demonstrated for CD4+ T cell clones and CD8+ short-term T cell lines expanded from PBMCs from JE-vaccinated subjects, rather than natural JEV exposure (Aihara et al., 1998; Konishi et al., 1998). Although we have not shown killing of virally infected target cells (itself a surrogate marker of protection), the data presented here make it highly likely that these responses would be capable of killing JEV-infected cells. T cell killing of JEV-infected cells, to our knowledge, has only been shown for m and never for human cytotoxic T lymphocytes (Murali-Krishna et al., 1994).
Previous work in the same location found a higher frequency of responses to NS3 (Kumar et al., 2004a,b), likely accounted for by the time interval between exposure and testing. The patients in our study were sampled on average 6 yr after JE, whereas in the studies by Kumar et al. (2004b), they were sampled ~6 mo after disease. In the case of DENV, when patients are sampled 2 wk after illness, the frequency of responses to NS3 reported is as high as 90% (Duangchinda et al., 2010). In recovered JE patients, IFN-γ ELISPOT responses to NS3 were small (unpublished data), and repeated attempts at in vitro expansion of responding T cells to NS3 consistently failed to produce responses in IFN-γ/TNF-α ICS assays. Collectively, our results are consistent with earlier findings that JE patients had defective IFN-γ but preserved proliferation responses to JEV NS3 amino acids 193–324 (Kumar et al., 2004b). Now, we have also shown that reduction in IFN-γ is a general phenomenon associated with a poor outcome from JE and occurs in response to all JEV proteins, not only NS3, increasing the likelihood that this is a biologically relevant result. In addition, we have extended these findings to show that an unbalanced CD4+ T cell response characterized by proinflammatory TNF-α production, in the absence of sufficient additional helper/antiviral IL2 and IFN-γ, is strongly associated with a poor outcome from JE.
We observed a striking degree of cross-reactivity in the NS protein–specific short-term CD8+ T cell lines from healthy JEV-exposed donors. The mean Shannon entropy of sequences recognized by this group was low, indicating they were highly conserved. A recent study on dengue also found that the majority of conserved CD8+ T cell epitopes are found in the NS proteins (Weiskopf et al., 2015). Short-term T cell lines from some healthy donors in our study recognized peptides from flaviviruses for which the chance of exposure was extremely remote, such as tick-borne encephalitis virus and St. Louis encephalitis virus (unpublished data), and some sequences were in common with geographically diverse viruses such as Murray Valley encephalitis virus and Zika virus, suggesting that this phenomenon goes beyond simply exposure and is largely accounted for by selective targeting of conserved regions. The dominant responses in recovered JE patients, however, cross reacted much less with variant peptides from DENV and WNV. DENV NAb assays revealed this was not simply caused by a lack of exposure. CD4+ T cells dominated the responses in recovered JE patients, and this cannot account for the lack of cross-reactivity because CD4+ T cells are known to mount cross-reactive responses to flaviviruses (Kurane et al., 1991; Aihara et al., 1998). Although the cross-reactivity of T cell responses appeared to differ between healthy JEV-exposed donors and recovered patients, the retrospective nature of this study and the likely difference in time between exposure and sampling for these different groups limit our ability to assign a protective role to these responses.
Both serotype-specific and serotype cross-reactive T cell responses are readily detectable after a single DENV infection (Friberg et al., 2011a), and over subsequent DENV exposures, the response may be skewed toward more conserved epitopes (Weiskopf et al., 2013). The exposure to DENV and the highly cross-reactive T cell responses in the healthy JEV-exposed donors in this study are consistent with these findings and suggest this phenomenon is not restricted to DENV. Given the protective effect of DENV on JEV (Edelman et al., 1975), one possibility is that exposure to DENV primes JEV cross-reactive T cell responses. Although there are well described examples of pathology mediated by DENV cross-reactive T cells (Mongkolsapaya et al., 2003), not all studies on dengue have confirmed the relationship between cross-reactive T cells and disease (Friberg et al., 2011a), and some observations imply a protective role for cross-reactive memory T cells (Hatch et al., 2011; Weiskopf et al., 2013). Similarly, animal models of sequential T cell priming and challenge can show either impaired or enhanced protection (Welsh et al., 2010). JE pathogenesis is distinct from some other viral infections in that viraemia is typically very low and often clinically inapparent; therefore, an early, modestly sized, cross-reactive CD8+ T cell response in the periphery may prevent dissemination to the central nervous system and limit the virus to a compartment where it is not particularly pathogenic. The retrospective nature of this study means we are unable to address this question directly, but this could be the subject of future prospective studies.
There are several possible reasons for the limited flavivirus cross-reactive T cell responses seen in recovered JE patients, even years after disease and despite DENV exposure. There may be intrinsically pathogenic responses in the context of disease that are different from those in health, and age at exposure or variation in innate immunity may affect the character of responses. Also, the order of priming may affect the cross-reactivity of T cell responses. Support for this is found in epidemiological observations that DENV infection is at least partially protective against JE (Edelman et al., 1975; Libraty et al., 2002) and that JEV NAbs (but not vaccination) has a detrimental effect on DENV (Anderson et al., 2011). However, a weakness of our study (common to many) is that we cannot determine the order in which different flaviviruses were encountered and therefore cannot provide direct evidence that JEV priming modifies DENV responses. Future prospective studies could address this question and investigate differences in the T cell response as a potential mechanism.
Ex vivo responses in recovered JE patients against the structural proteins and the pool containing NS1 were frequently observed but were rare in the healthy JEV-exposed donors. It is hypothesized based on observations in DF that structural proteins and the secreted protein NS1 interact with B cells and elicit CD4+ responses, whereas the remaining NS proteins elicit mostly CD8+ T cell responses (Rivino et al., 2013), consistent with what we have seen in this study. The reasons underlying the absence of responses to NS1 in the healthy JEV-exposed donors are not clear, but previous work in this region identified T cell responses against NS1 in healthy children (Kumar et al., 2004a), so anti-NS1 responses are likely not intrinsically pathogenic.
Although the dominant responses mapped in recovered JE patients were directed against less conserved viral proteins, we nevertheless identified several IFN-γ responses against NS3, NS4, and NS5. One CD8+ T cell response from a recovered JE patient (JE098/2) targeted a conserved epitope (ELGEAAAIFMTATPP; NS3 306–320), which showed a cross-reactive response in subject H013/3 (these two subjects shared HLA 02:01 and B40:01, as well as being closely matched at B35:01 and B35:03; Table 2). This response was not further tested in JE098/2 (because of insufficient cells), but it is likely that this response would have been cross-reactive. Although the predominance of JEV-specific responses in recovered JE patients is likely explained by variation in amino acid sequences between JEV and DENV, in some cases, single amino acid changes (aside from anchor residues) had very profound effects on responses. For example, changing Ala at position 5 of GEAAAIFMT to Gly completely abolished CD8+ T cell recognition in donor H013/3. In donor JE054/2, changing Leu to Ile (a very modest alteration) at position 12 of peptide AISGDDCVVKPLDDRF made no difference, whereas changing Asp to Glu (also similar amino acids) at position 14 eliminated the CD8+ T cell response. On the other hand, two amino acid changes from the HLA B*08:01–restricted epitope DLMCHATF (DENV) to DVMCHATL (JEV) gave rise to identical CD8+ T cell responses in donor H001/4 (who reported dengue illness), and a wide range of single amino acid variants from Gln at position 8 of the HLA B*58:01–restricted epitope MTTEDMLQVW (to Ser, Thr, Asp, or Glu) were also recognized by CD8+ T cells in donor H008/4. Therefore, the number of amino acid variants alone cannot explain the variation in cross-reactive responses seen here, and the reasons for this remain to be determined.
A limitation of our study is that the healthy JEV-exposed donors were older than the recovered JE patients. Practically, it is difficult to draw blood from children who have not been ill in this setting. Nevertheless, both groups were well matched for viral exposure and likely were exposed to JEV years earlier with ongoing exposure year to year, and we are not drawing direct comparisons between the two groups. Our within-group comparison of the outcome from JE does not suffer from this limitation. Ideally, the two groups should be matched for the time elapsed since first exposure to JEV, although within groups, we could not observe a signal indicating a change in responsiveness of cross-reactivity over time/age (unpublished data). The optimum way to address this question would be using a prospective study design, but given the high rate of asymptomatic JEV infection, relatively low incidence of JE, and existence of many other conditions that are hard to differentiate from JE, such a study would be impractical.
A second shortcoming is the small number of subjects with peptide mapping and cross-reactive T cell response data, despite the screening of larger numbers. Although the clinical setting introduces some limitations, our data demonstrate a clear lack of cross-reactive responses in recovered JE patients. A technical concern of our cross-reactivity experiments was that these were not conducted on optimally defined peptides, as optimal T cell epitopes located within a larger peptide can affect detection of the response (Draenert et al., 2004). The need to deconvolute the peptide pools in quick succession using single T cell lines, making maximal use of scarce samples, precluded minimal epitope mapping and HLA restriction in each case. Despite this, cross-reactive responses were readily detectable using library peptides. Because of the closed-ended nature of the HLA class I peptide-binding groove, this would reduce detection of cross-reactive CD8+ T cell responses more than CD4+, the converse of what we have observed. Lastly, we justified the use of short-term T cell lines by showing in a subset of donors that cross-reactive responses ex vivo correlated well with cross-reactive responses in T cell lines.
In summary, using the first full-breadth analysis of T cell responses to JEV in humans, we have demonstrated that a high quality IFN-γ–dominated CD4+ T cell response is associated with good recovery from JE in humans. JEV T cell responses are dominated by broadly cross-reactive CD8+ T cells in most healthy JEV-exposed donors. Cross-reactive responses in recovered JE patients, however, are much less frequent, though uncertainty remains regarding the true significance of this finding. In much of South and Southeast Asia, where JEV and DENV cocirculate, the effect of JEV immune responses could be significant in subsequent natural infection and in response to vaccines. Therefore, the sequence and timing of flavivirus infections and their effect on immunopathogenesis and potentially vaccine responses are worthy of further study. In the context of the current outbreak of Zika virus, this cross-reactivity at the T cell level, as we have identified here, could be of relevance to disease pathogenesis.
MATERIALS AND METHODS
Setting
The study was conducted at the Indian Institute of Science (IISc) and National Institute of Mental Health and Neurosciences (NIMHANS) in Bangalore and at VIMS, Bellary (all in Karanataka State, India). Karnataka is endemic for JE; the disease occurs in rural areas but is uncommon in districts around Bangalore. Recovered JE patients were therefore recruited at the VIMS medical center. For most experiments, blood samples were transported overnight by train for processing in a laboratory in Bangalore.
Study subjects
Healthy donors with no history of neurological illness were recruited at IISc, NIMHANS, and VIMS. Recovered JE patients were recruited at dedicated outpatient clinics held at the VIMS medical center. Healthy donors were taken from family members of patients and other members of the local community. JE patients were drawn from previous cohorts studied in this location (Lewthwaite et al., 2010b,c) and recruited based on hospital records of positive CSF JEV IgM samples and follow-up of acute JE cases recruited during the study period (October 2011 to March 2013). All recovered JE patients had been admitted to the VIMS medical center with a clinical illness compatible with encephalitis (fever plus one of the following: clouding of consciousness, seizures, or focal neurological signs) and a positive ELISA for JEV IgM in CSF. Patients had been admitted a median of 6 yr earlier (mean of 5.98 and range of 6 mo to 14 yr). A small cohort was also recruited in the UK. Age, sex, travel history/previous area of residence, flavivirus vaccine history, and medical history (focused on flavivirus illness) were collected from all participants. Recovered JE patients underwent detailed neurological examination and disability assessment using the Liverpool outcome score (Lewthwaite et al., 2010a).
Ethics statement
This study was performed in accordance with the principles of the declaration of Helsinki. The experimental studies in India were approved by the IISc Institutional Human Ethics Committee (ref 5/2011). All Indian studies were also approved by the Liverpool School of Tropical Medicine research ethics committee (ref 10.59). The component of the study conducted at the VIMS medical center was further reviewed by the VIMS ethics committee. The UK setup phase of the study was approved by the National Research Ethics Committees Northwest 7 (ref 10/H1008/23). All participants gave informed consent, or their parent/guardian gave consent if aged under 18. Minors age 12–18 were able to give assent in addition to the guardian’s consent if they wished.
Peptide libraries
All available JEV complete genome sequences in the NCBI nucleotide database were downloaded in April 2010. Open reading frames were translated and aligned, and a consensus amino acid sequence was generated using Se-AL carbon (v2.0a11). Because only JEV genotype III was identified as circulating in South India until April 2010, the sequence used for the JEV peptide library was the consensus sequence of JEV genotype III. However, genotype III sequences predominated in the database, and a consensus sequence generated using all the available sequences was identical. The JEV consensus sequence was used to generate a library of peptides 18 amino acids long overlapping by 10. In addition, if the C-terminal amino acid residue of the peptide was not an HLA class I anchor residue, the peptide was truncated until an “allowed” residue was reached or until a length of 15 amino acids (the overlap was kept constant at 10) to improve the sensitivity for CD8+ responses (Draenert et al., 2003, 2004). Peptides were synthesized by Mimotopes Ltd. as a PepSet (Table S1). Peptides were dissolved in DMSO and pooled as indicated. The following peptide sets were obtained through Biodefense and Emerging Infection Resources, the National Institute of Allergy and Infectious Diseases, and the National Institutes of Health: DENV1 Singapore/S275/1990 E (NR4551), NS1 (NR2751), NS3 (NR2752), NS5 (NR4203); DENV2 New Guinea C (NGC), C (NR505), prM (NR506), E (NR507), NS1 (NR508), NS2a (NR2747), NS2b (NR2748), NS3 (NR509), NS4a (NR2749), NS4b (NR2750), NS5 (NR2746); DENV3 Sleman 1978 E (NR511), DENV3 Philippines/H87/1956 NS1 (NR2753), NS3 (NR2754), NS5 (NR4204); DENV4 Dominica/814669/1981 E (NR512), DENV4 Singapore/8976/1995 NS1 (NR2755), NS3 (NR2756), NS5 (NR4205); WNV NY99-flamingo382-99 C (NR432), prM (NR433), M (NR434), E (NR435), NS1 (NR436), NS2a (NR437), NS2b (NR438), NS3 (NR439), NS4a (NR440), NS4b (NR441), and NS5 (NR442). Biodefense and Emerging Infection Resources peptides were dissolved using the same strategy as for JEV.
IFN-γ ELISPOT assay
ELISPOT assays were performed using anti–human IFN-γ capture and biotinylated detection antibodies from Mabtech according to the manufacturer’s instructions. Development was by detection with goat antibiotin-HRP and nitro-blue tetrazolium/BCIP 1-Step substrate (Thermo Fisher Scientific). PBMCs were plated at 2 × 105 per well and incubated overnight in 100 µl RPMI supplemented with 2 mM l-glutamine, 100 U/ml penicillin, and 0.1 mg/ml streptomycin with 10% FCS (R10) and peptides at a final concentration of 3–6 µg/ml. The positive control was concanavalin A at a final concentration of 5 µg/ml, and the negative control was DMSO at an equivalent concentration to the peptide pools.
ELISPOT assays were considered positive if there were at least 50 spot-forming cells/106 PBMC and double the background value. In cases close to the cut-off value, confirmation by another assay or mapping to a peptide was used. In seven JEV antibody–negative UK residents, no subject gave a response using these criteria, and studies on other members of the family Flaviviridae have used similar/identical thresholds (Appanna et al., 2007; Parsons et al., 2008; Barnes et al., 2012). Healthy donors were screened using 11 pools of peptides (one pool for C/prM, two for E, one each for NS1 and NS2, two for NS3, one for NS4, and three for NS5). Recovered JE patients, where available samples were smaller, were screened using six pools as indicated in Fig. 1 (A–C). For the healthy donors, responses to adjacent pools that were above the threshold were summed to create data comparative with the six pools used in recovered JE patients. Pools that did not reach the cut-off value were considered to be zero to prevent the creation of apparent responses by addition of two subthreshold values.
CFSE labeling and proliferation assays
PBMCs were used fresh or thawed and rested overnight. 8–10 million PBMCs were labeled with CFSE (Molecular Probes and Thermo Fisher Scientific) at a concentration of 1 µM in 1 ml of prewarmed PBS in the dark at 37°C for 10 min. Labeling was terminated by addition of five volumes of ice-cold R10 followed by incubation on ice for 3–4 min. Labeled cells were washed twice and then plated at 0.5 million cells per well in 250 µl streptomycin RPMI with 10% human serum (Valley Biomedical) and stimulated with peptide pools at a final concentration of 3 µg/ml. The positive control was concanavalin A at a final concentration of 2.5 µg/ml, and the negative control was DMSO at an equivalent concentration to the peptide pools. Cells were incubated for 8 d with removal and replacement of 75 µl of medium for cytokine assay after 48 h and 5 d. After incubation, the cells were stained with near infrared (IR) cell viability marker (Molecular Probes and Thermo Fisher Scientific), fixed in 2% formaldehyde in PBS, and then frozen at −80°C in 1% BSA and 10% DMSO in PBS until analysis by flow cytometry. Before analysis, the cells were stained with CD3 Alexa Fluor 700 (clone UCHT-1), CD4 PE (clone RPA-T4), CD8 APC (clone RPA-T8), and CD38 PE-Cy7 (clone HIT2; all from BD). Responses were considered positive if there were at least 1% of cells in the CD4+ or CD8+ gates were CFSElo/CD38hi and this value was at least double the background.
Ex vivo peptide stimulation and ICS
WB was stimulated with JEV peptide pools in the presence of 10 µg/ml brefeldin A (Sigma-Aldrich) and incubated for 6 h in a water bath at 37°C using the field-suitable method of Hanekom et al. (2004) that does not require a CO2 incubator. Power interruptions were frequent at VIMS, but the ambient temperature was 34–35°C, so the water bath maintained 36°C, within the normal range of human body temperature (Mackowiak et al., 1992). After 6 h, the stimulation was stopped by placing the tubes on ice followed by transport back to Bangalore the same night. Subjects studied in Bangalore followed an identical protocol with a 6-h stimulation terminated by placing the samples at 4°C overnight. Red blood cells were lysed the morning after. Freshly isolated PBMCs were stimulated with peptides overnight, and 10 µg/ml brefeldin A was added after 1 h. 25 ng/ml PMA and 250 ng/ml ionomycin (Sigma-Aldrich) served as the positive control; the negative control was the equivalent concentration of DMSO to the peptide pools. Cells stimulated by either method were stained with near IR cell viability marker and then fixed and cryopreserved pending analysis. Cells were permeabilized (Perm/Wash; BD) and stained with CD3 (clone UCHT-1), CD4 (clone RPA-T4), CD8 (clone RPA-T8), IFN-γ (clone 4S.B3), TNF-α (clone MAb11), IL2 (clone 5344.111), MIP-1β (clone D21-1351), and CD14 APC-Cy7 (clone MϕP9, dump channel). In some experiments, CD107a (clone H4A3) was added at the start of the stimulation to assess for degranulation (Betts et al., 2003). Anti–MIP-1β was from R&D Systems, and all other antibodies were from BD. Cells were acquired on a cytometer (Canto II; BD) with photomultiplier tube voltages set using a single batch of setup and tracking beads (BD) throughout. Data were analyzed using FlowJo (v8.8.6; Tree Star), and polyfunctional T cell distributions were analyzed and presented using Simplified Presentation of Incredibly Complex Evaluations (SPICE) software (v5.35), with preprocessing in Pestle (v1.7; Roederer et al., 2011). The SPICE statistical function compare pies was used for comparison of T cell function patterns (Fig. 6 C).
WB ICS responses were considered positive if the responding population was ≥0.02% of the parent gate and double the background (example data are given in Fig. 2 A). The background was subtracted before analysis.
Expansion of short-term T cell lines
Short-term T cell lines were expanded to JEV peptide pools or individual peptides showing ex vivo responses. Fresh or frozen rested PBMCs were stimulated with peptide pools at 3 µg/ml (up to a maximum of 1% DMSO). Where responding peptides were known, 10 µg/ml peptide was used to expand cells for cross-reactivity assays. Cells were cultured at two million cells per well in 24-well plates in 1 ml RPMI with 10% human serum, 10% natural T cell growth factor/IL2 (T-stim; Helvetica Health Care), and 20 ng/ml IL7 (R&D Systems) for 8–10 d. Before assay, the cells were rested overnight in R10 and then stimulated with peptides or pools of peptides for 6 h in the presence of brefeldin A. Cells were fixed, permeabilized, stained, and analyzed by flow cytometry by the same methods as ex vivo ICS.
Cytotoxicity assay
Cytotoxicity assays were conducted as previously described (Kurioka et al., 2015). In brief, appropriately HLA-matched peptide-loaded B cell lines were used as target cells and labeled with CFSE according to the manufacturer’s protocol. Cells were pulsed with peptide at the indicated concentrations for 1 h followed by three washes with R10. Nonpeptide-pulsed cells were labeled with Cell Trace violet (CTV; Molecular Probes). Pulsed and unpulsed cells were mixed in a 1:1 ratio and incubated with peptide-specific short-term T cell lines for 4 h at an effector/target ratio of 10:1 in duplicates. CD107a (clone H4A3) was added at the start of the stimulation. After incubation, cells were stained with near IR viability marker, CD3 (clone UCHT1; eBioscience), CD8 (clone RPA-T8; eBioscience), and CD19 (clone LT19; Miltenyi Biotec). The mean percent survival of CFSE-labeled cells in wells containing no effectors was used to calculate the expected frequency of target cells in each well: expected ratio (ER) was calculated as %CFSE+/%CTV+. The specific killing was then calculated as: % Specific killing = 100 × [(ER × %CTV+ cells) − %CFSE+ cells]/(ER × %CTV+ cells).
HLA typing
HLA typing was done using commercially available kits at the TTK Rotary blood bank(Bangalore, India) or the Churchill Hospital (Oxford, England, UK). Not all subjects in the cohort were HLA typed; typing focused on those subjects where peptides were mapped. HLA class I A and B and class II D-related typing was done on subjects who showed predominantly CD4 responses, and HLA class I A, B, and C typing was done on those showing predominantly CD8 responses.
HLA class I peptide tetramers
HLA class I peptide tetramers were supplied by the National Institutes of Health Tetramer Core Facility. JEV peptide tetramers were labeled with PE, and variant peptide tetramers were labeled with APC.
The reagents used were HLA B*58:01-MTTEDMLQVW-PE (JEV NS5 809–818), HLA B*58:01-MTTEDMLSVW-APC (DENV1 NS5 803–812), HLA B*58:01-MTTEDMLTVW-APC (DENV2/3 NS5 804–813), and HLA B*58:01-MTTEDMLEVW-APC (WNV NS5 808–817). Fresh viable PBMCs or T cell lines were stained for 30 min at 37°C before being stained with cell viability maker and other surface antibodies as described in the Ex vivo peptide stimulation and ICS section. Cells were fixed before analysis on a cytometer (LSR II; BD).
Bead array
Supernatants collected from proliferation assays on day 2 and day 5 were assayed using a human cytokine/chemokine kit (Bio-Plex 14-plex; Bio-Rad Laboratories) on the Bio-Plex 200 platform according to the manufacturer’s instructions. Supernatants were selected on the basis of the largest proliferation responses per subject. Responses were considered positive if the value secreted was at least double the amount detected from unstimulated cells, and the background was subtracted before analysis.
Viruses and cell lines
JEV 0423 (derived from SA14-14-2), DENV1 16007, DENV2 16681, DENV3 16562, DENV4 C0036/06 and C6/36, and LLC-MK2 cells for use in PRNT50 determination were supplied by the Armed Forces Research Institute of Medical Science virology department. JEV P20778, used for screening assays, and PS cells were grown in house at NIMHANS. LLC-MK2 cells were maintained in M199 medium, and C6/36 and PS cells were maintained in MEM. All media were supplemented with 2 mM l-glutamine, 10% FCS, and 100 U/0.1 mg/ml penicillin/streptomycin. LLC-MK2 cells and PS cells were maintained at 37°C with 5% CO2, and C6/36 cells were maintained at 28°C with 5% CO2. All viruses were passaged once before use in C6/36 cells, titered, and stored in aliquots at −80°C. Epstein-Barr virus–transformed HLA type B cell lines for HLA restriction experiments were provided by P. Goulder (University of Oxford, Oxford, England, UK).
PRNTs
The sera of healthy donors were screened for JEV NAbs by incubating twofold dilutions down to 1 in 16 of heat-inactivated sera with 100 PFUs of JEV P20778 for 1 h at 37°C. The mixtures were inoculated onto monolayers of PS cells with rocking at 37°C for 1 h to allow virus adsorption followed by removal of inoculum and addition of MEM with 2% FCS and 1% low melting point agarose. After 3 d, destruction of the monolayer was assessed by crystal violet staining after fixing with 10% formalin in saline for 30 min. PRNT50 were subsequently assayed in a subset by a standard method (Russell et al., 1967). In brief, heat-inactivated sera were diluted serially fourfold from 1 in 10 to 1 in 2,650, incubated with 30–50 PFUs of JEV or DENV1–4 at 37°C for 1 h, and then inoculated onto duplicate monolayers of LLC-MK2 cells with rocking at room temperature (25–28°C) for 1 h followed by removal of inoculum and addition of 1 ml phenol red-free overlay medium with 0.9% low melting point agarose. Plates were incubated for 4 d (JEV, DENV1, and DENV3), 6 d (DENV4), or 7 d (DENV2) before addition of a further 1 ml of overlay medium containing 4% neutral red. Plaques were visualized and counted after a further 24-h incubation. PRNT50 was calculated by probit regression.
Statistical and bioinformatic methods
All statistical analyses were done using R version 3.1.2. Differences between nonparametric unpaired variables were assessed with a two-sided Mann Whitney U (Wilcoxon rank sum) test. Categorical variables across groups were assessed by Fisher’s exact test. Correlations between ex vivo and in vitro expanded T cell responses were assessed by Spearman’s test. P-values <0.05 were considered significant. The Shannon entropy of aligned JEV and DENV polyprotein sequences was calculated using the protein variability server of the Immunomedicine Group (Universidad Complutense, Madrid; Garcia-Boronat et al., 2008). Sequence data were obtained from the National Institute of Allergy and Infectious Diseases Virus Pathogen Database and Analysis Resource online by searching for all JEV and DENV complete genomes restricted to India as the country of origin (Pickett et al., 2012). The polyprotein sequences of the JEV and Biodefense and Emerging Infection Resources DENV1–4 peptide libraries used were also included. Nucleic acid sequences were downloaded, edited to remove 5′ and 3′ untranslated regions, and translated using SeaView v4.4.2 (Gouy et al., 2010). Polyprotein alignments were performed using ClustalW (Larkin et al., 2007). HLA–peptide binding predictions were conducted using the software of the Bioinformatics and Molecular Analysis section of National Institutes of Health, SYFPEITHI, and HLArestrictor 1.2 server (Parker et al., 1994; Rammensee et al., 1999; Erup Larsen et al., 2011).
Online supplemental material
Table S1 shows peptides that were synthesized by Mimotopes Ltd. as a PepSet. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20151517/DC1.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge the National Institutes of Health Tetramer Core Facility (contract HHSN272201300006C) for provision of MHC class I tetramers and the Biodefense and Emerging Infections Research Resources Repository and the National Institute of Allergy and Infectious Diseases for provision of DENV and WNV peptide libraries. The Virus Pathogen Database and Analysis Resource has been wholly funded with federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, and Department of Health and Human Services under contract HHSN272201400028C. We thank Prof. Phillip Goulder for the gift of B cell lines. We thank the director, patients, relatives, and staff at VIMS Medical College Hospital and all the volunteers who took part.
This work was funded by a Wellcome Trust fellowship (087757/Z/08/Z) to L. Turtle (currently funded by the National Institute for Health Research, UK). P. Klenerman is a Wellcome Trust Senior Clinical Fellow (WT091663MA) funded through the National Institute for Health Research biomedical research center, Oxford, and the Oxford Martin School. T. Solomon is supported by the Health Protection Research Unit in Emerging Infections and Zoonoses of the National Institute for Health Research (HPRU-2012-10117).
The views expressed are those of the authors and not necessarily those of the National Health Service, the National Institute for Health Research, or the Department of Health or Public Health England.
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- CSF
- cerebrospinal fluid
- CTV
- Cell Trace violet
- DENV
- dengue virus
- DF
- dengue fever
- DHF
- dengue hemorrhagic fever
- ICS
- intracellular cytokine staining
- IQR
- interquartile range
- IR
- infrared
- JE
- Japanese encephalitis
- JEV
- JE virus
- NAb
- neutralizing antibody
- NS
- nonstructural
- prM
- premembrane protein
- PRNT
- plaque reduction neutralization titer
- WB
- whole blood
- WNV
- West Nile virus
References
- Aihara H., Takasaki T., Matsutani T., Suzuki R., and Kurane I.. 1998. Establishment and characterization of Japanese encephalitis virus-specific, human CD4+ T-cell clones: flavivirus cross-reactivity, protein recognition, and cytotoxic activity. J. Virol. 72:8032–8036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akondy R.S., Monson N.D., Miller J.D., Edupuganti S., Teuwen D., Wu H., Quyyumi F., Garg S., Altman J.D., Del Rio C., et al. . 2009. The yellow fever virus vaccine induces a broad and polyfunctional human memory CD8+ T cell response. J. Immunol. 183:7919–7930. 10.4049/jimmunol.0803903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson K.B., Gibbons R.V., Thomas S.J., Rothman A.L., Nisalak A., Berkelman R.L., Libraty D.H., and Endy T.P.. 2011. Preexisting Japanese encephalitis virus neutralizing antibodies and increased symptomatic dengue illness in a school-based cohort in Thailand. PLoS Negl. Trop. Dis. 5:e1311 10.1371/journal.pntd.0001311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appanna R., Huat T.L., See L.L., Tan P.L., Vadivelu J., and Devi S.. 2007. Cross-reactive T-cell responses to the nonstructural regions of dengue viruses among dengue fever and dengue hemorrhagic fever patients in Malaysia. Clin. Vaccine Immunol. 14:969–977. 10.1128/CVI.00069-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashok M.S., and Rangarajan P.N.. 1999. Immunization with plasmid DNA encoding the envelope glycoprotein of Japanese Encephalitis virus confers significant protection against intracerebral viral challenge without inducing detectable antiviral antibodies. Vaccine. 18:68–75. 10.1016/S0264-410X(99)00180-2 [DOI] [PubMed] [Google Scholar]
- Barnes E., Folgori A., Capone S., Swadling L., Aston S., Kurioka A., Meyer J., Huddart R., Smith K., Townsend R., et al. . 2012. Novel adenovirus-based vaccines induce broad and sustained T cell responses to HCV in man. Sci. Transl. Med. 4:115ra1 10.1126/scitranslmed.3003155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betts M.R., Brenchley J.M., Price D.A., De Rosa S.C., Douek D.C., Roederer M., and Koup R.A.. 2003. Sensitive and viable identification of antigen-specific CD8+ T cells by a flow cytometric assay for degranulation. J. Immunol. Methods. 281:65–78. 10.1016/S0022-1759(03)00265-5 [DOI] [PubMed] [Google Scholar]
- Betts M.R., Nason M.C., West S.M., De Rosa S.C., Migueles S.A., Abraham J., Lederman M.M., Benito J.M., Goepfert P.A., Connors M., et al. . 2006. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood. 107:4781–4789. 10.1182/blood-2005-12-4818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell G.L., Hills S.L., Fischer M., Jacobson J.A., Hoke C.H., Hombach J.M., Marfin A.A., Solomon T., Tsai T.F., Tsu V.D., and Ginsburg A.S.. 2011. Estimated global incidence of Japanese encephalitis: a systematic review. Bull. World Health Organ. 89:766–774. 10.2471/BLT.10.085233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturvedi U.C., Mathur A., Tandon P., Natu S.M., Rajvanshi S., and Tandon H.O.. 1979. Variable effect on peripheral blood leucocytes during JE virus infection of man. Clin. Exp. Immunol. 38:492–498. [PMC free article] [PubMed] [Google Scholar]
- Draenert R., Altfeld M., Brander C., Basgoz N., Corcoran C., Wurcel A.G., Stone D.R., Kalams S.A., Trocha A., Addo M.M., et al. . 2003. Comparison of overlapping peptide sets for detection of antiviral CD8 and CD4 T cell responses. J. Immunol. Methods. 275:19–29. 10.1016/S0022-1759(02)00541-0 [DOI] [PubMed] [Google Scholar]
- Draenert R., Brander C., Yu X.G., Altfeld M., Verrill C.L., Feeney M.E., Walker B.D., and Goulder P.J.. 2004. Impact of intrapeptide epitope location on CD8 T cell recognition: implications for design of overlapping peptide panels. AIDS. 18:871–876. 10.1097/00002030-200404090-00004 [DOI] [PubMed] [Google Scholar]
- Duangchinda T., Dejnirattisai W., Vasanawathana S., Limpitikul W., Tangthawornchaikul N., Malasit P., Mongkolsapaya J., and Screaton G.. 2010. Immunodominant T-cell responses to dengue virus NS3 are associated with DHF. Proc. Natl. Acad. Sci. USA. 107:16922–16927. 10.1073/pnas.1010867107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelman R., Schneider R.J., Chieowanich P., Pornpibul R., and Voodhikul P.. 1975. The effect of dengue virus infection on the clinical sequelae of Japanese encephalitis: a one year follow-up study in Thailand. Southeast Asian J. Trop. Med. Public Health. 6:308–315. [PubMed] [Google Scholar]
- Erup Larsen M., Kloverpris H., Stryhn A., Koofhethile C.K., Sims S., Ndung’u T., Goulder P., Buus S., and Nielsen M.. 2011. HLArestrictor—a tool for patient-specific predictions of HLA restriction elements and optimal epitopes within peptides. Immunogenetics. 63:43–55. 10.1007/s00251-010-0493-5 [DOI] [PubMed] [Google Scholar]
- Friberg H., Bashyam H., Toyosaki-Maeda T., Potts J.A., Greenough T., Kalayanarooj S., Gibbons R.V., Nisalak A., Srikiatkhachorn A., Green S., et al. . 2011a Cross-reactivity and expansion of dengue-specific T cells during acute primary and secondary infections in humans. Sci. Rep. 1:51 10.1038/srep00051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friberg H., Burns L., Woda M., Kalayanarooj S., Endy T.P., Stephens H.A., Green S., Rothman A.L., and Mathew A.. 2011b Memory CD8+ T cells from naturally acquired primary dengue virus infection are highly cross-reactive. Immunol. Cell Biol. 89:122–129. 10.1038/icb.2010.61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao N., Chen W., Zheng Q., Fan D.Y., Zhang J.L., Chen H., Gao G.F., Zhou D.S., and An J.. 2010. Co-expression of Japanese encephalitis virus prM-E-NS1 antigen with granulocyte-macrophage colony-stimulating factor enhances humoral and anti-virus immunity after DNA vaccination. Immunol. Lett. 129:23–31. 10.1016/j.imlet.2009.12.023 [DOI] [PubMed] [Google Scholar]
- Garcia-Boronat M., Diez-Rivero C.M., Reinherz E.L., and Reche P.A.. 2008. PVS: a web server for protein sequence variability analysis tuned to facilitate conserved epitope discovery. Nucleic Acids Res. 36:W35–W41. 10.1093/nar/gkn211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouy M., Guindon S., and Gascuel O.. 2010. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 27:221–224. 10.1093/molbev/msp259 [DOI] [PubMed] [Google Scholar]
- Halstead S.B., and Grosz C.R.. 1962. Subclinical Japanese encephalitis. I. Infection of Americans with limited residence in Korea. Am. J. Hyg. 75:190–201. [PubMed] [Google Scholar]
- Hanekom W.A., Hughes J., Mavinkurve M., Mendillo M., Watkins M., Gamieldien H., Gelderbloem S.J., Sidibana M., Mansoor N., Davids V., et al. . 2004. Novel application of a whole blood intracellular cytokine detection assay to quantitate specific T-cell frequency in field studies. J. Immunol. Methods. 291:185–195. 10.1016/j.jim.2004.06.010 [DOI] [PubMed] [Google Scholar]
- Hatch S., Endy T.P., Thomas S., Mathew A., Potts J., Pazoles P., Libraty D.H., Gibbons R., and Rothman A.L.. 2011. Intracellular cytokine production by dengue virus-specific T cells correlates with subclinical secondary infection. J. Infect. Dis. 203:1282–1291. 10.1093/infdis/jir012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoke C.H.A., Nisalak A., Sangawhipa N., Jatanasen S., Laorakapongse T., Innis B.L., Kotchasenee S., Gingrich J.B., Latendresse J., Fukai K., et al. . 1988. Protection against Japanese encephalitis by inactivated vaccines. N. Engl. J. Med. 319:608–614. 10.1056/NEJM198809083191004 [DOI] [PubMed] [Google Scholar]
- Konishi E., Kurane I., Mason P.W., Innis B.L., and Ennis F.A.. 1995. Japanese encephalitis virus-specific proliferative responses of human peripheral blood T lymphocytes. Am. J. Trop. Med. Hyg. 53:278–283. [DOI] [PubMed] [Google Scholar]
- Konishi E., Kurane I., Mason P.W., Shope R.E., Kanesa-Thasan N., Smucny J.J., Hoke C.H. Jr., and Ennis F.A.. 1998. Induction of Japanese encephalitis virus-specific cytotoxic T lymphocytes in humans by poxvirus-based JE vaccine candidates. Vaccine. 16:842–849. 10.1016/S0264-410X(97)00265-X [DOI] [PubMed] [Google Scholar]
- Konishi E., Yamaoka M., Khin-Sane-Win I. Kurane K. Takada, and Mason P.W.. 1999. The anamnestic neutralizing antibody response is critical for protection of mice from challenge following vaccination with a plasmid encoding the Japanese encephalitis virus premembrane and envelope genes. J. Virol. 73:5527–5534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P., Krishna V.D., Sulochana P., Nirmala G., Haridattatreya M., and Satchidanandam V.. 2004a Cell-mediated immune responses in healthy children with a history of subclinical infection with Japanese encephalitis virus: analysis of CD4+ and CD8+ T cell target specificities by intracellular delivery of viral proteins using the human immunodeficiency virus Tat protein transduction domain. J. Gen. Virol. 85:471–482. 10.1099/vir.0.19531-0 [DOI] [PubMed] [Google Scholar]
- Kumar P., Sulochana P., Nirmala G., Chandrashekar R., Haridattatreya M., and Satchidanandam V.. 2004b Impaired T helper 1 function of nonstructural protein 3-specific T cells in Japanese patients with encephalitis with neurological sequelae. J. Infect. Dis. 189:880–891. 10.1086/381768 [DOI] [PubMed] [Google Scholar]
- Kumar P., Sulochana P., Nirmala G., Haridattatreya M., and Satchidanandam V.. 2004c Conserved amino acids 193–324 of non-structural protein 3 are a dominant source of peptide determinants for CD4+ and CD8+ T cells in a healthy Japanese encephalitis virus-endemic cohort. J. Gen. Virol. 85:1131–1143. 10.1099/vir.0.19698-0 [DOI] [PubMed] [Google Scholar]
- Kurane I., Brinton M.A., Samson A.L., and Ennis F.A.. 1991. Dengue virus-specific, human CD4+ CD8− cytotoxic T-cell clones: multiple patterns of virus cross-reactivity recognized by NS3-specific T-cell clones. J. Virol. 65:1823–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurioka A., Ussher J.E., Cosgrove C., Clough C., Fergusson J.R., Smith K., Kang Y.H., Walker L.J., Hansen T.H., Willberg C.B., and Klenerman P.. 2015. MAIT cells are licensed through granzyme exchange to kill bacterially sensitized targets. Mucosal Immunol. 8:429–440. 10.1038/mi.2014.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larena M., Regner M., Lee E., and Lobigs M.. 2011. Pivotal role of antibody and subsidiary contribution of CD8+ T cells to recovery from infection in a murine model of Japanese encephalitis. J. Virol. 85:5446–5455. 10.1128/JVI.02611-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larena M., Regner M., and Lobigs M.. 2013. Cytolytic effector pathways and IFN-γ help protect against Japanese encephalitis. Eur. J. Immunol. 43:1789–1798. 10.1002/eji.201243152 [DOI] [PubMed] [Google Scholar]
- Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., et al. . 2007. Clustal W and Clustal X version 2.0. Bioinformatics. 23:2947–2948. 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- Lewthwaite P., Begum A., Ooi M.H., Faragher B., Lai B.F., Sandaradura I., Mohan A., Mandhan G., Meharwade P., Subhashini S., et al. . 2010a Disability after encephalitis: development and validation of a new outcome score. Bull. World Health Organ. 88:584–592. 10.2471/BLT.09.071357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewthwaite P., Perera D., Ooi M.H., Last A., Kumar R., Desai A., Begum A., Ravi V., Shankar M.V., Tio P.H., et al. . 2010b Enterovirus 75 encephalitis in children, southern India. Emerg. Infect. Dis. 16:1780–1782. 10.3201/eid1611.100672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewthwaite P., Shankar M.V., Tio P.H., Daly J., Last A., Ravikumar R., Desai A., Ravi V., Cardosa J.M., and Solomon T.. 2010c Evaluation of two commercially available ELISAs for the diagnosis of Japanese encephalitis applied to field samples. Trop. Med. Int. Health. 15:811–818. 10.1111/j.1365-3156.2010.02537.x [DOI] [PubMed] [Google Scholar]
- Libraty D.H., Nisalak A., Endy T.P., Suntayakorn S., Vaughn D.W., and Innis B.L.. 2002. Clinical and immunological risk factors for severe disease in Japanese encephalitis. Trans. R. Soc. Trop. Med. Hyg. 96:173–178. 10.1016/S0035-9203(02)90294-4 [DOI] [PubMed] [Google Scholar]
- Lubiniecki A.S., Cypess R.H., and Hammon W.M.. 1973. Passive immunity for arbovirus infection. II. Quantitative aspects of naturally and artificially acquired protection in mice for Japanese (B) encephalitis virus. Am. J. Trop. Med. Hyg. 22:535–542. [DOI] [PubMed] [Google Scholar]
- Mackowiak P.A., Wasserman S.S., and Levine M.M.. 1992. A critical appraisal of 98.6°F, the upper limit of the normal body temperature, and other legacies of Carl Reinhold August Wunderlich. JAMA. 268:1578–1580. 10.1001/jama.1992.03490120092034 [DOI] [PubMed] [Google Scholar]
- Mongkolsapaya J., Dejnirattisai W., Xu X.N., Vasanawathana S., Tangthawornchaikul N., Chairunsri A., Sawasdivorn S., Duangchinda T., Dong T., Rowland-Jones S., et al. . 2003. Original antigenic sin and apoptosis in the pathogenesis of dengue hemorrhagic fever. Nat. Med. 9:921–927. 10.1038/nm887 [DOI] [PubMed] [Google Scholar]
- Murali-Krishna K., Ravi V., and Manjunath R.. 1994. Cytotoxic T lymphocytes raised against Japanese encephalitis virus: effector cell phenotype, target specificity and in vitro virus clearance. J. Gen. Virol. 75:799–807. 10.1099/0022-1317-75-4-799 [DOI] [PubMed] [Google Scholar]
- Murali-Krishna K., Ravi V., and Manjunath R.. 1996. Protection of adult but not newborn mice against lethal intracerebral challenge with Japanese encephalitis virus by adoptively transferred virus-specific cytotoxic T lymphocytes: requirement for L3T4+ T cells. J. Gen. Virol. 77:705–714. 10.1099/0022-1317-77-4-705 [DOI] [PubMed] [Google Scholar]
- Pardo J., Bosque A., Brehm R., Wallich R., Naval J., Müllbacher A., Anel A., and Simon M.M.. 2004. Apoptotic pathways are selectively activated by granzyme A and/or granzyme B in CTL-mediated target cell lysis. J. Cell Biol. 167:457–468. 10.1083/jcb.200406115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker K.C., Bednarek M.A., and Coligan J.E.. 1994. Scheme for ranking potential HLA-A2 binding peptides based on independent binding of individual peptide side-chains. J. Immunol. 152:163–175. [PubMed] [Google Scholar]
- Parsons R., Lelic A., Hayes L., Carter A., Marshall L., Evelegh C., Drebot M., Andonova M., McMurtrey C., Hildebrand W., et al. . 2008. The memory T cell response to West Nile virus in symptomatic humans following natural infection is not influenced by age and is dominated by a restricted set of CD8+ T cell epitopes. J. Immunol. 181:1563–1572. 10.4049/jimmunol.181.2.1563 [DOI] [PubMed] [Google Scholar]
- Piazza P., McMurtrey C.P., Lelic A., Cook R.L., Hess R., Yablonsky E., Borowski L., Loeb M.B., Bramson J.L., Hildebrand W.H., and Rinaldo C.R.. 2010. Surface phenotype and functionality of WNV specific T cells differ with age and disease severity. PLoS One. 5:e15343 10.1371/journal.pone.0015343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickett B.E., Sadat E.L., Zhang Y., Noronha J.M., Squires R.B., Hunt V., Liu M., Kumar S., Zaremba S., Gu Z., et al. . 2012. ViPR: an open bioinformatics database and analysis resource for virology research. Nucleic Acids Res. 40:D593–D598. 10.1093/nar/gkr859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rammensee H., Bachmann J., Emmerich N.P., Bachor O.A., and Stevanović S.. 1999. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics. 50:213–219. 10.1007/s002510050595 [DOI] [PubMed] [Google Scholar]
- Rivino L., Kumaran E.A., Jovanovic V., Nadua K., Teo E.W., Pang S.W., Teo G.H., Gan V.C., Lye D.C., Leo Y.S., et al. . 2013. Differential targeting of viral components by CD4+ versus CD8+ T lymphocytes in dengue virus infection. J. Virol. 87:2693–2706. 10.1128/JVI.02675-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roederer M., Nozzi J.L., and Nason M.C.. 2011. SPICE: Exploration and analysis of post-cytometric complex multivariate datasets. Cytometry A. 79A:167–174. 10.1002/cyto.a.21015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman A.L. 2010. Cellular immunology of sequential dengue virus infection and its role in disease pathogenesis. Curr. Top. Microbiol. Immunol. 338:83–98. 10.1007/978-3-642-02215-9_7 [DOI] [PubMed] [Google Scholar]
- Russell P.K., Nisalak A., Sukhavachana P., and Vivona S.. 1967. A plaque reduction test for dengue virus neutralizing antibodies. J. Immunol. 99:285–290. [PubMed] [Google Scholar]
- Seder R.A., Darrah P.A., and Roederer M.. 2008. T-cell quality in memory and protection: implications for vaccine design. Nat. Rev. Immunol. 8:247–258. 10.1038/nri2274 [DOI] [PubMed] [Google Scholar]
- Solomon T. 2004. Flavivirus encephalitis. N. Engl. J. Med. 351:370–378. 10.1056/NEJMra030476 [DOI] [PubMed] [Google Scholar]
- Solomon T., Dung N.M., Kneen R., Thao T.T., Gainsborough M., Nisalak A., Day N.P., Kirkham F.J., Vaughn D.W., Smith S., and White N.J.. 2002. Seizures and raised intracranial pressure in Vietnamese patients with Japanese encephalitis. Brain. 125:1084–1093. 10.1093/brain/awf116 [DOI] [PubMed] [Google Scholar]
- Sumiyoshi H., Mori C., Fuke I., Morita K., Kuhara S., Kondou J., Kikuchi Y., Nagamatu H., and Igarashi A.. 1987. Complete nucleotide sequence of the Japanese encephalitis virus genome RNA. Virology. 161:497–510. 10.1016/0042-6822(87)90144-9 [DOI] [PubMed] [Google Scholar]
- Van Gessel Y., Klade C.S., Putnak R., Formica A., Krasaesub S., Spruth M., Cena B., Tungtaeng A., Gettayacamin M., and Dewasthaly S.. 2011. Correlation of protection against Japanese encephalitis virus and JE vaccine (IXIARO®) induced neutralizing antibody titers. Vaccine. 29:5925–5931. 10.1016/j.vaccine.2011.06.062 [DOI] [PubMed] [Google Scholar]
- Watt G., and Jongsakul K.. 2003. Acute undifferentiated fever caused by infection with Japanese encephalitis virus. Am. J. Trop. Med. Hyg. 68:704–706. [PubMed] [Google Scholar]
- Weiskopf D., Angelo M.A., de Azeredo E.L., Sidney J., Greenbaum J.A., Fernando A.N., Broadwater A., Kolla R.V., De Silva A.D., de Silva A.M., et al. . 2013. Comprehensive analysis of dengue virus-specific responses supports an HLA-linked protective role for CD8+ T cells. Proc. Natl. Acad. Sci. USA. 110:E2046–E2053. 10.1073/pnas.1305227110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiskopf D., Cerpas C., Angelo M.A., Bangs D.J., Sidney J., Paul S., Peters B., Sanches F.P., Silvera C.G., Costa P.R., et al. . 2015. Human CD8+ T-cell responses against the 4 Dengue virus serotypes are associated with distinct patterns of protein targets. J. Infect. Dis. 212:1743–1751. 10.1093/infdis/jiv289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh R.M., Che J.W., Brehm M.A., and Selin L.K.. 2010. Heterologous immunity between viruses. Immunol. Rev. 235:244–266. 10.1111/j.0105-2896.2010.00897.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter P.M., Dung N.M., Loan H.T., Kneen R., Wills B., Thu T., House D., White N.J., Farrar J.J., Hart C.A., and Solomon T.. 2004. Proinflammatory cytokines and chemokines in humans with Japanese encephalitis. J. Infect. Dis. 190:1618–1626. 10.1086/423328 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.