Pikman et al. demonstrate that the mitochondrial enzyme MTHFD2 is a potential therapeutic target in acute myeloid leukemia.
Abstract
Drugs targeting metabolism have formed the backbone of therapy for some cancers. We sought to identify new such targets in acute myeloid leukemia (AML). The one-carbon folate pathway, specifically methylenetetrahydrofolate dehydrogenase-cyclohydrolase 2 (MTHFD2), emerged as a top candidate in our analyses. MTHFD2 is the most differentially expressed metabolic enzyme in cancer versus normal cells. Knockdown of MTHFD2 in AML cells decreased growth, induced differentiation, and impaired colony formation in primary AML blasts. In human xenograft and MLL-AF9 mouse leukemia models, MTHFD2 suppression decreased leukemia burden and prolonged survival. Based upon primary patient AML data and functional genomic screening, we determined that FLT3-ITD is a biomarker of response to MTHFD2 suppression. Mechanistically, MYC regulates the expression of MTHFD2, and MTHFD2 knockdown suppresses the TCA cycle. This study supports the therapeutic targeting of MTHFD2 in AML.
It has been known for decades that cancer cells have an altered metabolism. As early as the 1920s, Otto Warburg observed that tumor cells consume glucose at a high rate and undergo fermentation even in the presence of oxygen (Warburg et al., 1927). Since then, drugs targeting metabolism have transformed the treatment of certain cancers. In the 1940s, the discovery and application of aminopterin, which was later found to target dihydrofolate reductase (DHFR), a cytoplasmic enzyme involved in one-carbon folate metabolism, yielded the first remission in a child with acute lymphoblastic leukemia (Farber et al., 1948). Other folate derivatives, such as methotrexate, were later developed. More recently, drugs such as 5-fluorouracil and pemetrexed that target thymidylate synthetase, another enzyme involved in one-carbon folate metabolism, were found to be effective therapies for some cancers (Locasale, 2013).
The discovery of germline and somatic mutations that alter metabolic proteins in cancer further supports the role of altered metabolism in cancer pathogenesis. Mutations in genes of the succinate dehydrogenase complex, critical for both the tricarboxylic acid (TCA) cycle and electron transport chain, have been implicated in the pathogenesis of hereditary paragangliomas (Baysal et al., 2000; Niemann and Müller, 2000), pheochromocytomas (Astuti et al., 2001), renal cell cancer (Vanharanta et al., 2004), and gastrointestinal stromal tumors (Janeway et al., 2011; Pantaleo et al., 2011). In addition, mutations in isocitrate dehydrogenases 1 and 2 (IDH1 and IDH2) have been found in subsets of gliomas (Yan et al., 2009; Brennan et al., 2013) and acute myeloid leukemia (AML; Paschka et al., 2010; Cancer Genome Atlas Research Network, 2013), among other malignancies. Drugs targeting these mutant proteins have entered the clinic with some successes in early phase trials (Stein et al. 2014. 56th Annual American Hematoligical Society Annual Meeting and Exposition. Abstract 115.). Moreover, as understanding of the metabolic derangements necessary to promote and maintain the malignant state continues to expand, so does the list of potential drug targets. For example, aerobic glycolysis is thought to enable the generation of the nucleotides, proteins, and lipids necessary to maintain the malignant proliferative state, in part through regulation of the glycolytic enzyme pyruvate kinase (Vander Heiden et al., 2010). Additionally, the discovery of the critical importance of glycine and serine in cancer metabolism has led to a resurgence in interest in better understanding the mechanistic relevance of one-carbon folate metabolism (Jain et al., 2012; Zhang et al., 2012; Labuschagne et al., 2014; Ye et al., 2014; Kim et al., 2015; Maddocks et al., 2016).
Although drugs targeting metabolism, such as methotrexate and asparaginase (a drug that reduces the availability of asparagine and glutamine), have been critical for the treatment of acute lymphoblastic leukemia, they are not used in therapy for AML, a hematopoietic malignancy where cure rates are still quite poor despite high-dose cytotoxic chemotherapy, including stem cell transplantation. This is especially true for patients with subtypes of AML characterized by high-risk features, such as the presence of FLT3-ITD mutations. New therapies are urgently needed for the treatment of these patients. In this study, we set out to define common mechanisms critical to the maintenance of AML cells to nominate novel, potentially targetable metabolic pathways for the treatment of this disease. We integrated gene expression signatures generated from the treatment of AML cells with multiple small molecules known to promote AML differentiation and death. Methylenetetrahydrofolate dehydrogenase 2 (MTHFD2), an NAD+-dependent enzyme with dehydrogenase and cyclohydrolase activity, which plays an essential role in mitochondrial one-carbon folate metabolism, was prioritized as a target relevant to AML cell growth and differentiation. Suppression of MTHFD2 impaired AML growth and induced differentiation in vitro and impaired disease progression in multiple mouse models of AML. Additionally, FLT3-ITD mutations are a biomarker of response to MTHFD2 suppression. Mechanistically, MYC directly regulates MTHFD2 expression, and suppression of MTHFD2 leads to marked alteration of the TCA cycle.
RESULTS
One-carbon folate metabolism is a core pathway altered with compounds active in AML cells
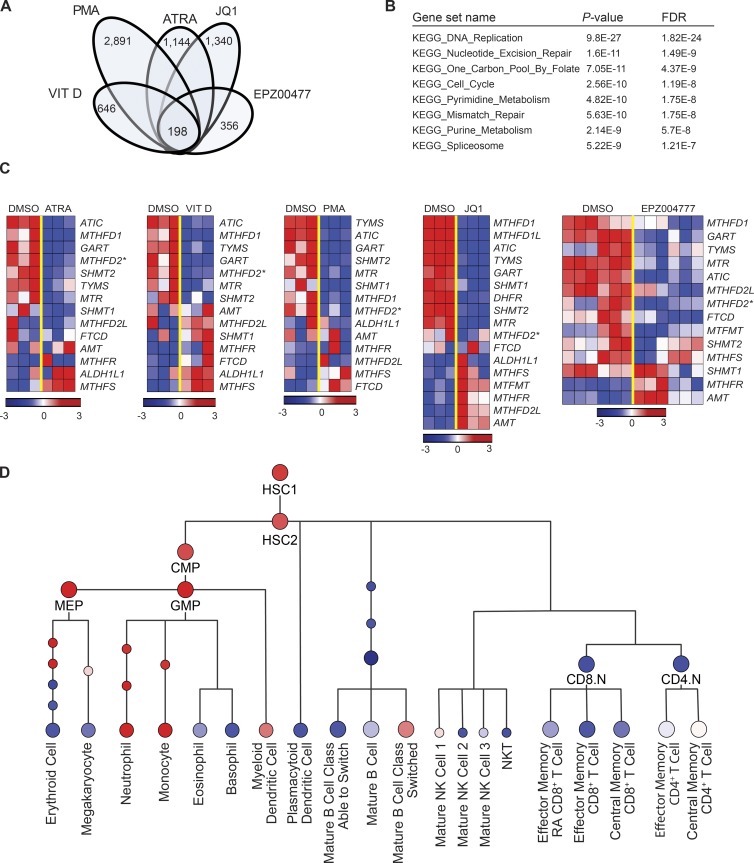
We analyzed gene expression signatures of HL-60 cells treated with 1,25-dihydroxy Vitamin D3 (Vitamin D), PMA, and all-trans retinoic acid (Stegmaier et al., 2004), along with gene expression signatures of THP-1 cells treated with the BET bromodomain inhibitor JQ1 (Zuber et al., 2011), and MV4-11 and MOLM-13 cells treated with the DOT1L inhibitor EPZ004777 (Daigle et al., 2011), compounds that cause AML differentiation and death. We identified a list of genes significantly down-regulated by the individual agents (fold change expression ≥ 1.5, P ≤ 0.05, Benjamini-Hochberg False Discovery Rate [FDR(BH)] ≤ 0.05) and found a core subset of 198 genes that were robustly down-regulated across all five datasets based on the selection criteria mean fold change of expression ≥ 1.5 across all datasets and (ii) significant fold change of expression ≥ 1.5 (P ≤ 0.05, FDR(BH) ≤ 0.05) in at least four out of the five datasets (Fig. 1 A and Table S1). We then used this core list of genes to probe for enrichment across the Kyoto Encyclopedia of Genes and Genomes (KEGG) curated collection of canonical pathways (Kanehisa et al., 2014), available from the Molecular Signature Database (MSigDB) v4.0 (Subramanian et al., 2005). Three metabolic pathways, One Carbon Pool by Folate, Purine Metabolism, and Pyrimidine Metabolism, along with five other pathways involved in cell cycle, splicing, and DNA mismatch repair, were significantly enriched in the core list of genes (FDR (q value) ≤ 10−4; Fig. 1 B). The role of folate-mediated one-carbon metabolism in cancer, especially in the mitochondrial compartment, has been a subject of renewed interest (Jain et al., 2012; Nilsson et al., 2014). A recent publication has highlighted the mitochondrial enzyme involved in one-carbon folate metabolism, MTHFD2, as the most differentially expressed enzyme in cancer cells compared with normal cells (Nilsson et al., 2014). Indeed, across all five treatments there was down-regulation of MTHFD2 expression (Fig. 1 C). We thus focused on the relevance of MTHFD2 to AML growth and maturation. We first determined the expression of MTHFD2 across normal myeloid differentiation using a publicly available dataset profiling the transcriptomes of hematopoietic cells across various stages of hematopoietic differentiation (Novershtern et al., 2011). MTHFD2 expression was enriched in the myeloid lineage during normal hematopoietic differentiation (Fig. 1 D).
Figure 1.
One-carbon folate metabolism is a core pathway altered with AML targeting compounds. (A) Intersection of genes down-regulated by five compounds that cause AML death and differentiation: Vitamin D, PMA, ATRA, JQ1, and EPZ004777. Reported is the number of significantly down-regulated genes by each agent (DMSO vs. agent: SNR ≥ 1.5; P ≤ 0.05; FDR[BH] ≤ 0.05). The core 198 genes are significantly down-regulated by each of the five agents (DMSO vs. agent: average SNR across all conditions ≥ 1.5 and SNR ≥ 1.5 in four out of five conditions). P-values calculated using the permutation test implemented in the comparative marker selection procedure, Gene Pattern v3.8.1. (B) Pathway enrichment analysis across KEGG canonical pathways showed eight significantly overlapping pathways. P-values calculated using the hypergeometric test for gene set overlapping analysis. (C) Heat maps showing expression for genes of one-carbon folate pathway across five treatments with differentiation agents. (D) Expression of MTHFD2 across the hematopoietic lineage.
MTHFD2 suppression impairs growth and promotes AML differentiation
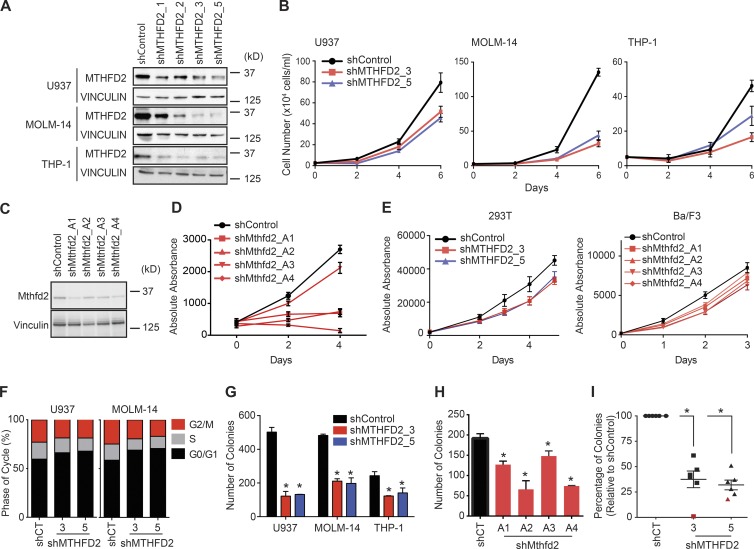
We generated multiple shRNAs targeting MTHFD2 and validated knockdown by Western blot (Fig. 2 A). Two shRNAs, shMTHFD2_3 and shMTHFD2_5, led to the greatest and most consistent suppression of MTHFD2 by Western blot, and we used these two shRNAs in subsequent experiments.
Figure 2.
MTHFD2 suppression impairs growth of AML. (A) Western blot evaluating knockdown of MTHFD2 with four unique shRNAs, shMTHFD2_1, shMTHFD2_2, shMTHFD2_3, and shMTHFD2_5. Vinculin is used as a loading control. (B) Viability measured by Trypan blue exclusion over a time course with MTHFD2-directed shRNA versus control shRNA. Shown are the mean ± SD of three replicates. Data show a representative experiment from at least three individual experiments. (C) pLKO.1 constructs expressing hairpins targeting Mthfd2 were used to infect mouse MLL-AF9 leukemia cells, and knockdown was confirmed by Western blot analysis. (D) Growth was assessed after Mthfd2 knockdown using an ATP-based assay. Data represent mean ± SD for six replicates. (E) Viability measured by an ATP-based assay over a time course with MTHFD2-directed shRNA versus control shRNA in 293T and Ba/F3 cells. Shown are the mean ± SD of 14 replicates. (F) Cell cycle analysis in U937 and MOLM-14 cells 4 d after MTHFD2 suppression. Shown is the mean of three separate experiments. (G) Colony formation in methylcellulose assessed 5 d after infection with shRNAs directed against MTHFD2 or a control shRNA in three AML cell lines. Data represent mean ± SD of two technical replicates. Data show a representative experiment from at least three individual experiments. *, P < 0.05 using unpaired Student’s t test with Welch’s correction. (H) Colony formation in methylcellulose was assessed 5 d after infection with shRNAs directed against Mthfd2 or a control. Data represent mean ± SD of two technical replicates. Data show a representative experiment of two individual experiments. *, P < 0.05 using unpaired Student’s t test with Welch’s correction. (I) Colony formation in methylcellulose in six primary patient samples. Red indicates patient sample with FLT3-ITD mutation. Data normalized to shControl for each sample. *, P < 0.05 using Mann-Whitney test.
Suppression of MTHFD2 in three human AML cell lines caused a decrease in growth (Fig. 2 B) that was also observed with Mthfd2 suppression in mouse MLL-AF9 leukemia cells (Fig. 2, C and D). MTHFD2 suppression in noncancer cell lines, human embryonic kidney cells 293T, and the mouse pro–B cell line Ba/F3, had minimal effect on cell growth (Fig. 2 E). MTHFD2 suppression accordingly led to a G0/G1 arrest (Fig. 2 F), and the formation of AML colonies in methylcellulose was decreased upon suppression of MTHFD2 in human AML cell lines (Fig. 2 G), in mouse MLL-AF9 AML cells (Fig. 2 H), and in primary AML patient samples (Fig. 2 I).
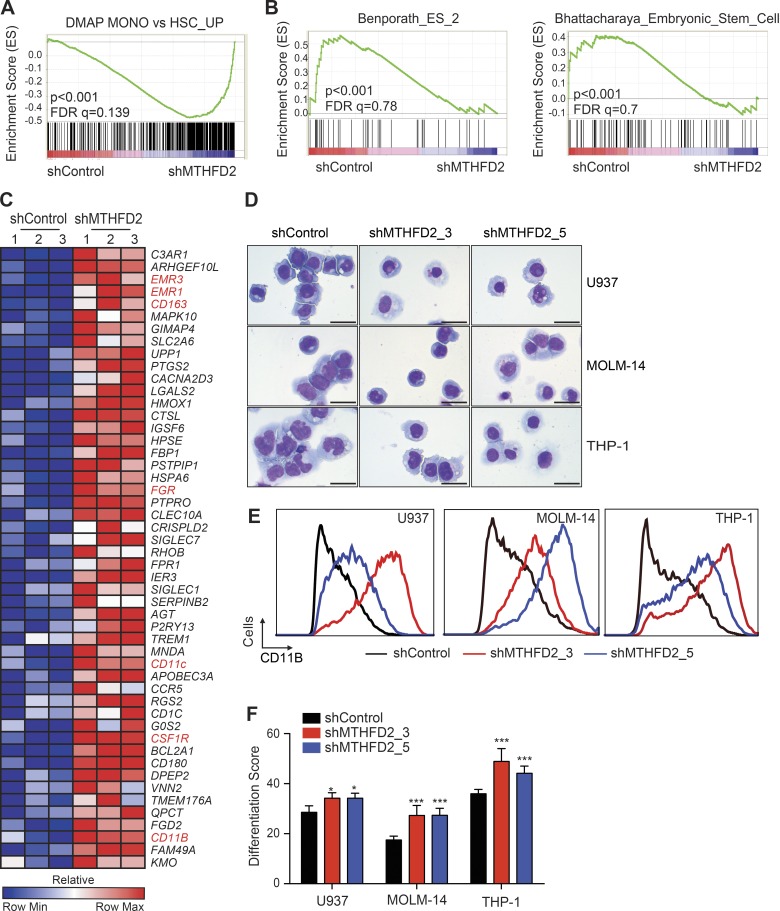
We next evaluated the effects of MTHFD2 suppression on differentiation using whole transcriptome profiling. shMTHFD2_5 was used to knockdown MTHFD2 in MOLM-14 cells and RNA sequencing was performed. Gene sets consistent with myeloid maturation were enriched in the transcriptional profiling of MOLM-14 cells with MTHFD2 suppression (Fig. 3 A), and gene sets consistent with hematopoietic stem cell signatures were repressed (Fig. 3 B). There was up-regulation of well-validated myeloid differentiation genes in the top 50 differentially expressed genes (Fig. 3 C). Moreover, MTHFD2 suppression resulted in morphological changes, such as nuclear condensation and cytoplasmic ruffling (Fig. 3 D), and the expression of surface CD11b (Fig. 3 E), both of which are consistent with myeloid maturation. We next evaluated the effects of MTHFD2 suppression on AML differentiation using our previously published 32-gene expression differentiation signature that can be measured by ligation-mediated amplification and a fluorescent, bead-based detection system (Stegmaier et al., 2004). Suppression of MTHFD2 resulted in induction of the myeloid differentiation signature in the three evaluated AML cell lines (Fig. 3 F).
Figure 3.
MTHFD2 suppression impairs AML differentiation. (A) GSEA plot showing the enrichment of the genes down-regulated by MTHFD2 knockdown in the DMAP monocyte differentiation signature. P < 0.001, computed using the permutation test for gene set enrichment implemented in the GSEA v2.0.14 platform. (B) GSEA plots showing enrichment of embryonic stem cell genes down-regulated with MTHFD2 suppression. P < 0.001, computed using the permutation test for gene set enrichment implemented in the GSEA v2.0.14 platform. (C) Heat map showing top 50 out of 229 leading edge genes for the shMTHFD2 genome-wide signature with respect to monocyte versus HSC signature from DMAP (SNR ≥ 1.5; P ≤ 0.05; FDR[BH] ≤ 0.05). P-value computed using the permutation test implemented in the comparative marker selection procedure, Gene Pattern v3.8.1. Exemplary genes known to be up-regulated with myeloid differentiation are highlighted in red. (D) May-Grunwald Giemsa staining of AML cell lines after MTHFD2 knockdown 9 d after infection. Bars, 25 µm. Shown are representative images from three different experiments. (E) CD11b staining by flow cytometry after MTHFD2 knockdown evaluated 9 d after infection with MTHFD2-targeting shRNAs. Shown are representative flow graphs from three different experiments. (F) Bar graphs depicting differentiation score (weighted summed score) for three AML cell lines after MTHFD2 knockdown. Error bars represent ± SD of eight replicates. *, P < 0.05; ***, P = 0.0002 using Mann-Whitney test, relative to control per cell line. Shown is a representative of two separate experiments.
MTHFD2 suppression impairs leukemia establishment and progression in vivo
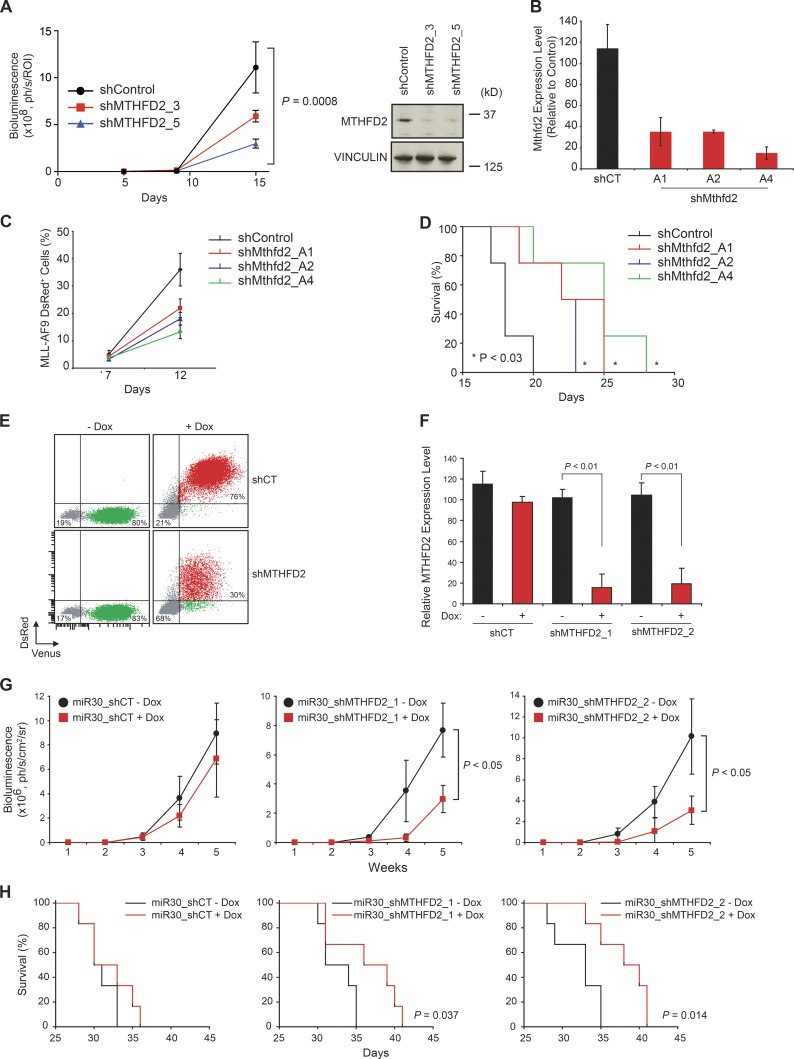
In vitro studies are critical for initial target validation, but do not completely recapitulate response in a vertebrate organism. Currently, there are no commercially available inhibitors of MTHFD2. We thus extended testing of MTHFD2-directed hairpins to a human AML orthotopic xenograft model. Here, U937 cells were labeled with luciferase (U937-LucNeo), infected with vectors bearing MTHFD2-directed shRNAs (shMTHFD2_3 and shMTHFD2_5), and selected with puromycin. Knockdown of MTHFD2 was then confirmed by Western immunoblotting before injection by tail vein into NOD-SCID IL2Rγnull (NSG) mice (Fig. 4 A). Loss of MTHFD2 impaired progression of this U937 orthotopic xenograft, as measured in vivo by bioluminescence (Fig. 4 A). Additionally, we used an MLL-AF9 mouse leukemia model to test the effect of Mthfd2 suppression. Leukemia cells were infected with vectors bearing Mthfd2-directed shRNAs (shMthfd2_A1, shMthfd2_A2, and shMthfd2_A4), and selected with puromycin. Knockdown of Mthfd2 was confirmed by RNA expression (Fig. 4 B). Loss of Mthfd2 decreased leukemia burden and prolonged survival in this aggressive mouse leukemia model (Fig. 4, C and D).
Figure 4.
MTHFD2 knockdown impairs leukemia establishment and progression in vivo. (A) Graph depicting growth in mice of U937-LucNeo cells infected with MTHFD2-directed shRNA versus LacZ (shControl). Bioluminescence was quantified as a measure of disease burden. Data are represented as a mean ± SEM of six mice per cohort. Western blot shows MTHFD2 levels at the time of injection of cells. P = 0.0008 using two-way ANOVA. (B) RNA was isolated from mouse MLL-AF9 cells, and Mthfd2 suppression was confirmed at time of cell injection. (C) Graph showing percentage of MLL-AF9–positive cells in mouse peripheral blood with or without Mthfd2 suppression. Shown is mean percentage ± SD for four mice. (D) Kaplan-Meier curve showing overall survival of mice with MLL-AF9 leukemia after suppression of MTHFD2 versus shControl (n = 4 per group). P < 0.03 calculated using log-rank test. (E) FACS plot showing induction of shRNA with doxycycline treatment (DsRed+/Venus+). (F) qRT-PCR showing relative amount of MTHFD2 after treatment with doxycycline. Bar graphs represent mean value ± SD for four replicates. P < 0.01, Mann-Whitney test. (G) Graph depicting growth of U937-LucNeo cells transduced with a doxycycline-inducible TRMPVIR vector containing either a scrambled miR30-shRNA (shCT) or shRNAs directed against MTHFD2 after injection into Nude mice and treatment with doxycycline to induce shRNA expression. Graphs represent mean bioluminescence ± SD for six mice per group. P < 0.05, Mann-Whitney test at week 5. (H) Kaplan-Meier curves showing overall survival of mice (n = 6 per group). P = 0.037 for shMTHFD2_1 and P = 0.014 for shMTHFD2_2, calculated using log-rank test.
To study the effects of MTHFD2 suppression after the development of AML in vivo, we deployed the doxycycline-inducible miR30 shRNA system directed against MTHFD2 using the two constructs that yielded efficient suppression of MTHFD2 (Fig. 4, E and F). U937-LucNeo cells bearing the doxycycline-inducible miR30 shRNA directed against MTHFD2 were injected into Nude mice. Leukemia establishment was confirmed by bioluminescence, and then mice were treated with doxycycline and monitored for disease progression and overall survival. Induction of MTHFD2 knockdown led to a decrease in leukemia burden and prolonged survival compared with control mice (Fig. 4, G and H).
FLT3-ITD is a biomarker of response to MTHFD2 suppression
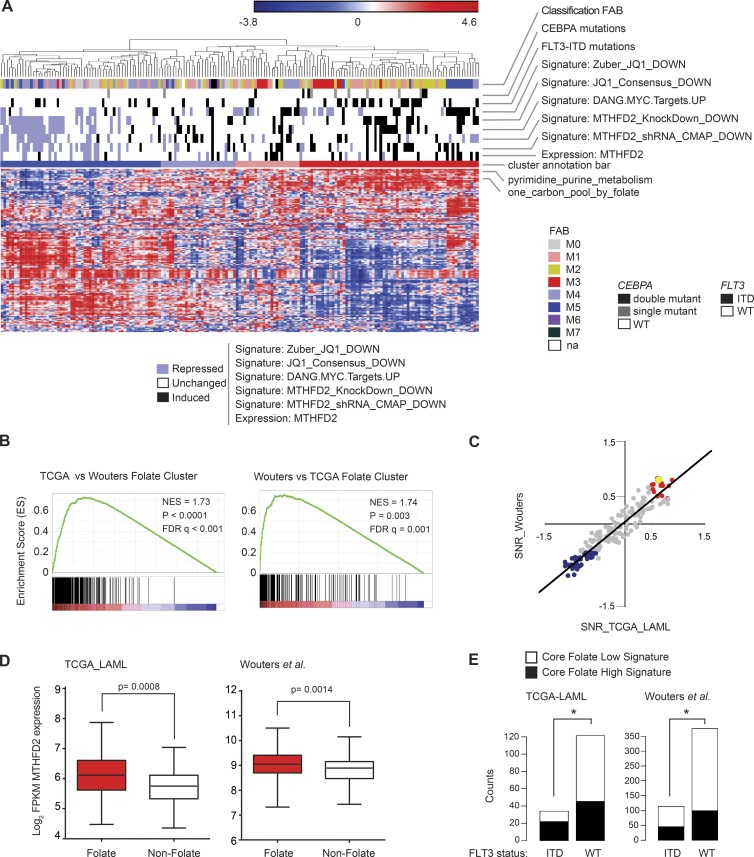
To identify potential biomarkers of response to MTHFD2 suppression, we performed single-sample Gene Set Enrichment Analysis (ssGSEA) in two large human primary patient AML gene expression datasets, TCGA LAML (Cancer Genome Atlas Research Network, 2013; 179 tumors) and a dataset described in Wouters et al. (2009); 526 tumors); 526 tumors) for enrichment across the collection of 186 canonical KEGG pathways. Consensus hierarchical clustering of samples in the ssGSEA projections identified a robust subset of n = 67 (out of 179) patients in the TCGA LAML data and n = 145 (out of 526) patients in the Wouters et al. (2009) data that showed enrichment in gene expression of members of the one-carbon folate pathway (Fig. 5 A; TCGA heat map shown). We called this the folate cluster. GSEA cross-validated the enrichment of the folate cluster in one dataset with the gene signature of the folate cluster in another dataset (Fig. 5 B). In addition, differential analysis for enrichment in KEGG canonical pathway signatures in the folate cluster versus other samples provided highly correlated results between the TCGA LAML and the Wouters et al. (2009) datasets (Fig. 5 C and Table S2). Thus, the folate cluster has consistent genomic and functional profiles across the primary AML datasets. Although MTHFD2 expression is high across all AML samples, the folate cluster had higher expression of MTHFD2 (Fig. 5 D). Additionally, it was enriched in genes that correlate with MTHFD2 signatures, a BET inhibitor (JQ1) signature in AML cell lines, and with a MYC target signature as defined by Zeller et al. (Zeller et al., 2003; based on two-tailed Fisher test with a cutoff P < 0.05; Fig. 5 A and Table S3). Comprehensive association tests in each of the two primary tumor datasets found that samples in the folate cluster were enriched for CEBPA and FLT3-ITD mutations (Fig. 5 A, TCGA data shown, and Fig. 5 E). MTHFD2 single gene expression, however, was not consistently associated with recurrent AML gene mutations, including CEBPA and FLT3-ITD.
Figure 5.
ssGSEA identifies a folate cluster and establishes FLT3-ITD as a sensitivity marker for MTHFD2 suppression. (A) Heat map of ssGSEA projection for the primary AML tumor dataset from TCGA on the collection of KEGG canonical pathways. Cluster annotation bar depicts consensus clusters and their degree of enrichment in the one-carbon folate pathway. Samples in the folate cluster are highlighted in red. Other annotation bars highlight enrichment in signatures of MTHFD2, JQ1 effect, MYC targets, mutations associated with folate cluster, and FAB subtypes. For each of these signatures, the enrichment was computed per primary tumor by applying the ssGSEA method. The level of enrichment was assessed based on the absolute cut-off value 1 applied to the ssGSEA z-scores: samples with ssGSEA z-scores ≥1 were labeled as significantly highly enriched in the signature and highlighted black, whereas samples with ssGSEA z-scores ≤−1 were labeled as significantly low enriched in the signature and highlighted blue. Samples that did not pass the significance cut-off were labeled as not significantly enriched and highlighted white. (B) GSEA cross-validation tests for consistency of the folate cluster signature across primary AML tumor datasets. Each GSEA plot shows enrichment of the folate cluster phenotype in one dataset with the gene signature of the folate cluster in the other dataset. P < 0.0001; P = 0.003, computed using the permutation test for gene set enrichment implemented in the GSEA v2.0.14 platform. (C) Scatter plot of differential KEGG pathway enrichment SNR scores for samples in the folate cluster versus all other samples in TCGA LAML versus Wouters et al. (2009) datasets. The regression line indicates the Pearson correlation coefficient R = 0.94 (P < 0.001, computed using the R v3.2 cor.test function, implemented in the library stats). Red dots correspond to KEGG pathways positively enriched in both TCGA LAML and Wouters et al. folate clusters. Blue dots correspond to KEGG pathways negatively enriched in both TCGA LAML and Wouters et al. folate clusters. The yellow highlighted dot corresponds to the one-carbon folate pathway. The complete descriptions of the KEGG pathways and of the differentially enriched scores in the two datasets are provided in Table S2. (D) Box plots showing MTHFD2 expression in the folate cluster, shown in red in A, compared with the nonfolate cluster, shown in dark blue in Panel A for TCGA and Wouters et al. datasets. P = 0.0008 for TCGA and P = 0.0014 for Wouters et al., Welch-corrected two-tailed Student’s t test. (E) Two-dimensional barplots depicting the results of two-tailed Fisher exact tests for enrichment of the folate cluster with FLT3-ITD mutations. *, P < 0.05, two-tailed Fisher’s exact test.
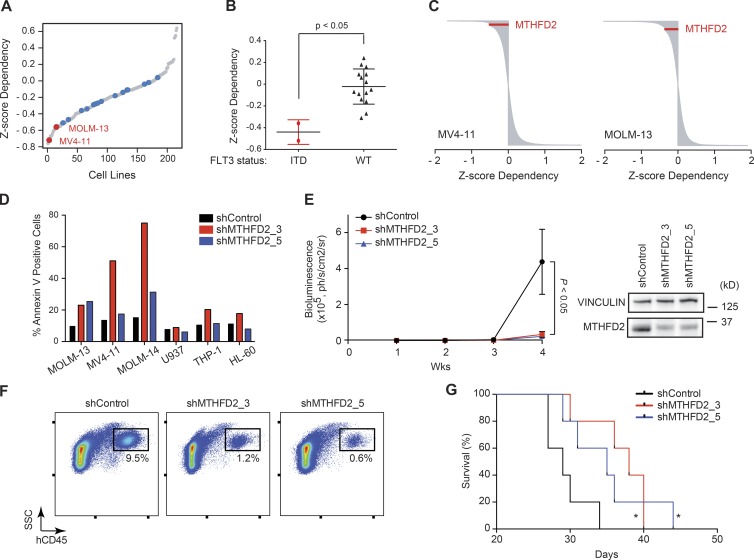
We next probed data from an shRNA screen of 56,903 shRNAs directed against 14,222 genes, out of which 11,946 genes have complete measurements (Achilles v2.4.3), performed in 216 cancer cell lines, including 17 AML cell lines (Cowley et al., 2014). AML cell lines characterized by a FLT3-ITD mutation, MV4-11 and MOLM-13, were more sensitive to MTHFD2 suppression compared with other AML cell lines (Fig. 6, A and B), and suppression of MTHFD2 was in the top 5% of dependencies in each individual cell line (Fig. 6 C). In support of this observation, suppression of MTHFD2 in FLT3-ITD positive cell lines significantly increased cell death compared with FLT3 wild-type cell lines (Fig. 6 D). Thus, targeting of MTHFD2 may be particularly efficacious in AML characterized by FLT3-ITDs.
Figure 6.
FLT3-ITD is a marker of sensitivity for MTHFD2 suppression. (A) Ranking of MTHFD2 depletion score across 216 cell lines in a shRNA screen. Red dots represent FLT3-ITD–mutated AML cell lines. Blue dots represent other AML cell lines. Gray dots represent non-AML cell lines. (B) Box plots showing mean MTHFD2 dependency for AML cell lines. P < 0.05, Mann-Whitney test. (C) MTHFD2 ranks as 191 out of 11,946 (top 1.6%) dependency in the MV4-11 cell line and 575 out of 11,946 (top 4.8%) in the MOLM-13 cell line. MTHFD2 is shown in red among all shRNA rankings for each individual cell line. (D) Suppression of MTHFD2 causes significant apoptosis in FLT3-ITD mutant cell lines. Shown is percent Annexin V–positive cells per cell line with shControl, shMTHFD2_3, or shMTHFD2_5. Data show a representative experiment from at least three individual experiments. (E) Graph depicting growth in mice of MV4-11-LucNeo cells infected with MTHFD2-directed shRNA versus LacZ (shControl). Bioluminescence was quantified as a measure of disease burden. Data are represented as a mean ± SEM of four mice per cohort. P < 0.05 at Week 4, Mann-Whitney test. Western blot shows MTHFD2 level at time of injection of cells. (F) FACS plot showing percent of human CD45+ cells in bone marrow in mice injected with MV4-11-LucNeo cells, 16 d after injection. (G) Kaplan-Meier curve showing overall survival of mice with MV4-11-LucNeo cells after suppression of MTHFD2 versus shControl (n = 5 per group). *, P < 0.05 relative to control, calculated using log-rank test.
To further study the effect of MTHFD2 suppression in a FLT3-ITD model, we extended our testing of MTHFD2-directed hairpins to a human AML orthotopic xenograft model, MV4-11. Here, MV4-11 cells were labeled with luciferase (MV4-11-LucNeo) infected with vectors bearing MTHFD2-directed shRNAs (shMTHFD2_3 and shMTHFD2_5), and selected with puromycin. Knockdown of MTHFD2 was then confirmed by Western immunoblotting before injection by tail vein into NSG mice (Fig. 6 E). Loss of MTHFD2 impaired leukemia progression of this MV4-11 orthotopic xenograft, as measured in vivo by bioluminescence (Fig. 6 E) and by flow cytometry (Fig. 6 F), and prolonged mouse survival (Fig. 6 G).
MYC regulates the mitochondrial one-carbon folate pathway
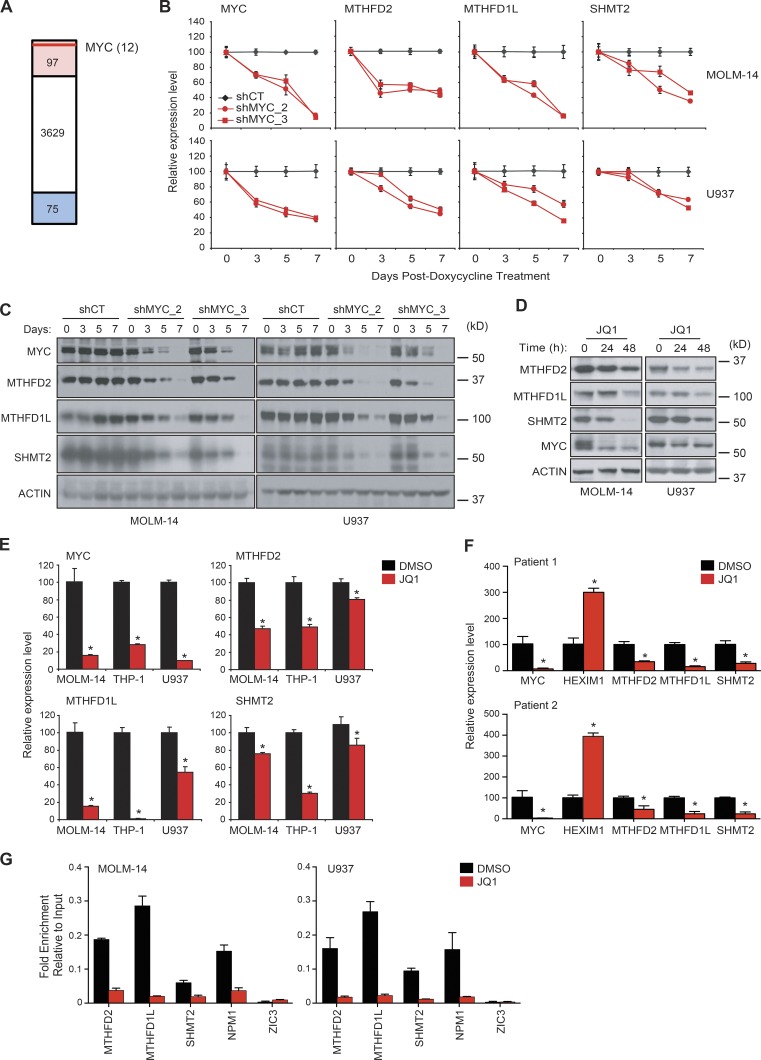
We next turned to the Connectivity Map, a large database profiling the effects of small molecules and shRNAs on gene expression in cancer cell lines (http://www.lincscloud.org) to evaluate whether suppression of other proteins can mimic MTHFD2 suppression. We used a gene expression signature of MTHFD2 suppression that was previously developed within the Connectivity Map and found that MYC knockdown scored as one of the top shRNA connections to the expression profile of MTHFD2 knockdown (Fig. 7 A and Table S4). MYC is a key regulator of cellular pathways and has been shown to control mitochondrial serine hydroxymethyltransferase 2 (SHMT2; Nikiforov et al., 2002). However, whether MYC directly binds and controls expression of genes in the mitochondrial one-carbon folate pathway in AML has not been reported. We first evaluated MYC knockdown with two unique doxycycline-inducible miR30 shRNAs. MYC suppression resulted in a decrease in MTHFD2 transcript and protein, in addition to decreased expression of other enzymes associated with the mitochondrial one-carbon folate pathway, such as MTHFD 1-like (MTHFD1L) and SHMT2 (Fig. 7, B and C). JQ1 is a small molecule inhibitor of the BET-bromodomain–containing protein BRD4 and decreases MYC expression in AML cells (Zuber et al., 2011). Treatment of AML cell lines with JQ1 recapitulated the findings of MYC suppression using shRNA (Fig. 7, D and E). Additionally, mononuclear cells from two primary patient AML samples were extracted and treated with 1 µM JQ1 for 24 h. qRT-PCR showed a decrease in MYC, MTHFD2, and the other evaluated mitochondrial proteins of the one-carbon folate pathway (Fig. 7 F).
Figure 7.
MYC controls one-carbon folate pathway. (A) MTHFD2 knockdown signature was used to probe the Connectivity Map for targets that mimic the effect of MTHFD2 knockdown. Red highlights 97 shRNAs that are significantly enriched (CMap consensus score > 0.90) and blue shows shRNAs that are anticorrelated (CMap consensus score < −0.90). Red line represents relative position of MYC hairpin enrichment. (B) Two AML cell lines were transduced with two different doxycycline inducible miR30 shRNAs targeting MYC (shMYC_2 and shMYC_3). A scrambled miR30 shRNA was used as control (shCT). Gene expression of MYC, MTHFD2, MTHFD1L, and SHMT2 was measured by qRT-PCR. Shown are the mean ± SD of four replicate measurements. (C) Western blot of the treated cell lines confirms a reduction in target protein after doxycycline treatment. (D) Western blot of two AML cell lines treated with 1 µM JQ1 showing a time-dependent decrease in proteins of the mitochondrial one-carbon folate pathway. (E) Three AML cell lines were treated with 1 µM JQ1 versus vehicle and qRT-PCR was performed. *, P < 0.05, Mann-Whitney test. (F) Mononuclear cells extracted from two AML patient samples were treated with 1 µM JQ1 for 24 h. Target gene expression was evaluated by qRT-PCR. Shown is the relative mean expression ± SD of 4 replicate measurements. HEXIM, a target known to increase with JQ1 treatment, was used as a control. *, P < 0.05, Mann-Whitney test. (G) ChIP-qPCR was performed on two AML cell lines after treatment with 1 µM JQ1 for 48 h. Shown is fold enrichment over input. Error bars represent four calculations based on two replicates. Binding at NPM1 promoter region was measured as a positive control and binding at ZIC3 promoter region as a negative control. Data show a representative experiment of two individual experiments.
We next used publicly available chromatin immunoprecipitation sequencing (ChIP-Seq; Lin et al., 2012) and ENCODE data to assess whether MYC binds at promoter regions of SHMT2, MTHFD1L, and MTHFD2. MYC binds at SHMT2, MTHFD2, and MTHFD1L promoters in Burkitt lymphoma, small cell lung cancer, glioblastoma multiforme, and multiple myeloma cell lines (Table S5; data for MTHFD2 shown). MYC also binds at the MTHFD2, SHMT2, and MTHFD1L promoters of K562, a chronic myeloid leukemia cell line. Interestingly, the relative signal score is highest in K562 cells compared with H1-hESC (normal human embryonic stem cells) and GM12878 (normal B lymphocytes) for MTHFD2 (Table S5). To evaluate whether MYC directly binds to promoters of genes of the mitochondrial one-carbon folate pathway in AML, we treated two AML cell lines, MOLM-14 and U937, with JQ1 and performed ChIP-quantitative PCR (ChIP-qPCR). After MYC immunoprecipitation and qPCR, we found a significant decrease in MYC binding at MTHFD2, MTHFD1L, and SHMT2 promoters (Fig. 7 G). Collectively, these data suggest that the expression of genes involved in mitochondrial one-carbon folate metabolism is directly regulated by MYC.
Suppression of MTHFD2 causes metabolic alterations
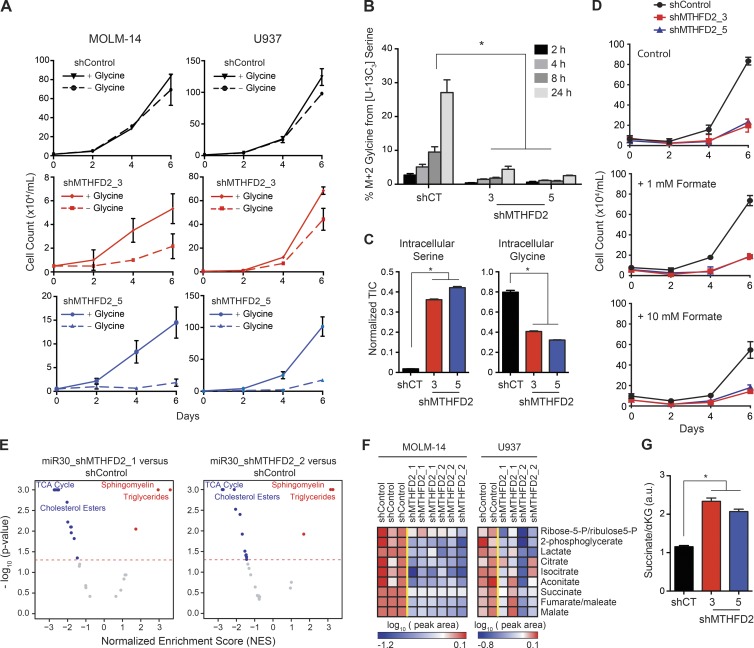
Loss of MTHFD2 has been reported to result in glycine auxotrophy in murine fibroblasts (Patel et al., 2003). In line with this observation, suppression of MTHFD2 in two AML cell lines resulted in increased dependence on exogenous glycine (Fig. 8 A). We next measured production of labeled glycine from [U-13C3]serine over time using Gas Chromatography-Mass Spectrometry (GC-MS) in media after MTHFD2 suppression in MOLM-14 cells. MTHFD2 suppression led to an inability to produce glycine from serine (Fig. 8 B). Measurement of intracellular metabolite levels confirmed an accumulation of serine and a decrease in intracellular glycine in MOLM-14 cells after MTHFD2 suppression (Fig. 8 C).
Figure 8.
Suppression of MTHFD2 increases glycine dependence, depletes intermediates of the TCA cycle and alters the succinate to α-ketoglutarate ratio. (A) MTHFD2 knockdown using shRNA resulted in an increased dependence on exogenous glycine in two AML cell lines. Graphs depict cell number as measured by trypan blue exclusion. Shown are the means ± SD of 3 replicates. (B) MTHFD2 suppression results in failure to produce glycine from serine in MOLM-14 cells. After MTHFD2 suppression, MOLM-14 cells were cultured in glycine free media in the presence of [U-13C3]Serine (all three carbons labeled with 13C). Media was extracted at the time points shown and M+2 glycine (both carbons labeled with 13C) was measured by GC-MS. (*) represents P < 0.05 at each time point for hairpins shMTHD2_3 and shMTHFD2_5 relative to shControl, Wilcoxon rank sum test. Shown are the mean ± SD of three replicates. (C) Amounts of intracellular serine and glycine (total ion counts, TIC) from MOLM-14 cells cultured in complete media for 24 h after MTHFD2 suppression, as measured by GC-MS, and then normalized to norvaline as an internal standard. Shown are the mean ± SD of 3 replicates. (*) represents P < 0.05 for hairpins shMTHD2_3 and shMTHFD2_5 relative to shControl, Wilcoxon rank sum test with continuity correction. (D) Suppression of MTHFD2 affects growth of AML cells that are not rescued by formate supplementation in the MOLM-14 cell line. Viability measured by Trypan blue exclusion over a time course with MTHFD2-directed shRNA versus control shRNA. Shown are the mean ± SD of three replicates. (E) MSEA showing metabolic pathways differentially altered by MTHFD2 suppression. Shown is data for MOLM-14 cell line. Blue dots represent pathways that are significantly depleted and red dots represent pathways that are significantly enriched (P < 0.05, permutation test in MSEA). (F) Heat map showing changes in glycolysis intermediates, lactate, and TCA cycle intermediates. Metabolites were selected if they were significantly (P < 0.05, Student’s t test) decreased in MOLM-14 cell line. (G) Ratio of intracellular succinate to α-ketoglutarate (a.u.) in MOLM-14 cells cultured in complete media for 24 h after MTHFD2 suppression, as measured by GC-MS. Shown is the mean ± SEM of three replicates. *, P < 0.05 for hairpins shMTHD2_3 and shMTHFD2_5 relative to shControl, Wilcoxon rank sum test with continuity correction.
MTHFD2 catalyzes conversion of methylene tetrahydrofolate to formyl tetrahydrofolate via its dehydrogenase and cyclohydrolase activities. Formyl THF is converted to formate by MTHFD1L (Christensen and Mackenzie, 2008). Thus, formate supplementation might be expected to rescue MTHFD2 suppression. Addition of 1 and 10 mM formate to media, however, did not rescue the viability effect of MTHFD2 suppression in the MOLM-14 cell line (Fig. 8 D). Thus, the metabolic alterations leading to decreased AML cell growth with suppression of MTHFD2 are likely distinct from any reduction in formate production.
We next turned to an unbiased approach with metabolic profiling using Liquid Chromatography and Mass Spectroscopy (LC-MS) to discover other metabolic alterations caused by MTHFD2 suppression. MOLM-14 and U937 cells were profiled after MTHFD2 suppression for 3 d. Suppression of MTHFD2 significantly depleted TCA cycle intermediates and cholesterol esters (Fig. 8, E and F). Additionally, there was up-regulation of sphingomyelin and triglyceride production (Fig. 8 E). A metabolic hallmark of naive embryonic stem cells is their ability to maintain pluripotency by using high levels of α-ketoglutarate with alterations in the succinate to α-ketoglutarate ratio, leading to changes in the stemness state of the cell (Carey et al., 2015). In AML cells, MTHFD2 suppression led to an increase in the succinate to α-ketoglutarate ratio (Fig. 8 G) consistent with the increase in differentiation and decrease in stem cell signatures in AML cell lines perturbed by MTHFD2-directed shRNAs.
DISCUSSION
In the last two decades, little advancement has been made in cure rates of AML, the most common cause of acute leukemia in adults. Thus, new therapies are needed for this disease. We set out to identify pathways that are consistently suppressed in AML with AML-directed drugs. The one-carbon folate pathway emerged at the intersection of genes significantly altered by these agents. Given the recent interest in this pathway in cancer metabolism, we focused on its role in AML, with a specific study of MTHFD2, a mitochondrial enzyme involved in folate metabolism that is consistently down-regulated across all of the probed datasets and highly differentially expressed in cancer compared with normal cells.
One-carbon folate metabolism is a complex network involving several amino acids, nucleotide synthesis, support of cellular methylation reactions via methionine and s-adenosylmethionine production, and support of lipid metabolism (Locasale, 2013). In the cytoplasm, serine is converted to formate by the trifunctional enzyme MTHFD1. In the mitochondria, the dehydrogenase and cyclohydrolase activities are performed by MTHFD2, whereas the synthetase activities are performed by MTHFD1L (Christensen and Mackenzie, 2008). Recent studies have led to an increased appreciation for metabolic reprogramming and its contribution to cancer pathogenesis. Although there is a long history of targeting folate metabolism in the treatment of patients with cancer, the precise role of glycine/serine and one-carbon folate metabolism in mitochondria and its potential contribution to cancer metabolic reprogramming, have only recently come to light. Glycine metabolism has been correlated with rates of cancer cell proliferation, and antagonizing glycine synthesis through suppression of SHMT2 was shown to decrease cancer cell growth (Jain et al., 2012). In the same year, glycine decarboxylase and other glycine/serine-related enzymes, including SHMT2, were found to promote cellular transformation and tumorigenesis in lung cancer models (Zhang et al., 2012). Additionally, SHMT2 was found to drive glioma cell proliferation in the presence of ischemia, making the cells sensitive to inhibition of the glycine cleavage system (Kim et al., 2015). In an analysis of differentially expressed mRNA in cancer tissues, Nilsson et al. (2014) identified enzymes of the one-carbon folate pathway to be among the most differentially expressed metabolic enzymes in cancer compared with normal cells, with MTHFD2 ranking at the top of the list. In addition, MTHFD2 expression correlated with poor survival in patients with breast cancer (Liu et al., 2014; Nilsson et al., 2014). The combination of its high expression in cancer cells and restricted pattern of expression in normal tissues made MTHFD2 an attractive target for further exploration.
The exact mechanistic role of MTHFD2 in cancer is still a topic of debate. The mitochondrial one-carbon folate pathway and MTHFD2 have been suggested to provide mitochondrial-derived one-carbon units in the form of formate for purine biosynthesis (Christensen and Mackenzie, 2008). However, we could not rescue the effects of MTHFD2 suppression in AML cell lines with the addition of up to 10 mM formate concentrations, a similar observation to that of Nilsson et al. (2014). More recently, several publications supported the role of mitochondrial one-carbon metabolism in redox homeostasis and NADPH production (Fan et al., 2014; Ye et al., 2014). Fan et al. (2014) demonstrated a decrease in the NADPH/NADP+ ratio with MTHFD2 knockdown in HEK293T cells, and Ye et al. (2014) demonstrated a decrease in the NADPH/NADP+ ratio with SHMT2 knockdown only in hypoxic conditions in neuroblastoma cells (Fan et al., 2014; Ye et al., 2014). Given that MTHFD2 is an NAD+ dependent enzyme, we focused on measuring NAD+/NADH ratio in AML cells after MTHFD2 suppression and did not find a consistent difference in this ratio in the AML cell line setting (unpublished data), possibly due in part to the fact that the whole cell NAD+/NADH ratio is dominated by cofactor levels in the cytosol, and MTHFD2 knockdown is expected to affect the mitochondrial NAD+/NADH ratio. Tracing compartmentalized NADPH metabolism (Lewis et al., 2014) was also not possible because MTHFD2 suppression resulted in a profound decrease in cellular α-ketoglutarate levels. In fact, levels of all TCA cycle metabolites were suppressed, potentially because loss of MTHFD2 alters mitochondrial redox metabolism. Decreases in TCA intermediates were also observed with impaired mitochondrial NAD+/NADH metabolism (Sullivan et al., 2015).
Despite a fall in all TCA cycle intermediates, there was a significant increase in the succinate to α-ketoglutarate ratio in MOLM-14 cells after MTHFD2 suppression, a possible metabolic mechanism for the inhibition of stem cell signatures and the induction of differentiation of AML cells. Recent work from Carey et al. (2015) suggests that naive embryonic stem cells maintain an elevated α-ketoglutarate to succinate ratio that promotes pluripotency through histone and DNA demethylation. It will be important, in future studies, to explore the effects of MTHFD2 suppression on the epigenetic state of AML cells.
The limited expression of MTHFD2 in normal adult tissues, and high level of expression in cancer cells, may in part be explained by the regulation of MTHFD2 expression by MYC. MYC is a master regulator of cell proliferation, affecting regulation of immortality, cell cycle progression, genetic instability, apoptosis and metabolism (Dang, 2012). In cancer, pathological activation of MYC commonly plays a key role in disease pathogenesis. MYC stimulates expression of many mitochondrial genes that are encoded in the nucleus and even regulates mitochondrial biogenesis itself (Li et al., 2005). In the Connectivity Map, an MTHFD2 knockdown signature recapitulated changes in expression associated with knockdown of MYC. Using publicly available ChIP-Seq data, we identified MYC binding at the promoter sites of SHMT2, MTHFD2, and MTHFD1L and preferential binding of MYC at MTHFD2 promoter sites in cancer cells compared with nonmalignant cells. We then performed ChIP-qPCR and confirmed this finding in AML cell lines. Previously, MYC has been implicated in controlling the one-carbon folate pathway, especially in the presence of hypoxia (Nikiforov et al., 2002; Ye et al., 2014), though this has not been previously studied in AML. In future studies, it may also be informative to perform metabolic profiling with MYC suppression by BET bromodomain inhibitors alone and in combination with MTHFD2-specific suppression.
Based on the expression level of MTHFD2 in hematopoietic stem cells, MTHFD2 suppression would be expected to have some bone marrow toxicity. Indeed, we observed highly variable suppression of colony formation in normal CD34+ cells with MTHFD2-directed shRNAs in four independent experiments (unpublished data). The variability likely reflects the challenges of transducing CD34+ cells, experimental differences in levels of knockdown, and variability in donor CD34+ cells. However, given the low expression of MTHFD2 in other adult tissues, our expectation is that an MTHFD2 inhibitor would be more selective than other inhibitors of the one-carbon folate pathway, such as methotrexate, on normal adult tissues. Accordingly, MTHFD2 suppression has minimal effect on the growth of 293T or Ba/F3 cells.
Predictors of response are essential to successful cancer drug development. FLT3-ITD emerged as a biomarker of response to MTHFD2 suppression at the intersection of an shRNA screen and in silico query of genome-wide expression profiling of primary patient AML cells. FLT3-ITD is a gain-of-function mutation and is one of the most common alterations in AML, accounting for ∼20% of adult AML (Marcucci et al., 2011). AML with FLT3-ITD is characterized by higher rates of relapse, leading to poor overall survival of as low as 25% (Thiede et al., 2002). Inhibitors of FLT3 have recently been reported to improve survival in combination with chemotherapy in patients with FLT3 mutant AML (Stone et al., 2015). It will thus be important to test FLT3 inhibitors in combination with MTHFD2 suppression in future studies. Although the precise mechanism for increased sensitivity to MTHFD2 suppression in FLT3-ITD AML is uncertain, one might speculate that the increased production of reactive oxygen species by internal tandem duplications of FLT3 may render greater sensitivity (Sallmyr et al., 2008; Godfrey et al., 2012; Stanicka et al., 2015).
This work argues that MTHFD2 inhibitors are needed to further examine targeting this enzyme as a novel treatment strategy in AML. All of the previously published work and the studies presented here have relied on shRNA-directed suppression of MTHFD2, a limitation to the studies. It will be critical to validate that enzymatic inhibition of the target reproduces the effects seen with loss of total protein. Moreover, small-molecule probes will empower the mechanistic dissection of this target as the kinetics of shRNA suppression are slower than acute inhibition with a compound.
In summary, this study defines a critical role for MTHFD2 in AML and nominates FLT3-ITD as a biomarker of response to MTHFD2 suppression. With differential expression of MTHFD2 in cancer tissues, and its preferential expression in embryonic tissues, targeting of MTHFD2 may offer a superior therapeutic window for inhibition in comparison with drugs in current use that target the one-carbon folate metabolism more broadly.
MATERIALS AND METHODS
Cell culture and primary AML cells
U937, HL-60, and 293T cells were purchased from ATCC. Ba/F3 cells were purchased from DSMZ. THP-1, MV4-11, and MOLM-14 cells were provided by Scott Armstrong. MOLM-13 cells were provided by Benjamin Ebert. All cell lines were maintained in RPMI 1640 (Cellgro) supplemented with 1% penicillin/streptomycin (PS; Cellgro) and 10% FBS (Sigma-Aldrich) at 37°C with 5% CO2. The 293T cells were maintained in Dulbecco’s modified Eagle’s medium (Invitrogen) supplemented with 10% FBS (Invitrogen) and 1% PS. Ba/F3 cells were maintained in RPMI-1640 supplemented with 1% PS, 10% FBS, and IL-3 (PeproTech).
Primary patient AML blasts were collected from peripheral blood or bone marrow aspirate after obtaining informed consent under Dana-Farber Institute Internal Review Board–approved protocols. Mononuclear cells were isolated using Ficoll-Paque Plus (GE Healthcare), and red blood cells were lysed (eBioscience). AML blasts from patients were maintained in SFEM (Stemcell Technologies) with SCF, FLT3L, IL-3, IL-6, and GCSF (PeproTech). For drug treatment of primary AML cells, RPMI-1640 with PS and FBS were used.
Murine MLL-AF9–expressing leukemia cells were obtained from a quaternary bone marrow transplant model from Benjamin Ebert’s laboratory. Cells were maintained in SFEM media (Stemcell Technologies) with murine IL-3, IL-6, and SCF cytokines (PeproTech).
Compounds
JQ1 was provided by J. Bradner (Dana-Farber Cancer Institute, Boston, MA). JQ1 was diluted in DMSO.
Plasmids and shRNA constructs
Oligonucleotides encoding human and murine shRNAs were obtained from The RNAi Consortium (TRC) and were cloned into pLKO.1 vectors as described previously (Moffat et al., 2006). Sequences targeted by each of these hairpins are listed in Table S6. shRNA constructs targeting MTHFD2 with miR30 expression cassettes were designed and delivered via a TRMPVIR vector as previously described (Puissant et al., 2014). For virus production, 12 µg of the aforementioned vector with 6 µg pCMV-GAG/POL and pCMV-VSVG (for retroviral infection of human cells) or 6 µg pCMV8.9 and pCMV-VSVG (for lentiviral infection of human or murine cells, respectively) packaging vectors were transfected into the 293 packaging cell line using X-tremeGENE 9 (Roche), and the resulting viral supernatants were harvested as previously described (Banerji et al., 2012). The lentiviral viruses were concentrated using PEG-it Virus Precipitation Solution (SBI Biosciences).
Morphological evaluation
Cellular morphology was evaluated on day 7 after infection of AML cell lines. Cytospins were prepared and stained with May-Grunwald Giemsa (Sigma-Aldrich) to evaluate cellular morphology. Images were acquired by light microscopy under oil at 1,000× magnification with a Bx41 microscope (Olympus) and SPOT software.
Viability assays
For assessment of growth, cells were plated in triplicate and manually counted with Trypan blue exclusion on the indicated days. Additionally, viability was evaluated using the CellTiter-Glo Luminescent Cell Viability Assay (Promega). Luminescence was measured using FLUOstar Omega from BMG Labtech. For the glycine deprivation assay, cells were washed in PBS, and then resuspended in custom-made glycine-free RPMI media (GIBCO) with or without glycine. Glycine solution was added at 140 µM. Dialyzed FBS was used to minimize the contribution of glycine from FBS.
Flow cytometry
The cell surface marker Cd11b (IM0530U; Beckman Coulter) was measured at day 7 after infection with the pLKO.1 vectors. For cell cycle analysis, AML cells were harvested at the indicated time points, washed, and fixed in ethanol, and then resuspended in 49 µg/ml propidium iodide (Sigma-Aldrich) and 100 µg/ml of RNase A (QIAGEN). Cell death was assessed using flow cytometric analysis of Annexin V and propidium iodide staining according to the manufacturer’s instructions (eBioscience). Samples were analyzed on a Fortessa analyzer. Data analysis was completed using ModFit Software for cell cycle analysis and FlowJo software for the remainder of flow cytometry data.
Immunoblotting
Cells were lysed in Cell Signaling Lysis Buffer (Cell Signaling Technology), as previously reported (Banerji et al., 2012), and resolved by gel electrophoresis using Novex 4–12% Bis-Tris Gel (Invitrogen), transferred to nitrocellulose membranes (Bio-Rad Laboratories), and blocked for 1 h in 5% BSA (Sigma-Aldrich). Blots were incubated in primary antibody to MTHFD2 (ab56772; Abcam), MYC (5605; Cell Signaling Technology), MTHFD1L (16113–1-AP; Protein Tech), SHMT2 (155230; Abcam), Actin (MS-1295-P; Neomarkers), and Vinculin (ab18058; Abcam), followed by the secondary antibodies anti–rabbit HRP (NA9340V; GE Healthcare) or anti–mouse HRP (NA9310V; GE Healthcare). Bound antibody was detected using Western Lightning Chemiluminescence Reagents (Perkin Elmer).
Methylcellulose colony-forming assay
U937, MOLM-14, and THP-1 cells were infected with shRNAs directed against LacZ (shControl) or MTHFD2. Puromycin was added for selection after 2 d. After 3 d of selection, cells were counted by Trypan blue exclusion, and 3 × 104 cells were plated at 1:10 (vol/vol) in methylcellulose (ClonaCell-TCS Medium) with 1% PS and 1 µg/ml puromycin.
Primary patient AML blasts were obtained as described above. These cells were maintained in StemSpan SFEM (StemCell Technologies) medium supplemented with 20 ng/ml IL-3 (PeproTech), 20 ng/ml IL-6 (PeproTech), 20 ng/ml GCSF (PeproTech), 100 ng/ml FLT3-Ligand (PeproTech), and 100 ng/ml SCF (PeproTech). They were serially transduced with concentrated lentivirus, with shRNA directed against LacZ or MTHFD2 by two rounds of spin infection (6 h at 1,800 g) each at 24-h intervals using plates coated with 50 µg/ml retronectin (Takara Bio Inc.) before a 48-h antibiotic selection. Viable cells (1.5 × 103 cells/ml) were then plated into semisolid methylcellulose medium (MethoCult; StemCell Technologies) supplemented with selection antibiotics and cytokines. After at least 10 d, the colony number was evaluated after MTT staining of the colonies (R&D Systems).
Gene expression–based high-throughput screening (GE-HTS)
The GE-HTS assay, using ligation-mediated amplification (LMA) and a fluorescent bead-based detection, was performed as previously described (Banerji et al., 2012). The weighted summed score was used to compare differentiation signature induction, as previously described (Banerji et al., 2012).
In vivo studies
U937 or MV4-11 cells were transduced with pMMP-LucNeo retrovirus and selected with neomycin at 1 mg/ml. U937-LucNeo or MV4-11-LucNeo cells were transduced with lentiviruses encoding shRNAs targeting MTHFD2 (shMTHFD2_3 and shMTHFD2_5) or LacZ (shControl). Transduced cells were selected with 1 µg/ml puromycin for 48 h, and viable cells were counted using trypan blue exclusion. For U937-LucNeo cells, 5 million cells per mouse were injected via tail vein into 7-wk-old NSG mice (The Jackson Laboratory). For MV4-11-LucNeo cells, 1.5 million cells per mouse were injected via tail vein into 6–8-wk-old NSG mice (The Jackson Laboratory). Leukemia burden was serially assessed using noninvasive bioluminescence imaging by injecting mice with 75 mg/kg i.p. d-Luciferin (Promega), anesthetizing them with 2–3% isoflurane, and imaging them on an IVIS Spectrum (Caliper Life Sciences). A standardized region of interest (ROI) encompassing the entire mouse was used to determine total body bioluminescence, with data expressed as photons/s/ROI (ph/s/ROI). Additionally, leukemia burden was assessed in the MV4-11-LucNeo model using flow cytometry analysis of the percent of human CD45+ leukemic cells within mice bone marrow at 16 d after injection.
The U937-LucNeo study was conducted under the auspices of protocols approved by the Dana-Farber Cancer Institute Animal Care and Use Committee. The MV4-11-LucNeo study was reviewed and approved by the Massachusetts Institute of Technology Committee on Animal Care (Cambridge, MA).
U937 cells were transduced with the pMMP-LucNeo retrovirus and selected with neomycin at 1 mg/ml. After selection, luciferase-expressing U937 cells were transduced with retroviral doxycycline-inducible miR30-shRNA constructs, either control or constructs expressing two MTHFD2-targeting shRNAs. 1.5 million cells were subsequently injected via tail vein into 6–8-wk-old nude mice (The Jackson Laboratory) and mice were imaged weekly on an IVIS Spectrum (Caliper Life Sciences) to assess the bioluminescence level. At a significant detection level of bioluminescence (∼7–10 d after cell injection), mice were treated daily by i.p injection with 20 µg/ml doxycycline to quickly induce hairpin expression, and drinking water was supplemented with 1 mg/ml doxycycline and 5% sucrose throughout the experiment.
MLL-AF9-DsRed L-GMP cells were transduced with lentivirus encoding shRNAs targeting Mthfd2 (shMthfd2_A1, shMthfd2_A2, and shMthfd2_A4) or LacZ (shControl) and selected with 1 µg/ml puromycin for 24 h before tail vein injection into sublethally irradiated 6-wk-old C57BL/6 recipient mice. Leukemia burden was assessed using flow cytometry analysis of the percent of DsRed-positive MLL-AF9 leukemic cells within mice bone marrow at 7 and 12 d after injection. The Massachusetts Institute of Technology Committee on Animal Care reviewed and approved these mouse experiments.
RNA extraction and qRT-PCR
RNA was extracted from cells with an RNeasy kit (QIAGEN). Probes for qRT-PCR were obtained from Applied Biosciences and are listed in Table S7. Data were collected in technical quadruplicate, analyzed using the ΔΔCT method, and plotted as the percentage of transcript compared with the negative control condition.
ChIP-qPCR
ChIP-qPCR was performed as previously described by Bernt et al. (2011). In brief, cross-linking was performed in cell culture medium containing 1% formaldehyde with gentle rotation for 5 min in room temperature. Fixation was stopped by the addition of glycine (125 mM final concentration). Fixed cells were washed twice in ice-cold PBS, and then resuspended in SDS lysis buffer (1% SDS, 10 mM EDTA, 50 mM and Tris-HCl, pH 8.1, supplemented with Complete mini-protease inhibitor cocktail [Roche]). Chromatin was sheared to ∼200-bp fragments by Covaris ultrasonication. MYC (Santa Cruz Biotechnology, Inc.) antibody was used for ChIP. Precipitates were washed sequentially with ice cold low-salt wash (0.1% SDS, 1% Triton-X-100, 2 mM EDTA, 20 mM Tris-HCl, pH 8.1, and 150 mM NaCl), high salt wash (0.1% SDS, 1% Triton-X-100, 2 mM EDTA, 20 mM Tris-HCl, pH 8.1, and 500 mM NaCl), LiCl wash (0.25 M LiCl, 1% IGEPAL CA-630, 1% deoxycholic acid, 1 mM EDTA, and 10 mM Tris-HCl, pH 8.1), and TE wash (1 mM EDTA and 10 mM Tris-HCl, pH 8.1) and eluted in elution buffer (1% SDS and 0.1 M NaHCO3). Eluted DNA fragments were analyzed by qPCR. Primers used for ChIP-qPCR are as follows: MTHFD2 (forward: 5′-TCGGACTTCGCTTACGTTTAATAA-3′ reverse: 5′-GCCACTGGCTGTCTGTTGAG-3′), MTHFD1L (forward: 5′-GTGGGCGTGGGCATCTC-3′ reverse: 5′-GGCGCGATTGCATCAGA-3′), SHMT2 (forward: 5′-TGTCCTCATTCTGGCGATCA-3′ reverse: 5′-TCCCCTAATGCCACGTGAAA-3′), NPM1 (forward: 5′-GTTGGAGCGGGTAGATGCA-3′ reverse: 5′-GCACGCGAGGTAAGTCTACGT-3′), and ZIC3 (forward: 5′-CCTTTCCGACTACGGCACTTC-3′ reverse: 5′-TCATGGGTGGGACGTTCTG-3′).
Global metabolite profiling
Metabolite profiles of lipid and polar metabolite extracts from cultured cells and spent media were measured using several liquid chromatography tandem mass spectrometry (LC-MS) methods. Lipids were extracted from cells cultured in 6-well plates using 800 µl of isopropanol and from 10 µl of media using 190 µl of isopropanol. Lipid extracts were directly analyzed (10 µl) using BEH C8 chromatography method operated on Nexera X2 U-HPLC (Shimadzu) coupled to an Exactive Plus Orbitrap mass spectrometer (Thermo Fisher Scientific), as previously described (Mascanfroni et al., 2015). Polar metabolites were extracted from cells cultured in 6-well plates using 800 µl of 80% methanol and analyzed using two LC-MS methods and from 10 µl of media using 190 µl of isopropanol. Lipid extracts were directly analyzed using a BEH C8 chromatography method operated on Nexera X2 U-HPLC (Shimadzu) coupled to an Exactive Plus Orbitrap mass spectrometer (Thermo Fisher Scientific) as previously described (Mascanfroni et al., 2015). Polar metabolites were extracted from cultured cells using 800 µl of 80% methanol and from media using either four volumes of 80% methanol for negative ion mode analyses or nine volumes of 74.9:24.9:0.2 vol/vol/vol acetonitrile/methanol/formic acid containing stable isotope-labeled internal standards (valine-d8, Isotec; and phenylalanine-d8, Cambridge Isotope Laboratories) for positive ion mode analyses. Using previously described hydrophilic interaction liquid chromatography (HILIC) methods, negative ion mode analyses of polar metabolites, including central metabolites, were conducted using an AQUITY UPLC (Waters) coupled to a 5500 QTRAP (SCIEX; Townsend et al., 2013), and positive ion mode profiles of polar metabolites, including amino acids, were acquired using a Nexera X2 U-HPLC (Shimadzu) coupled to a Q Exactive Orbitrap (Thermo Fisher Scientific; Mascanfroni et al., 2015).
Measuring succinate to α-ketoglutarate ratio
Polar metabolites were extracted from cells after MTHFD2 knockdown for 4 d using 60% methanol. Metabolites were back extracted with an equal volume of chloroform, dried, and then analyzed by gas chromatography mass spectrometry (GC-MS) as previously described (Lewis et al., 2014).
Measuring intracellular serine and glycine
After MTHFD2 knockdown for 4 d, cells were cultured for 24 h in full media and polar metabolites were extracted and analyzed by GC-MS as described for the measurement of the succinate to α-ketoglutarate ratio.
Stable isotope tracing
Glycine production from serine was measured by culturing MTHFD2 knockdown cells in glycine-free media containing 0.4 mM [U-13C3]serine (Cambridge Isotope Laboratories) supplemented with 10% dialyzed FBS for 24 h. Polar metabolites from spent media were extracted using acetone (300 µl acetone, 20 µl media), dried, and analyzed by GC-MS, as described for the measurement of the succinate to α-ketoglutarate ratio.
Computational biology methods
Development of a core differentiation signature for AML
A core differentiation signature for AML was created by intersecting the lists of genes down-regulated by five prodifferentiation small molecules in studies for which gene expression data are available at the National Center for Biotechnology Information’s Gene Expression Omnibus (GEO) database: HL-60 cell line treated with 1,25-dihydroxy Vitamin D3 (Vitamin D), PMA, and all-trans retinoic acid versus DMSO (Stegmaier et al., 2004; GEO accession no. GSE995; Affymetrix HG-U133A); THP-1 cell line treated with JQ1 versus DMSO (Zuber et al., 2011; GEO accession no. GSE29799; Affymetrix HuGene-1.0 ST Array), and MV4-11 and MOLM-13 cell lines treated with the DOT1L inhibitor EPZ004777 versus DMSO (Daigle et al., 2011; GEO accession no. GSE29828; Affymetrix HG-U133_Plus_2).
Data processing
Each expression dataset was log2 transformed and all probe sets with the maximum log2 expression across samples less than four were filtered. After filtering, the number of probe sets remained for each dataset included: 21,620 out of the 22,215 (GSE995), 26,672 out of the 32,321 (GSE29799), and 26,422 out of the 54,613 (GSE29828). Each filtered dataset was then collapsed to nonredundant genes with distinct HUGO symbols by assigning to each gene the probe set with maximum average intensity. The collapsed datasets were described by 12,891 genes (GSE995), 20,201 genes (GSE29799), and 11,727 genes (GSE29828).
Comparative marker analysis
The individual genes that were differentially down-regulated between samples treated with a differentiation agent versus DMSO were identified by applying the Comparative Marker Selection module from the GenePattern v3.8.1 platform (Reich et al., 2006). A 2-sided signal-to-noise ratio (SNR) test followed by 1,000 permutations of phenotype labels was performed for each analysis. The settings for the SNR parameters were log-transformed-data: yes, complete: no, balanced: no, smooth p-values: yes. A permutation P ≤ 0.05, a FDR(BH) ≤ 0.05, accounting for multiple hypothesis testing, and an absolute fold-change (FC) ≥1.5 served as a cutoff for significant genes. The genes in the core differentiation signature were selected based on two criteria: mean FC of expression between DMSO and each differentiation agent across all datasets ≥1.5 and significant fold change of expression ≥ 1.5 (P ≤ 0.05; FDR[BH] ≤ 0.05) between DMSO and a differentiation agent in at least four out of the five experiments.
Overlap of the core differentiation signature with the collection of KEGG canonical pathways
The core differentiation signature was analyzed for significant overlap with the collection of 186 canonical KEGG pathways (Kanehisa et al., 2014) available from the Molecular Signature Database v4.0 (Subramanian et al., 2005). Computations were performed by applying the hypergeometric test implemented in the MSigDB module Investigate Gene Sets, with the significance cutoff of 10−4 for the FDR q-value.
MTHFD2 knockdown (MTHFD2 KD) RNA-Seq data analysis
MOLM-14 cells were transduced with lentivirus encoding shRNAs targeting MTHFD2 (shMTHFD2_5 hairpin, denoted M5) and control (LacZ, shControl). RNA from six samples, biological duplicates (LacZ1, LacZ2; M5-1, M5-2), and a technical replicate (LacZ3, M5-3) were sequenced as 50+50 bp paired-end reads using Illumina TruSeq strand specific library. The total number of reads for individual samples ranged from 44 to 48 Mb. Quality control tests for the unmapped reads were performed using the FASTQC software (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). The reads were aligned to the GRCh37 (hg19) GENCODE v17 annotated human reference genome by using Bowtie v1.0.0 (Langmead et al., 2009). The average percentage of uniquely mapped reads in the aligned data were 85.11%, with a standard deviation <5%. FPKM scores for genes were computed based on the RSEM R software package (Li and Dewey, 2011). Expression data were estimated as log2(FPKM). The significance of the differential expression between the LacZ and M5 phenotypes was estimated by using the EBSeq method implemented in the EBSeq R library (Leng et al., 2013), based on the significance cutoff (P < 0.05) for the posterior probability. The RNASeq data for the MTHFD2 KD experiment is available for download from GEO under accession no. GSE81062.
MTHFD2 KD GSEA
The GSEA software (v2.0.14; Mootha et al., 2003; Subramanian et al., 2005) was used to identify associations of the MTHFD2 KD phenotype with hematopoietic lineage differentiation gene signatures based on the Differentiation Map data (Novershtern et al., 2011), available in the GEO database as GSE24759. The gene signatures for hematopoietic lineage differentiation were created as described in Novershtern et al. (2011) based on the Student’s t test for differential expression and corrected for multiple hypotheses with the Bonferroni correction (significance cutoffs, P = 0.05 and FDR = 0.05).
GSEA analyzes the list of differentially expressed genes between two experimental classes, ranked by the correlation with the class distinction. The goal of GSEA is to identify groups of genes sharing common biological function (gene sets) that are distributed at the top or at the bottom of the ranked list of differentially expressed genes. GSEA assigns to each gene set an enrichment score (ES) calculated as a running sum statistic by walking down across the ranked list of differentially expressed genes, increasing the sum when encountering genes in the gene set and decreasing it when encountering genes not in the gene set. The significance of the ES is estimated based on a permutation p-value and adjusted for multiple hypotheses testing through FDR. Gene sets with an FDR q-value ≤ 0.25 and a nominal P ≤ 0.05 were considered significant. The gene ranking metric in the weighted enrichment score was the two-sided SNR, and the p-values were calculated using 1,000 permutations of the phenotype.
MTHFD2 Connectivity Map (CMap) gene signature
The MTHFD2 hairpin knockdown signature (MTHFD2-trt.sh) was downloaded from the LINCS-CMap/L1000 database (Lamb et al., 2006; Lamb, 2007). The Library of Integrated Cellular Signatures (LINCS)-Connectivity Map (LINCS-CMap) is a catalog of gene expression data collected from human cells treated with chemical compounds and genetic reagents. The LINCS is a National Institutes of Health program (U54 HG006093) that supports the generation of profiles across multiple cell and perturbation types, as well as assay read-outs. The L1000 assay is a novel mRNA expression profiling technique based on a reduced representation of the genome whereby a collection of 1,000 selected transcripts are monitored and used to computationally infer the remainder of the transcriptome. This approach utilizes Luminex beads and provides a high-throughput (384-well based), low-cost method that can enable large-scale profiling efforts.
The CMap MTHFD2 trt-sh signature was defined based on five hairpins (TRCN0000290804, TRCN0000036550, TRCN0000036551, TRCN0000036553, and TRCN0000290805) in >90 experiments across a panel of cell lines (HEPG2, VCAP, HCC515, HT29, A549, A375, MCF7, PC3, HA1E, and NPC). The genetic perturbation connections of the CMap MTHFD2-trt.sh signature were queried with the Gene Digest Look Up module.
In silico identification of a Folate cluster in primary AML expression data
Primary human AML expression data are as follows: the TCGA LAML RNASeq data were created for 179 primary AML samples obtained from adult de novo patients and were previously described in Cancer Genome Atlas Research Network (2013). Processed FPKM gene level data were downloaded from the TCGA GDAC website (http://gdac.broadinstitute.org). RPKM data were log2 transformed and the genes expressed below the noise threshold RPKM < 1 across all the samples were filtered out. The filtered data were described by 19,796 genes identified through unique Human Genome Organisation (HUGO) names. Clinical annotation, mutation profiles, and survival information were available for download from Cancer Genome Atlas Research Network (2013) and http://gdac.broadinstitute.org.
Wouters AML data are as follows: gene expression profiling on Affymetrix HG-U133_Plus_2 chips for 526 samples of de novo AML was previously described (Wouters et al., 2009) and is available for download from the GEO repository under accession no. GSE14468. Expression data were log2 transformed, and the probe sets with log2 expression below 4 across all the samples were filtered out. The remaining probe sets were collapsed to 16,593 genes described by unique HUGO names by selecting for each gene the probe set with the maximum average expression across all the samples.
ssGSEA projection of primary AML samples on collection of canonical KEGG pathways
ssGSEA is an unsupervised gene enrichment method that calculates a separate enrichment score for each pairing of sample and gene set, independent of sample phenotype labeling (Subramanian et al., 2005; Barbie et al., 2009). ssGSEA is implemented in the ssGSEA Projection module of GenePattern v3.9.0 (Subramanian et al., 2005; Barbie et al., 2009). The method is also implemented in the GenePrerank module of the GSEA v2.0.14 platform. The ssGSEA method assigns to each individual sample—represented as a genome-wide ranked list of genes—an ES with respect to each gene set in a given collection of pathways. ssGSEA provides an unsupervised representation of the gene expression data in the space of gene sets, and thus offers potentially more biological interpretability. The ssGSEA gene set representation can be further analyzed with unsupervised or supervised machine learning and statistical tools. ES is calculated as described for MTHFD2 KD GSEA analysis. A positive ES denotes a significant overlap of the signature gene set with groups of genes at the top of the ranked list, whereas a negative ES denotes a significant overlap of the signature gene set with groups of genes at the bottom of the ranked list. For each sample, the ES was further transformed into a z-score by subtracting the mean of the ES’s assigned to all other samples and by dividing the result to their standard deviation. ssGSEA projection on the collection of 186 KEGG canonical pathways, collections of MYC target gene set signatures, MTHFD2 signatures and JQ1 treatment effect signatures was applied separately to each of the primary AML transcript datasets–TCGA_LAML and Wouters (Cancer Genome Atlas Research Network, 2013). ssGSEA projection on gene set signatures for MTHFD2 KD, CMap MTHFD2 trt-sh, MTHFD2 expression z-score, JQ1 treatment (Zuber et al., 2011), Consensus_JQ1 signature (Puissant et al., 2014), and MYC target gene signatures (MSigDB c2.v04 collection) were performed in each primary tumor dataset. A ssGSEA z-score cutoff 1 was used to assess if a tumor sample is highly enriched in any of the gene signatures.
The associations between samples in the folate cluster and samples enriched in the MTHFD2 expression signature, JQ1 effect, MYC target signature, or with genetic or clinical covariates, were estimated based on a two-tailed Fisher exact test.
Consensus hierarchical clustering for the identification of the folate cluster
Consensus hierarchical clustering based on the mean linking method (Monti et al., 2003), implemented in the R package ConsensusClustering Plus (Wilkerson and Hayes, 2010) was used for clustering the ssGSEA projections on the collection of 186 canonical KEGG pathways for each of the TCGA LAML and Wouters et al. (2009) datasets. Consensus clustering is an unsupervised machine learning approach aimed at identifying robust groups of samples that remain clustered together irrespective of small stochastic perturbations, either in the data or in the clustering procedure. For a preselected number of clusters, k = 2, 3, consensus clustering applies iteratively (e.g., n = 1,000 times), a clustering method (e.g., hierarchical clustering) specific to perturbed instances of the data (e.g., after subsampling 80% of samples and/or 80% of genes), and then splits the data into k clusters based on an agreement matrix that describes the robustness of the clustering solutions along the N iterations. The agreement matrix is defined as a matrix with rows and columns corresponding to samples that record for each pair of samples the fraction of times (out of the N iterations) the samples are clustered together. The agreement matrix is further clustered into k consensus clusters. For each consensus cluster, the core is defined by the subset of samples in the cluster with high pairwise agreement scores (e.g., above the cutoff 0.75), and the border is defined by the samples with low pairwise agreement scores (e.g., below the cutoff 0.25). The overall quality of a clustering solution is measured by silhouette scores defined based on the pairwise similarity of samples within each cluster and by the pairwise dissimilarity of samples within different clusters. The optimal number k of clusters in the data are assessed based on the maximum quality score for various clustering solutions (k = 2, 3, etc.) estimated by a consensus cumulative distribution function (Monti et al., 2003). In each of the primary AML datasets, the samples in the two core clusters identified by applying the consensus clustering approach at a level k = 2, were found to show very high mean pairwise agreement scores (>0.90) and to remain unsplit as core groups in any subsequent clustering into k = 3, 4, 5 clusters. In each primary AML dataset, one of these two core clusters was found to be significantly enriched in the one-carbon pool folate, purine metabolism, and pyrimidine metabolism pathways, and it was called the folate cluster. Gene signatures for the folate cluster were created separately in the TCGA LAML and the Wouters et al. (2009) expression datasets based on the top 300 genes that are significantly up-regulated (or down-regulated) in the folate cluster versus the other cluster (absolute FC of expression ≥ 1.5; P ≤ 0.05; FDR[BH] ≤ 0.05). Heat map projections were created using the GENE-E software.
shRNA screen analysis
The screen was performed from the Achilles v2.4.3 dataset (Cowley et al., 2014) derived from next-generation sequencing deconvolution of 216 cell lines from 22 cancer types, with an shRNA library of 56,903 barcoded shRNAs in lentiviral vectors targeting 14,222 genes, out of which 11,946 genes have complete measurements.
Dependency z-scores for each gene were identified by using the previously described Analytic Technique for Assessment of RNAi by Similarity (ATARiS; Shao et al., 2013). The MTHFD2 dependency z-scores across the 216 lines were computed based on the three MTHFD2 hairpins with reliable consistency scores for on-target gene suppression: TRCN0000290805, TRCN0000290881, and TRCN0000290880. The differences in MTHFD2 dependency z-scores in AML-FLT3.ITD versus AML FLT3.WT cell lines in Achilles data were evaluated based on the Mann-Whitney nonparametric test with the cut-off 0.05 applied to the p-value. The genome-wide gene level dependency z-scores within the individual FLT3-ITD AML cell lines, MOLM-13 and MV4-11, were illustrated through waterfall barplots.
Statistics
Statistical significance was determined by Mann-Whitney, Wilcoxon rank test, or unpaired Student’s t test for pairwise comparison of groups as indicated in figure legends. Log-rank test was used for survival curves. Two-way ANOVA was used for comparisons of two or more groups over time. Permutation test was used for GSEA and MSEA statistical analysis.
Online supplemental material
Table S1 lists the 198 differentially expressed after treatment of AML cells with five AML-targeting agents. Table S2 lists the differentially enriched KEGG pathways in the folate cluster versus other tumors. Table S3 shows the Fisher tests for significant functional correlations with the folate cluster in the TCGA LAML and Wouters et al. (2009) datasets. Table S4 lists 97 genetic perturbations that are positively correlated with MTHFD2 suppression in the connectivity map. Table S5 shows analysis of MYC binding at promoter regions of genes of mitochondrial one-carbon folate pathway in Lin et al. (2012) and ENCODE datasets. Table S6 lists the shRNAs used in this study. Table S7 lists the TaqMan probes used in this study. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20151574/DC1.
Supplementary Material
Acknowledgments
We are grateful to Dr. Elizaveta Freinkman for help with LC-MS experiments. We thank the Bradner laboratory for providing their small-molecule inhibitor JQ1.
This research was supported by National Cancer Institute grants R01 CA140292; K. Stegmaier) and R21 CA198028 (M. Vander Heiden), National Heart, Lung, and Blood Institute grant 5T32 HL07574-32 (Y. Pikman), the National Institute of Child Health and Human Development grant 5K12HD052896-09 (Y. Pikman), the Lauri Strauss Leukemia Foundation (Y. Pikman), Lady Tata Memorial Trust International Award (Y. Pikman), Boston Children’s Hospital Career Development Award (Y. Pikman), Children’s Leukemia Research Association (K. Stegmaier), Cubans Curing Children’s Cancers (4C’s Fund; K. Stegmaier), Project Cupid (K. Stegmaier), When Everyone Survives (K. Stegmaier), Post MD NCIC/Terry Fox Research Fellowship (V. Banerji), CancerCare Manitoba Foundation, the University of Mannitoba (V. Banerji),and the Library of Integrated Cellular Signatures grants U54HG006093 and U54HL127366. K. Stegmaier is a Leukemia and Lymphoma Society Scholar, A. Puissant is a Leukemia and Lymphoma Society Fellow, and Y. Pikman an Alex’s Lemonade Stand Young Investigator Award Recipient.
M. Vander Heiden is a consultant and SAB member for Agios Pharmaceuticals. The authors declare no additional competing financial interests.
Footnotes
Abbreviations used:
- AML
- acute myeloid leukemia
- ChIP
- chromatin immunoprecipitation
- DHFR
- dihydrofolate reductase
- GC-MS
- gas chromatography-mass spectrometry
- GSEA
- Gene Set Enrichment Analysis
- IDH
- isocitrate dehydrogenase
- KEGG
- Kyoto Encyclopedia of Genes and Genomes
- LC-MS
- liquid chromatography-mass spectrometry
- MTHFD2
- methylenetetrahydrofolate dehydrogenase-cyclohydrolase 2
- SHMT2
- serine hydroxymethyltransferase 2
- TCA
- tricarboxylic acid
References
- Astuti D., Latif F., Dallol A., Dahia P.L., Douglas F., George E., Sköldberg F., Husebye E.S., Eng C., and Maher E.R.. 2001. Gene mutations in the succinate dehydrogenase subunit SDHB cause susceptibility to familial pheochromocytoma and to familial paraganglioma. Am. J. Hum. Genet. 69:49–54. 10.1086/321282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerji V., Frumm S.M., Ross K.N., Li L.S., Schinzel A.C., Hahn C.K., Kakoza R.M., Chow K.T., Ross L., Alexe G., et al. 2012. The intersection of genetic and chemical genomic screens identifies GSK-3α as a target in human acute myeloid leukemia. J. Clin. Invest. 122:935–947. 10.1172/JCI46465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbie D.A., Tamayo P., Boehm J.S., Kim S.Y., Moody S.E., Dunn I.F., Schinzel A.C., Sandy P., Meylan E., Scholl C., et al. 2009. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 462:108–112. 10.1038/nature08460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baysal B.E., Ferrell R.E., Willett-Brozick J.E., Lawrence E.C., Myssiorek D., Bosch A., van der Mey A., Taschner P.E., Rubinstein W.S., Myers E.N., et al. 2000. Mutations in SDHD, a mitochondrial complex II gene, in hereditary paraganglioma. Science. 287:848–851. 10.1126/science.287.5454.848 [DOI] [PubMed] [Google Scholar]
- Bernt K.M., Zhu N., Sinha A.U., Vempati S., Faber J., Krivtsov A.V., Feng Z., Punt N., Daigle A., Bullinger L., et al. 2011. MLL-rearranged leukemia is dependent on aberrant H3K79 methylation by DOT1L. Cancer Cell. 20:66–78. 10.1016/j.ccr.2011.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan C.W., Verhaak R.G.W., McKenna A., Campos B., Noushmehr H., Salama S.R., Zheng S., Chakravarty D., Sanborn J.Z., Berman S.H., et al. TCGA Research Network . 2013. The somatic genomic landscape of glioblastoma. Cell. 155:462–477. 10.1016/j.cell.2013.09.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network 2013. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 368:2059–2074. 10.1056/NEJMoa1301689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey B.W., Finley L.W.S., Cross J.R., Allis C.D., and Thompson C.B.. 2015. Intracellular α-ketoglutarate maintains the pluripotency of embryonic stem cells. Nature. 518:413–416. 10.1038/nature13981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen K.E., and Mackenzie R.E.. 2008. Mitochondrial methylenetetrahydrofolate dehydrogenase, methenyltetrahydrofolate cyclohydrolase, and formyltetrahydrofolate synthetases. Vitam. Horm. 79:393–410. 10.1016/S0083-6729(08)00414-7 [DOI] [PubMed] [Google Scholar]
- Cowley G.S., Weir B.A., Vazquez F., Tamayo P., Scott J.A., Rusin S., East-Seletsky A., Ali L.D., Gerath W.F., Pantel S.E., et al. 2014. Parallel genome-scale loss of function screens in 216 cancer cell lines for the identification of context-specific genetic dependencies. Sci. Data. 1:140035 10.1038/sdata.2014.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigle S.R., Olhava E.J., Therkelsen C.A., Majer C.R., Sneeringer C.J., Song J., Johnston L.D., Scott M.P., Smith J.J., Xiao Y., et al. 2011. Selective killing of mixed lineage leukemia cells by a potent small-molecule DOT1L inhibitor. Cancer Cell. 20:53–65. 10.1016/j.ccr.2011.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang C.V. 2012. MYC on the path to cancer. Cell. 149:22–35. 10.1016/j.cell.2012.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J., Ye J., Kamphorst J.J., Shlomi T., Thompson C.B., and Rabinowitz J.D.. 2014. Quantitative flux analysis reveals folate-dependent NADPH production. Nature. 510:298–302. 10.1038/nature13236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farber S., Diamond L.K., Mercer R.D., Sylvester R.F., and Wolff J.A.. 1948. Temporary remissions in acute leukemia in children produced by folic acid antagonist, 4-aminopteroyl-glutamic acid. N. Engl. J. Med. 238:787–793. 10.1056/NEJM194806032382301 [DOI] [PubMed] [Google Scholar]
- Godfrey R., Arora D., Bauer R., Stopp S., Müller J.P., Heinrich T., Böhmer S.-A., Dagnell M., Schnetzke U., Scholl S., et al. 2012. Cell transformation by FLT3 ITD in acute myeloid leukemia involves oxidative inactivation of the tumor suppressor protein-tyrosine phosphatase DEP-1/PTPRJ. Blood. 119:4499–4511. 10.1182/blood-2011-02-336446 [DOI] [PubMed] [Google Scholar]
- Jain M., Nilsson R., Sharma S., Madhusudhan N., Kitami T., Souza A.L., Kafri R., Kirschner M.W., Clish C.B., and Mootha V.K.. 2012. Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science. 336:1040–1044. 10.1126/science.1218595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janeway K.A., Kim S.Y., Lodish M., Nosé V., Rustin P., Gaal J., Dahia P.L.M., Liegl B., Ball E.R., Raygada M., et al. NIH Pediatric and Wild-Type GIST Clinic . 2011. Defects in succinate dehydrogenase in gastrointestinal stromal tumors lacking KIT and PDGFRA mutations. Proc. Natl. Acad. Sci. USA. 108:314–318. 10.1073/pnas.1009199108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Goto S., Sato Y., Kawashima M., Furumichi M., and Tanabe M.. 2014. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 42(D1):D199–D205. 10.1093/nar/gkt1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Fiske B.P., Birsoy K., Freinkman E., Kami K., Possemato R.L., Chudnovsky Y., Pacold M.E., Chen W.W., Cantor J.R., et al. 2015. SHMT2 drives glioma cell survival in ischaemia but imposes a dependence on glycine clearance. Nature. 520:363–367. 10.1038/nature14363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labuschagne C.F., van den Broek N.J.F., Mackay G.M., Vousden K.H., and Maddocks O.D.K.. 2014. Serine, but not glycine, supports one-carbon metabolism and proliferation of cancer cells. Cell Reports. 7:1248–1258. 10.1016/j.celrep.2014.04.045 [DOI] [PubMed] [Google Scholar]
- Lamb J. 2007. The Connectivity Map: a new tool for biomedical research. Nat. Rev. Cancer. 7:54–60. 10.1038/nrc2044 [DOI] [PubMed] [Google Scholar]
- Lamb J., Crawford E.D., Peck D., Modell J.W., Blat I.C., Wrobel M.J., Lerner J., Brunet J.-P., Subramanian A., Ross K.N., et al. 2006. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 313:1929–1935. 10.1126/science.1132939 [DOI] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., and Salzberg S.L.. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leng N., Dawson J.A., Thomson J.A., Ruotti V., Rissman A.I., Smits B.M.G., Haag J.D., Gould M.N., Stewart R.M., and Kendziorski C.. 2013. EBSeq: an empirical Bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics. 29:1035–1043. 10.1093/bioinformatics/btt087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis C.A., Parker S.J., Fiske B.P., McCloskey D., Gui D.Y., Green C.R., Vokes N.I., Feist A.M., Vander Heiden M.G., and Metallo C.M.. 2014. Tracing compartmentalized NADPH metabolism in the cytosol and mitochondria of mammalian cells. Mol. Cell. 55:253–263. 10.1016/j.molcel.2014.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., and Dewey C.N.. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 12:323 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Wang Y., Zeller K.I., Potter J.J., Wonsey D.R., O’Donnell K.A., Kim J.-W., Yustein J.T., Lee L.A., and Dang C.V.. 2005. Myc stimulates nuclearly encoded mitochondrial genes and mitochondrial biogenesis. Mol. Cell. Biol. 25:6225–6234. 10.1128/MCB.25.14.6225-6234.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.Y., Lovén J., Rahl P.B., Paranal R.M., Burge C.B., Bradner J.E., Lee T.I., and Young R.A.. 2012. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 151:56–67. 10.1016/j.cell.2012.08.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F., Liu Y., He C., Tao L., He X., Song H., and Zhang G.. 2014. Increased MTHFD2 expression is associated with poor prognosis in breast cancer. Tumour Biol. 35:8685–8690. 10.1007/s13277-014-2111-x [DOI] [PubMed] [Google Scholar]
- Locasale J.W. 2013. Serine, glycine and one-carbon units: cancer metabolism in full circle. Nat. Rev. Cancer. 13:572–583. 10.1038/nrc3557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddocks O.D.K., Labuschagne C.F., Adams P.D., and Vousden K.H.. 2016. Serine metabolism supports the methionine cycle and DNA/RNA methylation through de novo ATP synthesis in cancer cells. Mol. Cell. 61:210–221. 10.1016/j.molcel.2015.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcucci G., Haferlach T., and Döhner H.. 2011. Molecular genetics of adult acute myeloid leukemia: prognostic and therapeutic implications. J. Clin. Oncol. 29:475–486. 10.1200/JCO.2010.30.2554 [DOI] [PubMed] [Google Scholar]
- Mascanfroni I.D., Takenaka M.C., Yeste A., Patel B., Wu Y., Kenison J.E., Siddiqui S., Basso A.S., Otterbein L.E., Pardoll D.M., et al. 2015. Metabolic control of type 1 regulatory T cell differentiation by AHR and HIF1-α. Nat. Med. 21:638–646. 10.1038/nm.3868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffat J., Grueneberg D.A., Yang X., Kim S.Y., Kloepfer A.M., Hinkle G., Piqani B., Eisenhaure T.M., Luo B., Grenier J.K., et al. 2006. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 124:1283–1298. 10.1016/j.cell.2006.01.040 [DOI] [PubMed] [Google Scholar]
- Monti S., Tamayo P., Mesirov J.P., and Golub T.R.. 2003. Consensus clustering: a resampling-based method for class discovery and visualization of gene expression microarray data. Mach. Learn. 52:91–118. 10.1023/A:1023949509487 [DOI] [Google Scholar]
- Mootha V.K., Lindgren C.M., Eriksson K.-F., Subramanian A., Sihag S., Lehar J., Puigserver P., Carlsson E., Ridderstråle M., Laurila E., et al. 2003. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34:267–273. 10.1038/ng1180 [DOI] [PubMed] [Google Scholar]
- Niemann S., and Müller U.. 2000. Mutations in SDHC cause autosomal dominant paraganglioma, type 3. Nat. Genet. 26:268–270. 10.1038/81551 [DOI] [PubMed] [Google Scholar]
- Nikiforov M.A., Chandriani S., O’Connell B., Petrenko O., Kotenko I., Beavis A., Sedivy J.M., and Cole M.D.. 2002. A functional screen for Myc-responsive genes reveals serine hydroxymethyltransferase, a major source of the one-carbon unit for cell metabolism. Mol. Cell. Biol. 22:5793–5800. 10.1128/MCB.22.16.5793-5800.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson R., Jain M., Madhusudhan N., Sheppard N.G., Strittmatter L., Kampf C., Huang J., Asplund A., and Mootha V.K.. 2014. Metabolic enzyme expression highlights a key role for MTHFD2 and the mitochondrial folate pathway in cancer. Nat. Commun. 5:3128 10.1038/ncomms4128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novershtern N., Subramanian A., Lawton L.N., Mak R.H., Haining W.N., McConkey M.E., Habib N., Yosef N., Chang C.Y., Shay T., et al. 2011. Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell. 144:296–309. 10.1016/j.cell.2011.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantaleo M.A., Astolfi A., Indio V., Moore R., Thiessen N., Heinrich M.C., Gnocchi C., Santini D., Catena F., Formica S., et al. 2011. SDHA loss-of-function mutations in KIT-PDGFRA wild-type gastrointestinal stromal tumors identified by massively parallel sequencing. J. Natl. Cancer Inst. 103:983–987. 10.1093/jnci/djr130 [DOI] [PubMed] [Google Scholar]
- Paschka P., Schlenk R.F., Gaidzik V.I., Habdank M., Krönke J., Bullinger L., Späth D., Kayser S., Zucknick M., Götze K., et al. 2010. IDH1 and IDH2 mutations are frequent genetic alterations in acute myeloid leukemia and confer adverse prognosis in cytogenetically normal acute myeloid leukemia with NPM1 mutation without FLT3 internal tandem duplication. J. Clin. Oncol. 28:3636–3643. 10.1200/JCO.2010.28.3762 [DOI] [PubMed] [Google Scholar]
- Patel H., Pietro E.D., and MacKenzie R.E.. 2003. Mammalian fibroblasts lacking mitochondrial NAD+-dependent methylenetetrahydrofolate dehydrogenase-cyclohydrolase are glycine auxotrophs. J. Biol. Chem. 278:19436–19441. 10.1074/jbc.M301718200 [DOI] [PubMed] [Google Scholar]
- Puissant A., Fenouille N., Alexe G., Pikman Y., Bassil C.F., Mehta S., Du J., Kazi J.U., Luciano F., Rönnstrand L., et al. 2014. SYK is a critical regulator of FLT3 in acute myeloid leukemia. Cancer Cell. 25:226–242. 10.1016/j.ccr.2014.01.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich M., Liefeld T., Gould J., Lerner J., Tamayo P., and Mesirov J.P.. 2006. GenePattern 2.0. Nat. Genet. 38:500–501. 10.1038/ng0506-500 [DOI] [PubMed] [Google Scholar]
- Sallmyr A., Fan J., Datta K., Kim K.-T., Grosu D., Shapiro P., Small D., and Rassool F.. 2008. Internal tandem duplication of FLT3 (FLT3/ITD) induces increased ROS production, DNA damage, and misrepair: implications for poor prognosis in AML. Blood. 111:3173–3182. 10.1182/blood-2007-05-092510 [DOI] [PubMed] [Google Scholar]
- Shao D.D., Tsherniak A., Gopal S., Weir B.A., Tamayo P., Stransky N., Schumacher S.E., Zack T.I., Beroukhim R., Garraway L.A., et al. 2013. ATARiS: computational quantification of gene suppression phenotypes from multisample RNAi screens. Genome Res. 23:665–678. 10.1101/gr.143586.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanicka J., Russell E.G., Woolley J.F., and Cotter T.G.. 2015. NADPH oxidase-generated hydrogen peroxide induces DNA damage in mutant FLT3-expressing leukemia cells. J. Biol. Chem. 290:9348–9361. 10.1074/jbc.M113.510495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmaier K., Ross K.N., Colavito S.A., O’Malley S., Stockwell B.R., and Golub T.R.. 2004. Gene expression-based high-throughput screening(GE-HTS) and application to leukemia differentiation. Nat. Genet. 36:257–263. 10.1038/ng1305 [DOI] [PubMed] [Google Scholar]
- Stone R.M., Mandrekar S., Sanford B.L., Geyer S., Bloomfield C.D., Döhner K., Thiede C., Marcucci G., Lo-Coco F., Klisovic R., et al. 2015. The multi-kinase inhibitor midostaurin (M) prolongs survival compared with placebo (P) in combination with daunorubicin (D)/cytarabine (C) induction (ind), high-dose c consolidation (consol), and as maintenance (maint) Therapy in Newly Diagnosed Acute Myeloid Leukemia (AML) Patients (pts) Age 18-60 with FLT3 Mutations (muts): an international prospective randomized (rand) P-controlled double-blind trial (CALGB 10603/RATIFY [Alliance]). Am. Soc. Hematol. Annu. Meet. Plenary Session. [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., and Mesirov J.P.. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 102:15545–15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan L.B., Gui D.Y., Hosios A.M., Bush L.N., Freinkman E., and Vander Heiden M.G.. 2015. Supporting aspartate biosynthesis is an essential function of respiration in proliferating cells. Cell. 162:552–563. 10.1016/j.cell.2015.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiede C., Steudel C., Mohr B., Schaich M., Schäkel U., Platzbecker U., Wermke M., Bornhäuser M., Ritter M., Neubauer A., et al. 2002. Analysis of FLT3-activating mutations in 979 patients with acute myelogenous leukemia: association with FAB subtypes and identification of subgroups with poor prognosis. Blood. 99:4326–4335. 10.1182/blood.V99.12.4326 [DOI] [PubMed] [Google Scholar]
- Townsend M.K., Clish C.B., Kraft P., Wu C., Souza A.L., Deik A.A., Tworoger S.S., and Wolpin B.M.. 2013. Reproducibility of metabolomic profiles among men and women in 2 large cohort studies. Clin. Chem. 59:1657–1667. 10.1373/clinchem.2012.199133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden M.G., Locasale J.W., Swanson K.D., Sharfi H., Heffron G.J., Amador-Noguez D., Christofk H.R., Wagner G., Rabinowitz J.D., Asara J.M., and Cantley L.C.. 2010. Evidence for an alternative glycolytic pathway in rapidly proliferating cells. Science. 329:1492–1499. 10.1126/science.1188015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanharanta S., Buchta M., McWhinney S.R., Virta S.K., Peçzkowska M., Morrison C.D., Lehtonen R., Januszewicz A., Järvinen H., Juhola M., et al. 2004. Early-onset renal cell carcinoma as a novel extraparaganglial component of SDHB-associated heritable paraganglioma. Am. J. Hum. Genet. 74:153–159. 10.1086/381054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg O., Wind F., and Negelein E.. 1927. The metabolism of tumors in the body. J. Gen. Physiol. 8:519–530. 10.1085/jgp.8.6.519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkerson M.D., and Hayes D.N.. 2010. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 26:1572–1573. 10.1093/bioinformatics/btq170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wouters B.J., Löwenberg B., Erpelinck-Verschueren C.A.J., van Putten W.L.J., Valk P.J.M., and Delwel R.. 2009. Double CEBPA mutations, but not single CEBPA mutations, define a subgroup of acute myeloid leukemia with a distinctive gene expression profile that is uniquely associated with a favorable outcome. Blood. 113:3088–3091. 10.1182/blood-2008-09-179895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H., Parsons D.W., Jin G., McLendon R., Rasheed B.A., Yuan W., Kos I., Batinic-Haberle I., Jones S., Riggins G.J., et al. 2009. IDH1 and IDH2 mutations in gliomas. N. Engl. J. Med. 360:765–773. 10.1056/NEJMoa0808710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J., Fan J., Venneti S., Wan Y.-W., Pawel B.R., Zhang J., Finley L.W.S., Lu C., Lindsten T., Cross J.R., et al. 2014. Serine catabolism regulates mitochondrial redox control during hypoxia. Cancer Discov. 4:1406–1417. 10.1158/2159-8290.CD-14-0250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeller K.I., Jegga A.G., Aronow B.J., O’Donnell K.A., and Dang C.V.. 2003. An integrated database of genes responsive to the Myc oncogenic transcription factor: identification of direct genomic targets. Genome Biol. 4:R69 10.1186/gb-2003-4-10-r69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W.C., Shyh-Chang N., Yang H., Rai A., Umashankar S., Ma S., Soh B.S., Sun L.L., Tai B.C., Nga M.E., et al. 2012. Glycine decarboxylase activity drives non-small cell lung cancer tumor-initiating cells and tumorigenesis. Cell. 148:259–272. 10.1016/j.cell.2011.11.050 [DOI] [PubMed] [Google Scholar]
- Zuber J., Shi J., Wang E., Rappaport A.R., Herrmann H., Sison E.A., Magoon D., Qi J., Blatt K., Wunderlich M., et al. 2011. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 478:524–528. 10.1038/nature10334 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.