Kubes et al. show that methicillin-resistant Staphylococcus aureus (MRSA) survive and proliferate inside Kupffer cells. Intracellular MRSA is resistant to neutrophil-killing and antibiotics treatment and, when released into the circulation, can infect other organs.
Abstract
Methicillin-resistant Staphylococcus aureus (MRSA) bacteremia is reaching epidemic proportions causing morbidity, mortality, and chronic disease due to relapses, suggesting an intracellular reservoir. Using spinning-disk confocal intravital microscopy to track MRSA-GFP in vivo, we identified that within minutes after intravenous infection MRSA is primarily sequestered and killed by intravascular Kupffer cells (KCs) in the liver. However, a minority of the Staphylococci overcome the KC’s antimicrobial defenses. These bacteria survive and proliferate for many days within this intracellular niche, where they remain undetected by recruited neutrophils. Over time, the KCs lyse, releasing bacteria into the circulation, enabling dissemination to other organs such as the kidneys. Vancomycin, the antibiotic of choice to treat MRSA bacteremia, could not penetrate the KCs to eradicate intracellular MRSA. However, based on the intravascular location of these specific macrophages, we designed a liposomal formulation of vancomycin that is efficiently taken up by KCs and diminished the intracellular MRSA. Targeting the source of the reservoir dramatically protected the liver but also dissemination to other organs, and prevented mortality. This vancomycin formulation strategy could help treat patients with Staphylococcal bacteremia without a need for novel antibiotics by targeting the previously inaccessible intracellular reservoir in KCs.
Staphylococcus aureus causes much morbidity and collectively, in North America, is responsible for more annual deaths than those caused by HIV and influenza combined (Magill et al., 2014). Skin and soft tissue infections are widespread and are usually treatable; however, in several patients these infections disseminate, leading to bacteremia and sepsis (Lowy, 1998). Furthermore, as a result of the large increase in invasive procedures requiring catheterization, as well as increases in the frequency of heart valve and joint replacement, the incidence of S. aureus bacteremia is increasing (Saginur and Suh, 2008). High mortality rates of up to 30% are documented when S. aureus enters the blood stream, making it one of the most common serious infections worldwide (Mermel et al., 2009; Thwaites et al., 2011). This is compounded by the lack of a staphylococcal vaccine (Fowler and Proctor, 2014) and the emergence of the methicillin-resistant S. aureus (MRSA) strains, making vancomycin one of the few remaining useful antibiotics (Klevens et al., 2006, 2007). Vancomycin requires i.v. administration for 2–6 wk; however, vancomycin use can be limited by its nephrotoxicity. In addition, repeated vancomycin treatment regimens are problematic in as much as they increase the risk of developing resistance to this critical antibiotic.
The need for a 2–6 wk vancomycin administration protocol contrasts with the very effective staphylococcal killing that is observed for this antibiotic in test tubes. This, and the fact that clinical infection relapse is not uncommon, raises the possibility that there is an in vivo staphylococcal reservoir which is poorly accessed by vancomycin (Kullar et al., 2011). Intracellular persistence of S. aureus has been proposed to be an immune evasion mechanism that may concomitantly serve to circumvent antibiotic treatment. Studies dating back over half a century have shown that S. aureus can survive in vitro in endothelial cells, epithelial cells, osteoclasts, and even in professional phagocytes such as neutrophils and macrophages (Rogers, 1956; Gresham et al., 2000; Koziel et al., 2009; Tuchscherr et al., 2011). In fact, it was recently shown that MRSA placed inside cultured cells can survive intracellularly when those cultured cells were injected into mice, and required expensive antibody-based immunotherapy to be eradicated (Lehar et al., 2015). However, there is still no real direct evidence that demonstrates that S. aureus can hide, and perhaps even thrive, intracellularly in vivo, or that this is the reason why antibiotic treatment often fails. Though, in vitro data have undoubtedly demonstrated that MRSA can live inside cultured cells, an in vivo systematic assessment of the pathogenesis of S. aureus within a mammalian host is desperately needed.
We set out to identify a reservoir of MRSA in vivo, using spinning-disk intravital microscopy (SD-IVM) that allowed us to visualize, in real-time, the very rapid movement of cells responding to bacteria within the blood stream. In this study, we repeat that liver-resident macrophages, Kupffer cells (KCs), are the cell type that immediately and predominantly catches MRSA out of the circulation. More importantly, the pathogen survives and grows in a small percentage of these cells and ultimately escapes to colonize other tissues. Based on the first two findings, a simple, inexpensive and rational way to target and eradicate the pathogen was further uncovered. This immunotherapeutic approach could help treat patients with MRSA bacteremia by increasing effectiveness of antibiotics and decreasing the length of administration.
RESULTS AND DISCUSSION
KCs represent an intracellular reservoir
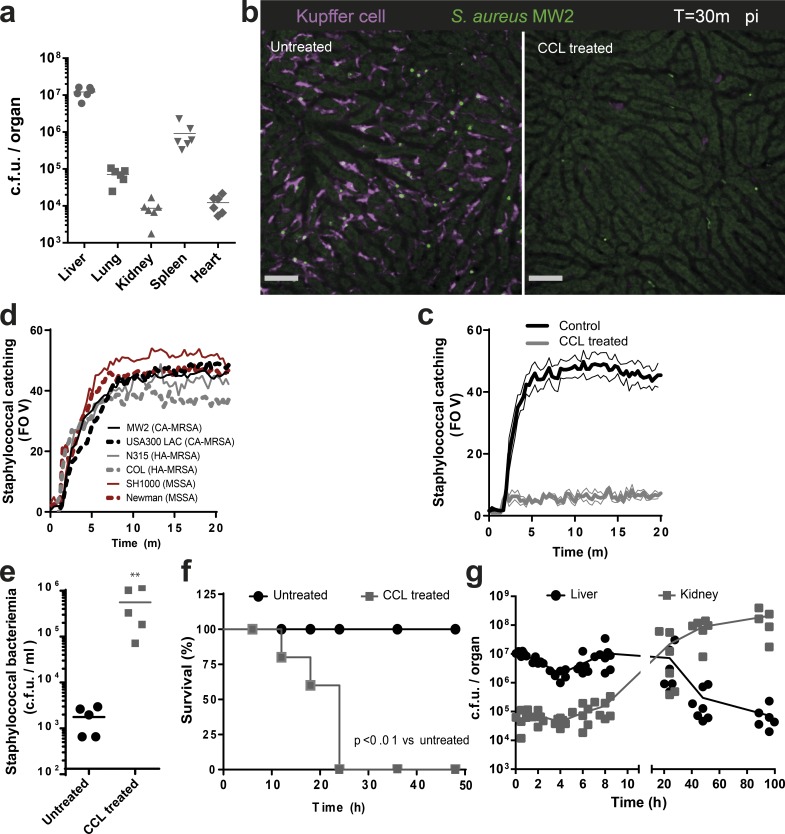
Bacteremia was induced by i.v. injection of MRSA. The liver was the dominant organ; it sequestered 90% of MRSA from the circulation and was 10-fold more efficient than the spleen and 1,000-fold more efficient than the lung, kidney, or heart (Fig. 1 a). SD-IVM showed an F4/80+ macrophage population known as KCs, which reside within the capillary network (sinusoids) of the liver. These intravascular cells were stationary but often had pseudopods in more than one sinusoid (Video 1). Intravenous injection of MRSA-GFP caused rapid capture by F4/80+ KCs (Fig. 1, b and c; and Video 2) but not by endothelium, dendritic cells, hepatocytes, stellate cells, or any other liver cells. The capacity for bacterial sequestration was not fully saturated as the injection of a fourfold higher dose further increased MRSA capture (unpublished data). The KCs were equally capable of catching 6 different methicillin-resistant or susceptible S. aureus strains (Fig. 1 d). In the rest of our experiments, the clinically important community-associated MRSA strain MW2 was used as the model organism and USA300 was used to confirm results. Removal of KCs with clodronate liposomes (CCL) reduced sequestration of MRSA in the liver (Fig. 1, b and c; and Video 2) leading to persistent bacteremia and 100% mortality. At this inoculum of bacteria, no mortality was observed in control mice (Fig. 1, e and f), indicating a crucial role for KCs in clearing bloodborne MRSA.
Figure 1.
KCs are essential for capturing MRSA from circulation. (a) Staphylococcal dissemination to different organs 30 min after i.v. infection (n = 6 mice; data were pooled from two independent experiments). (b) SD-IVM image of staphylococcal (MW2-GFP; green) catching by KCs (F4/80; purple) in WT (left) or KC depleted liver (right; Video 2). Bars, 50 µm. (c) Enumeration of staphylococcal catching by KCs in the livers of WT mice or CCL-treated mice. (b and c) n = 4 mice; thin lines, SEM. Data were pooled from two independent experiments. (d) Quantification of staphylococcal catching by KCs in mice infected with various S. aureus strains. Black, CA-MRSA strains (MW2 and USA300); gray, HA-MRSA strains (Col and N315); red, MSSA strains (Newman and SH1000). n = 3 per condition. Data were pooled from three independent experiments. (e) Staphylococcal bacteremia 4 h after i.v. infection with MRSA (MW2) in untreated or KC-depleted mice by CCL treatment. n = 5 per treatment group; **, P < 0.01, Student’s t test. (f) Survival of MRSA (MW2)-infected WT mice or CCL-treated mice. n = 5 mice per treatment group; log-rank test. (g) Staphylococcal CFU at various time points after i.v. infection with MRSA (MW2) in liver and kidney. n = 6–8 mice per time point. Data shown are compiled from three independent experiments.
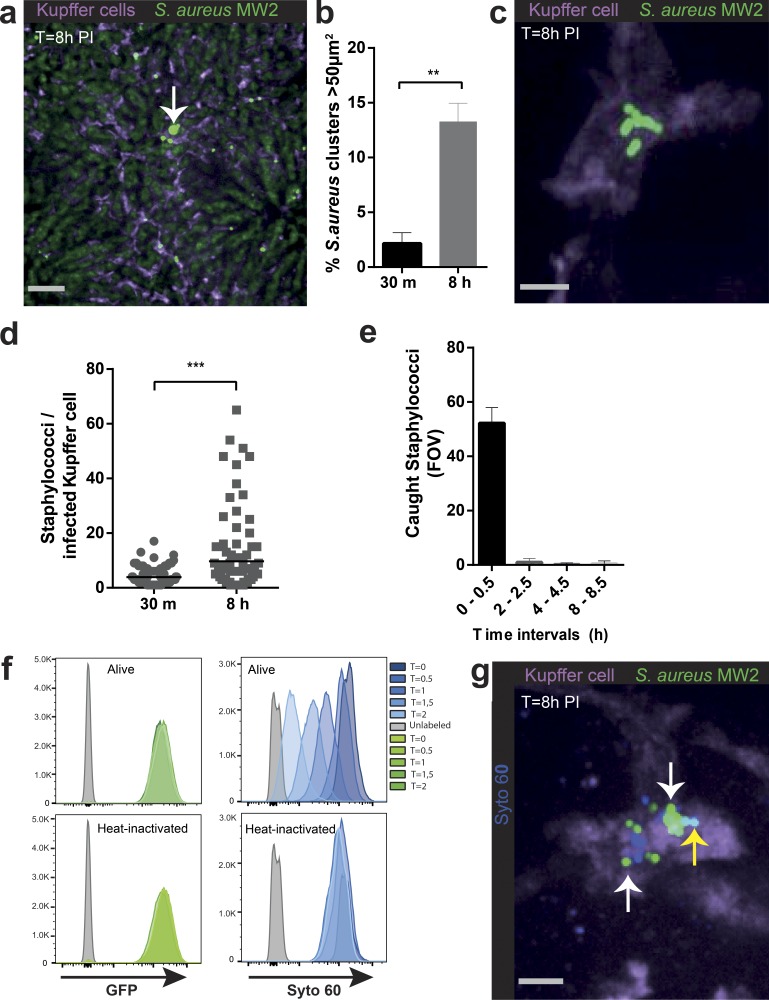
We observed an early fivefold decrease in staphylococcal numbers at 4 h after i.v. injection in the liver. MRSA numbers rapidly rebounded at 8 h and remained high for 24 h, then dropped but persisted for at least 100 h (Fig. 1 g). Interestingly, MRSA was initially near or below the level of detection in kidneys, but the decrease in liver MRSA levels after 24 h coincided with an increase in levels in the kidneys (Fig. 1 g). This suggests that MRSA must overcome an innate immune mechanism to disseminate to other organs (McVicker et al., 2014). Examining the rebound time point in the liver (8 h), revealed that 10–15% of the KCs harbored large clusters of bacteria (>50 µm2; Fig. 2, a and b). These clusters of bacteria were intracellular, as indicated by three dimensional reconstruction showing MRSA contained inside the KCs (Fig. 2 c and Video 3). Quantification of the entire volume of 100 infected KCs revealed that by 8 h, 10% of these macrophages had accumulated >20 MRSA-GFP (Fig. 2 d). Tracking KC sequestration of MRSA-GFP at different time intervals for 8 h revealed minimal new capture events occurring after 30 min (Fig. 2 e). This is consistent with near complete eradication of bacteria from blood (unpublished data). To examine whether MRSA was replicating inside the KCs, a fluorescence based proliferation assay was developed where MRSA-GFP was labeled in vitro with the dye Syto60. This label became more dilute with each subsequent MRSA division, whereas the Syto60 signal remained static in heat-killed bacteria (Fig. 2 f). In vivo, these reporter bacteria were double positive for GFP and Syto60 during initial capture (not depicted), whereas at 8 h, a dilutional spectrum of the Syto60 signal inside KCs was observed (Fig. 2 g). Additionally, the GFP-signal was rapidly degraded when mice were injected with heat-inactivated bacteria (unpublished data), further supporting the belief that the GFP clusters were viable staphylococci growing inside KCs.
Figure 2.
MRSA replicates in KCs. (a) SD-IVM image of liver 8 h after i.v. infection with MRSA (MW2-GFP). Arrow, large cluster of bacteria co-localizing with KCs (purple). Bar, 50 µm. (b) Quantification of SD-IVM images for GFP-clusters >50 µm2/FOV assessed at 30 m and 8 h after i.v. infection with MRSA (MW2-GFP). (a and b) n = 5 per time point. Error bars, SEM; **, P < 0.01, Student’s t test. Data were pooled from two independent experiments. (c) 3D SD-IVM image reconstruction of a staphylococcal cluster (MW2-GFP) co-localizing with KCs (F4/80, purple) 8 h after i.v. infection. Bar, 5 µm (Video 3). (d) Number of MRSA (MW2-GFP) per infected KCs at 30 m or 8 h after i.v. infection. (c and d) n = 100 KCs compiled from eight mice per condition. Error bars, SEM; Student’s t test. (e) Quantification of captured Staphylococci (MW2-GFP) over different time intervals after i.v. infection. n = 3 per time interval. Data were pooled from two independent experiments. (f) Flow cytometric analysis of Syto 60 dye upon culturing MRSA (MW2-GFP) WT or heat-inactivated for different time points. One representative out of three independent experiments is shown. (g) SD-IVM image of dilution of Syto 60 by replicating MRSA within KCs. KCs (F4/80, purple) in mouse liver 8 h after infection with MRSA (MW2-GFP; green) in vitro labeled with Syto 60 (blue). White arrow indicates staphylococci that have lost Syto 60 dye as a result of replication. Yellow arrow indicates MRSA that still have Syto 60. Bar, 5 µm. Shown is one representative image out of five independent experiments.
MRSA replicate inside the phagolysosomes of KCs
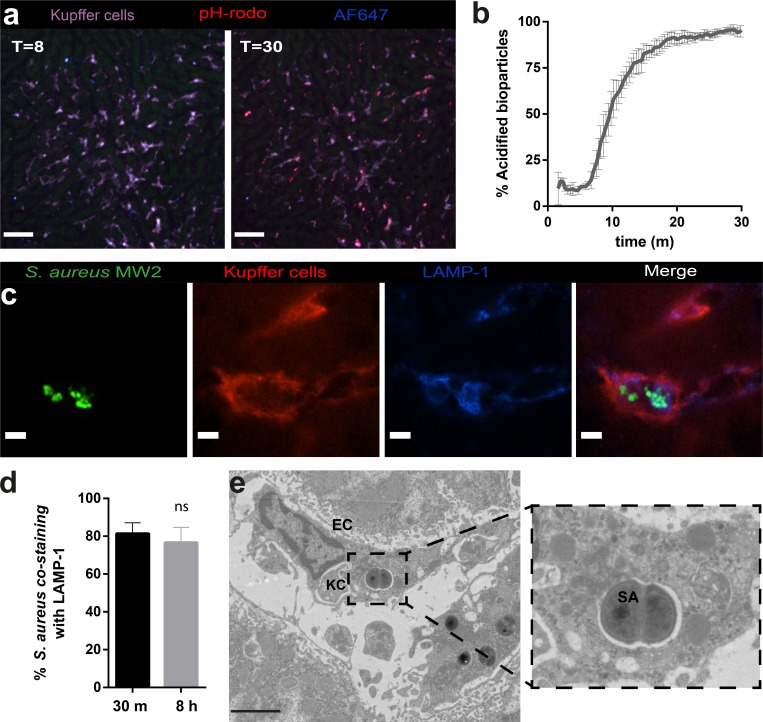
There is conflicting in vitro data regarding the precise intracellular compartment in which staphylococci are replicating. The cytosol (Grosz et al., 2014) and the phagolysosome (Kubica et al., 2008; Flannagan et al., 2016) are the two locations where this has been proposed to occur. To tackle this question in vivo, we used AF647-labeled pH-rodo S. aureus bioparticles as an indicator system for the acidification of phagolysosomes. These bioparticles are methanol-killed staphylococci that were labeled with AF647 as reference color (blue) and pH-rodo that increases in red fluorescence upon acidification. As expected, 95% of initial uptake of S. aureus bioparticles were rapidly internalized in low pH compartments, namely in phagolysosomes (Fig. 3, a and b; and Video 4). Immunofluorescence showed that in KCs, 80% of the intracellular staphylococci were proliferating inside LAMP-1–positive phagolysosomes for at least the first 8 h (Fig. 3, c and d), and electron microscopy confirmed the presence of dividing MRSA inside phagolysosomes of KCs (Fig. 3 e).
Figure 3.
MRSA replicate inside phagolysosomes of KCs. (a) SD-IVM image of mouse livers injected with pH-rodo S. aureus bioparticles (red) additionally labeled with AF647 (blue) as reference fluorophore, showing no acidification at 8 m. Almost all bioparticles were acidified (as indicated by the shift to red staining) at 30 m after infection (Video 4). Bar, 50 µm. (b) Quantification of intracellular acidification of pH-rodo S. aureus bioparticles in KCs over time. Data represent percentage of acidified S. aureus bioparticles compiled from five separate FOV per time point. n = 4 mice. Error bars, SEM. (c) Immunofluorescence image of intracellular localization of replicating Staphylococci. Green, MRSA (MW2-GFP) co-localizes with LAMP-1 (blue) inside KCs (F4/80, purple). Bars, 5 µm. (d) Quantification of co-localization of MRSA with LAMP-1 in KCs at 30 m and 8 h after infection. Data represent percentage of 50 analyzed KCs per mice. n = 3. Error bars, SEM; Student’s t test. (e) Electron microscopy image of dividing intracellular MRSA (MW2) at 8 h after infection. EC, endothelial cell; SA, S. aureus. Bar, 2.5 µm. Shown is one representative image out of three independent experiments.
MRSA clearance in KCs depends on reactive oxygen species (ROS) production
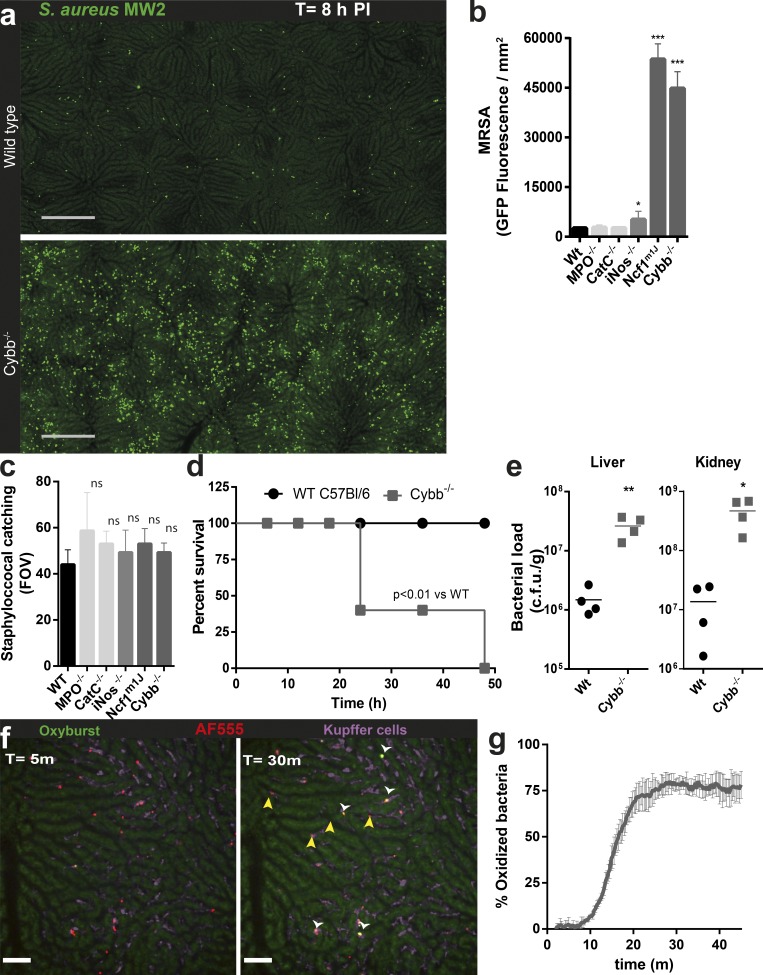
Computer-generated stitched images revealed a significant number of large colonies evenly distributed across the entire liver in WT mice. Loss of phagocyte NADPH oxidase (Ncf1m1j and Cybb−/− mice) revealed a dramatic increase (20–30-fold) in MRSA-GFP inside KCs (Fig. 4 a), but no increase was observed in inducible nitric oxide synthase (iNOS)−/−, myeloperoxidase (MPO)−/−, or cathepsin (cat) c−/− mice (Fig. 4 b). Sequestration of bacteria appeared to be similar in KCs in all aforementioned strains of mice (Fig. 4 c). Early in infection, the increased load of bacteria in Cybb−/− mice was restricted to the liver and spleen (unpublished data). At 24 h, CFU numbers in both liver and kidneys dramatically increased, eventually resulting in 100% mortality at 48 h (Fig. 4, d and e). Collectively, the data strongly suggest that KCs mainly use high levels of ROS to control staphylococcal infection, whereas MPO, cathepsin C (Cat C), and iNOS seem to play limited roles in eradicating MRSA. This is consistent with observations that chronic granulomatous disease patients lacking the capacity to make oxidants have a high risk of staphylococcal liver abscesses (Lublin et al., 2002), and that humans lacking MPO or Cat C deficiency do not suffer from staphylococcal diseases.
Figure 4.
Phagolysosomal ROS production within KCs controls intracellular MRSA replication. (a) Stitched image of mouse livers by SD-IVM from WT and ROS-deficient Cybb−/− mice at 8 h after i.v. infection with MRSA (MW2-GFP; green). Bar, 250 µm. Quantification of Staphylococcal accumulation in the liver (c) 15 min and (b) 8 h after infection in WT, MPO−/−, CatC−/− iNOS−/−, Ncf1m1j, and Cybb−/− mice. Sum of MRSA-GFP fluorescence calculated from 2 mm2 stitched liver images. n = 4 per group. Error bars, SEM. *, P < 0.05; ***, P < 0.001, versus WT by one-way ANOVA with Bonferroni’s posttest. (a–c) Data were pooled from two independent experiments. (d) Survival of MRSA (MW2)-infected WT mice or Cybb−/− mice. n = 5 mice per group; log-rank test. (e) Staphylococcal accumulation in liver and kidney 24 h after i.v. infection with MRSA (MW2) in WT or Cybb−/− mice measured by CFU. n = 4 per condition. *, P < 0.005; **, P < 0.01, Student’s t test. SD-IVM imaging of in vivo ROS production. (f) WT mice infected with oxidation-reporter MRSA (MW2 labeled with AF647 (blue) and OxyBURST (green). White arrow heads indicate MRSA that have been oxidized. Yellow arrowheads show MRSA where no oxidation was detected. Images were taken at 5 and 30 min after infection (Video 5). (g) Percentage of intracellularly oxidized MRSA within KCs over time. Data represent mean of five separate FOV, per time point. n = 4 mice. Error bars, SEM.
Coating MRSA with both the oxidant-sensitive dye OxyBURST and AF555 as reference fluorophores consistently revealed that the majority of MRSA was oxidized by KCs in WT mice, whereas no bacteria were oxidized in Cybb−/− mice (Video 5). Interestingly, ∼20–25% of staphylococci was never oxidized in KCs of WT mice (Fig. 4, f and g). Previous studies have shown functionally distinct KC populations by flow cytometry (Kinoshita et al., 2010; Tacke and Zimmermann, 2014), yet we could not distinguish heterogeneous populations when examining CD11b, CD64, CD115, CD19, Ly6G, and Siglec1 staining by intravital microscopy (unpublished data). In addition, every KC had the capacity to acidify the phagolysosomal compartments. Despite this, we found heterogeneity in the ability of individual phagolysosomes to generate superoxide, both between and within individual KCs (Video 5), suggesting heterogeneous NAPDH oxidase activity in individual phagolysosomes (Tian et al., 2008; Schlam et al., 2013). Although, ROS production is essential for controlling intracellular MRSA replication, some MRSA in phagolysosomes are never oxidized and could thus provide an intracellular niche for MRSA to replicate.
Intracellular localization of MRSA in KCs prevents recognition by neutrophils
It has been suggested that intracellular replication is an immune evasion mechanism in itself. MRSA infection recruits a large number of neutrophils to the liver (Kolaczkowska et al., 2015). Interestingly, these neutrophils patrolled the sinusoids but often crawled past KCs, seemingly unable to detect the large colonies of intracellular bacteria (Video 6). At no point did these neutrophils try to fuse, enter, or phagocytose these infected KCs. However, KCs with very large colonies of MRSA eventually underwent lysis, taking up propidium iodide and releasing MRSA into the vasculature (Video 7). In such instances, one could see neutrophils swarm to these now extracellular bacteria, which were rapidly phagocytosed.
Liposomal-formulated vancomycin, but not free vancomycin, eliminates the intracellular MRSA reservoir in KCs
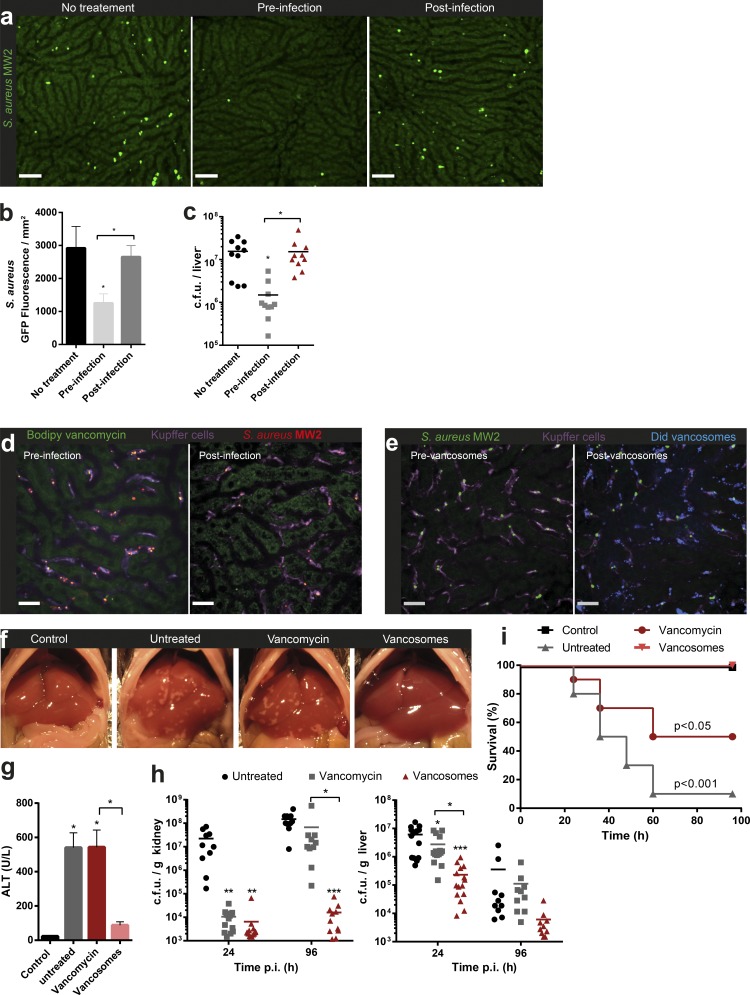
Vancomycin is the most available and most commonly used antibiotic to treat MRSA bacteremia on a global basis. Treatment regiments involve daily intravenous administrations for 2–6 wk, dependent on the source of the infection and interruption in therapy may result in relapse of disease. Even with adequate treatment courses, relapses of S. aureus infection have been well documented (Tong et al., 2015). Pretreatment of mice with vancomycin before infection revealed effective eradication of MRSA-GFP. However administration of vancomycin just 1 h after infection had no impact on the number or size of growing intracellular colonies of MRSA-GFP when examined at 8 h (Fig. 5, a–c). In fact, injecting mice with fluorescent Bodipy-labeled vancomycin 30 min before injecting MRSA-AF555 resulted in binding of vancomycin to the bacteria (Fig. 5 d and Video 8). In contrast, once MRSA was inside KCs, subsequent administration of Bodipy-vancomycin resulted in no color change in intracellular MRSA-AF555, showing that vancomycin could not access intracellular staphylococci (Fig. 5 d and Video 9). This data are in line with clinical studies where prophylactic vancomycin treatment of patients diminishes infection rates (Jensen et al., 1985).
Figure 5.
Intracellular vancomycin delivery targets hepatic MRSA reservoir clearance and suppresses bacterial dissemination. (a) SD-IVM images from mouse livers from untreated mice or mice treated with 50 mg/kg vancomycin i.v. 1 h before or after infection with MRSA (MW2–GFP; green). Images were taken 8 h after infection. Bar, 50 µm. Quantification of stitched SD-IVM images (b) or liver bacterial load (c) of mice treated with 50 mg/kg vancomycin 1 h before or after i.v. infection with MRSA (MW2-GFP). (b) n = 4 or (c) n = 10 per condition. Error bars, SEM. *, P < 0.05, one-way ANOVA with Bonferroni’s posttest. Data shown are compiled from two independent experiments. (d) SD-IVM image of Bodipy-vancomycin staining (green) of MRSA (MW2-AF555, red) injected 15 m before infection (left) or 15 m after infection (right) in mouse (Videos 8 and 9). (e, left) SD-IVM image of mouse liver infected with MRSA (MW2-AF555, red) co-localizing with KC (F4/80, purple) and (right) 15 min after injection of Did-labeled vancosomes (blue; Corresponding Video 10). (d and e) Bar, 25 µm. (f) Representative images of the healthy uninfected liver (control), MRSA-infected liver (24 h; untreated), and MRSA-infected liver treated 1 h after infection with 50 mg/kg vancomycin or vancosomes i.v. (g) Serum ALT levels at 24 h after infection (n = 8 per treatment group, error bars, SEM; *, P < 0.05, one-way ANOVA with Bonferroni’s posttest). (h) CFU levels at 24 and 96 h in mice i.v. infected with MRSA (MW2) with or without treatment 1 h after infection with 50 mg/kg vancomycin or vancosomes. n = 10–15 per treatment group. *, P < 0.05; **, P < 0.05; ***, P < 0.001, one-way ANOVA with Bonferroni’s posttest). Data shown are compiled from 3 independent experiments. (i) Survival of a lethal dose of MRSA (108 CFU MW2)-infected mice with or without treatment 1 h after infection with 50 mg/kg vancomycin or vancosomes. n = 10 per treatment group, Log-rank test versus vancosomes. Data shown are compiled from two independent experiments.
Because vancomycin had poor penetration into KCs, a better vancomycin delivery system was developed to target these cells. There is evidence that vancomycin encapsulated in liposomes (herein termed vancosomes) enables better intracellular killing of MRSA (Pumerantz et al., 2011) and that liposomes are efficiently captured by KCs, raising a possible effective vancomycin delivery system.
Injection of Did-labeled vancosomes resulted in their rapid capture and internalization by KCs (Fig. 5 e and Video 10). Co-localization of large numbers of vancosomes in the same KCs that were harboring MRSA-GFP was observed.
KCs capture the majority of circulating bacteria, perhaps in an attempt to protect other organs with lower regenerative capacity. MRSA infection leads to foci of ischemic nonperfused areas with necrotic hepatocytes, as well as elevated serum alanine transaminase (ALT) levels at 24 h, but damage appeared to resolve within 72 h (Kolaczkowska et al., 2015). Mice that received vancosomes after treatment had no notable ischemic liver areas or elevated serum ALT levels (Fig. 5, f and g) and had a log-fold reduction in CFU levels 24 h after infection compared with untreated controls and vancomycin-treated animals (Fig. 5 h). Treatment of MRSA at the reservoir source by vancosomes prevented dissemination. Indeed, bacterial dissemination to the kidneys was decreased at 24 h by both vancosomes and vancomycin, but by day 4, only vancosome treatment prevented MRSA dissemination to kidneys. This demonstrates that targeting the intracellular MRSA reservoir is far more effective in preventing late-stage dissemination. Most importantly, although vancomycin only rescued 50% of mice treated with a lethal dose of MRSA, treatment with vancosomes allowed for 100% survival of the mice (Fig. 5 i).
Many in vitro studies have demonstrated that S. aureus can survive inside endothelial cells, epithelial cells, fibroblasts, neutrophils, and macrophages, and have predicted an intracellular reservoir of S. aureus in vivo. Lehar et al. (2015) recently described a novel anti–S. aureus antibody conjugated to an antibiotic that is activated only after it is released in the proteolytic environment of the phagolysosome to be more efficacious than conventional antibiotics. This suggests that intracellular S. aureus represents an important component of invasive infections. Here, we report that the major sequestration of MRSA from blood occurs in the F4/80+ KCs that reside in the liver sinusoids. We observed no sequestration in sinusoidal endothelium, hepatocytes, or other liver cells, presumably because hepatocytes have no direct access to the blood and endothelium or other cells lack complement receptors such as CRIg, which is essential for catching S. aureus under shear conditions (Helmy et al., 2006). MRSA survived and replicated in KCs for at least 100 h. Hidden from innate immune mechanisms MRSA, have the propensity to eventually lyse host cells, causing infection relapses, which are a major clinical problem with this kind of infection. In support of this, when livers from MRSA-infected patients were used for organ donation, whole genome sequencing detected the transmission of the donor MRSA strain into the subsequent organ recipient (Altman et al., 2014). This is consistent with liver functioning as a reservoir for this pathogen in humans.
Although the liver reservoir of MRSA was not accessible to vancomycin, our study proposes an alternative way of treating these patients using liposome-based therapy. Indeed, we show that liposomal vancomycin formulations were taken up very efficiently by KCs greatly increasing the intracellular vancomycin thereby killing the intracellular bacteria. This reduced local tissue damage and most importantly reduced dissemination and mortality. Although it is tempting in the clinic to simply use higher doses of vancomycin, this is problematic because it increases renal toxicity. Harnessing the KC’s tremendous capacity to catch foreign particles delivers vancomycin to the previously inaccessible intracellular reservoir of MRSA. As a form of immunotherapy, this could greatly reduce relapses of S. aureus bacteremia, and thus decrease the global disease burden caused by S. aureus.
MATERIALS AND METHODS
Mice
Animal experiments were performed with male adult mice (6–10-wk old), and all experimental animal protocols were approved by the University of Calgary Animal Care Committee and were in compliance with the Canadian Council for Animal Care Guidelines. WT C57BL/6J, Cybb-deficient (Cybb−/−), neutrophil cytosolic factor 1 (Ncf1m1J) spontaneous mutant, iNos−/−, and MPO −/− mice were purchased from The Jackson Laboratory. Cathepsin C–deficient (CatC−/−) mice were a gift from GlaxoSmithKline (Philadelphia, PA). All animals were maintained in a specific pathogen–free environment at the University of Calgary Animal Resource Centre. Mice were housed under standardized conditions of temperature (21–22°C) and illumination (12 h light/12 h darkness) with access to tap water and pelleted food ad libitum.
Antibodies and reagents
Antibodies against CD11b (M1/70), CD68 (FA-11), CD115 (AFS98), and F4/80 (BM8) were obtained from eBioscience. Antibodies against CD64 (X54-5/7), LAMP-1 (1D4B), Siglec-1 (3D6.112), and Ly6G (1A8) were obtained from BioLegend. Anti-CD31 (390) was obtained from Fitzgerald Industries and conjugated with Alexa Fluor 647 protein labeling kit as per the manufacturer’s instructions (Thermo Fisher Scientific). Propidium iodide, OxyBURST Green H2DCFDA SE ester, pHrodo Red S. aureus BioParticles, pH-rodo, Alexa Fluor 647, and 555 NHS Ester, Vancomycin BODIPY, Vybrant DiD Cell-Labeling Solution, and Syto 60 were obtained from Thermo Fisher Scientific.
SD-IVM
A tail vein catheter was inserted into mice after anesthetization with 200 mg/kg ketamine (Bayer Animal Health) and 10 mg kg−1 xylazine (Bimeda-MTC). Surgical preparation of the liver for intravital imaging was performed as previously described (Wong et al., 2011). Mouse body temperature was maintained at 37°C with a heated stage. Image acquisition was performed using Olympus IX81 inverted microscope, equipped with an Olympus focus drive and a motorized stage (Applied Scientific Instrumentation) and fitted with a motorized objective turret equipped with 4×/0.16 UPLANSAPO, 10×/0.40 UPLANSAPO, and 20×/0.70 UPLANSAPO objective lenses and coupled to a confocal light path (WaveFx; Quorum Technologies) based on a modified Yokogawa CSU-10 head (Yokogawa Electric Corporation). Target cells were visualized using fluorescently stained antibodies or fluorescent reporter bacteria. Typically, KCs and neutrophils were stained by i.v. injection of 2.5 µg anti–F4-80 or 3.5 µg anti-ly6G fluorescent conjugated mAbs. Laser excitation wavelengths 491, 561, 642, and 730 nm (Cobolt) were used in rapid succession, together with the appropriate band-pass filters (Semrock). A back-thinned EMCCD 512 × 512 pixel camera was used for fluorescence detection (Hamamatsu). Volocity software (Perkin Elmer) was used to drive the confocal microscope and for 3D rendering, acquisition, and analysis of images. For bacterial catching/acidification and ROS production, five random fields of view (FOV) with 10× objective were selected before injection of bacteria. Acquisition of images was 3 m−1, after 1 m of acquiring background images bacteria/bioparticles were i.v. injected in mice. Find objects function in Volocity software was used to identify individual captured bacteria, bioparticles, and reporter bacteria by KCs (F4/80+ cells in liver) and to determine the amount of fluorescence per particle. Percentage acidified particles or oxidized MRSA within F4/80+ cells was calculated from the total number of identified particles/bacteria from the reference fluorophore (when appropriate, autofluorescent spots were subtracted) and the particles/bacteria that over time became positive (2× background fluorescence) for either acidification (pH-rodo,) or oxidation (OxyBURST).
Staphylococcal strains and culture conditions
Staphylococcus aureus strains COL, N315, SH1000, Newman, MW2, and USA300 were obtained from NARSA (Network on Antimicrobial Resistance in Staphylococcus aureus). These strains, mutants were transformed with pCM29 (Pang et al., 2010) to constitutively express high levels of GFP (Surewaard et al., 2012). Bacteria were grown in brain heart infusion at 37°C while shaking. When appropriate, chloramphenicol (10 µg ml−1) was added for overnight maintenance of the plasmids. For infection experiments, S. aureus strains were subcultured without antibiotics until exponential phase (OD660nm, 1.0) washed with saline once, resuspended in saline, and injected i.v. into the tail vein at 5 × 107 CFU in 200 µl (unless otherwise indicated). Killed S. aureus was prepared either by heating cultures at 65°C for 20 min or by incubation with 1% paraformaldehyde for 30 min at room temperature, followed by washing in saline.
Labeling of Staphylococcal strains
Fresh S. aureus-GFP cultures were washed twice with saline, and labeled at 5 × 108 CFU in 500 µl saline with 10 µM Syto 60 for 30 min at room temperature. Cultures were washed with saline twice, checked for similar Optic density, resuspended, and analyzed by flow cytometry or, alternatively, injected i.v. in mice for replication experiments. Generation of reporter bacteria for lysosome acidification was carried out as follows: pHrodo Red S. aureus BioParticles were labeled at 2 mg ml−1 with 50 µg ml−1 AF647 NHS ester in 100 mM bicarbonate, pH 8.3, buffered saline for 30 min at room temperature under vigorous agitation. Labeled BioParticles were washed twice with PBS and checked for labeling efficiency by flow cytometry or injected i.v. into mice. AF647-labeled pH-rodo BioParticles could be stored for up to 1 wk at 4°C without loss of signal. Generation of reporter bacteria for oxidation or imaging was carried out as follows: fresh staphylococcal cultures or methanol-killed cells were washed twice in saline, resuspended at 5 × 108 CFU in 500 µl in carbonate, pH 8.3, buffered saline, and labeled for 30 min with 20 µg ml−1 AF647 or AF555 NHS ester and/or 60 µg ml−1 OxyBURST Green H2DCFDA SE (DMSO stock) under vigorous agitation. Activation of OxyBURST was accomplished by adding 250 µl 1.5 M hydroxylamine, pH 8.5, and incubating for 30 min on ice. Reporter bacteria were washed twice with PBS and checked for labeling efficiency by flow cytometry or injected i.v. into mice.
Mouse infections and in vivo treatments
For infection experiments, S. aureus strains were subcultured without antibiotics until exponential phase (OD660nm, 1.0), washed with saline once, and then resuspended in saline and injected i.v. into the tail vein at 5 × 107 CFU in 200 µl (unless otherwise indicated). Mice were monitored and sacrificed at various time-points according to experimental design. KC depletion was performed by intravenous injection of 200 µl per mouse (0.69 mol/l) clodronate liposome 48 h before the experiment. 50 mg Kg−1 vancomycin (Sigma-Aldrich) or liposomal encapsulated vancomycin (vancosomes; Sande et al., 2012) was administered i.v. 1 h before or after infection. Alternatively, 6 mg vancomycin liposomes were labeled with 5 µl Did solution in 1 ml saline. Liposomes were washed twice and injected at 10 mg/kg per mouse 30 min after infection of S. aureus while imaging the liver. Mice were prepared for IVM and injected with 50 µg Bodipy FL-vancomycin i.v. 30 min before infection with unlabeled S. aureus MW2, or mice were infected with AF555-labeled S. aureus MW2 30 min before i.v. injection of 50 µg Bodipy FL-vancomycin.
Bacteriological analysis
Anesthetized mice were washed with 70% ethanol under sterile conditions. Blood was collected in a heparinized syringe by cardiac puncture. Samples were then centrifuged at 400 g for 10 min for the retrieval of plasma. Alanine transaminase in the plasma was analyzed by Calgary Laboratory Services. The lungs, liver, heart, kidneys, and spleen were removed after thoracotomy, weighed, and homogenized. For determination of colony forming units (CFU), 30 µl of tissue homogenate or blood was serially diluted, plated onto brain-heart infusion agar plates, and incubated at 37°C for 18 h, and then bacterial colonies were counted.
Immunohistochemistry
Livers of infected animals were dissected, incubated at room temperature in 10% formalin for 16 h, immersed in 30% sucrose solution for 4 h, and then embedded in Tissue-Tek OCT and frozen at −80°C. Samples were cryosectioned (30-µm cuts), mounted on slides, and stored at −80°C. Before staining, slides were warmed to room temperature for 30 min and permeabilized with 0.1% Triton in PBS for 10 min. Slides were washed three times with PBS containing 0.1% Tween, blocked with 10% normal goat serum (NGS) in PBS, washed three times with PBS containing 0.1% Tween, stained with anti-F4/80 (1:100) and anti-LAMP1 (1:500) in antibody diluent (Dako) containing 10% NGS and then washed three times with PBS containing 0.1% Tween. The slides were washed once more with PBS, mounted with fluorescent mounting media (Dako), and viewed with the Olympus IX81 inverted spinning-disc confocal microscope.
Electron microscopy
The cultured cells were processed in situ for fixation, dehydration, infiltration, and embedding in the culture dish. The cells were prefixed with 1.6% paraformaldehyde and 2.5% glutaraldehyde in 0.1 M cacodylate buffer, pH 7.3, for 1 h and post-fixed with cacodylate-buffered 1% osmium tetroxide for 1 h at room temperature. Cells were then dehydrated through graded ethanol and embedded in epon mixture. After polymerizing, the hardened epon layer containing the embedded cells was separated from the plastic culture dish. Under a light microscope, a representative area was selected, trimmed, and glued to resin stub for sectioning. Ultrathin sections were cut with a diamond knife on an ultramicrotome (Ultracut E; Reichert-Jung) and collected on single-hole grids with Formvar supporting film. The sections were stained with aqueous uranyl acetate and Reynolds’s lead citrate and observed under a Hitachi H7650 TEM at 80 kV. The images were acquired with an AMT16000 digital camera mounted on the microscope.
For TEM analyses, mice were i.v. infected with MRSA as described above. Liver tissue sections were processed in situ for fixation, dehydration, infiltration, and embedding on slides. The sections were prefixed with 1.6% paraformaldehyde and 2.5% glutaraldehyde in 0.1 M cacodylate buffer, pH 7.3, for 1 h and postfixed with cacodylate-buffered 1% osmium tetroxide for 1 h at room temperature. Sections were then dehydrated through graded ethanol and embedded in epon mixture. After polymerizing, the hardened epon layer containing the tissue sections was separated from the slide. Under a light microscope, a representative area was selected, trimmed, and glued to resinstub for sectioning. Ultrathin sections were cut with a diamond knife on an ultramicrotome (Ultracut E; Reichert-Jung) and collected on single-hole grids with Formvar supporting film. The sections were stained with aqueous uranyl acetate and Reynolds’s lead citrate and observed under a Hitachi H7650TEM at 80 kV. The images were acquired with an AMT16000 digital camera mounted on the microscope.
Statistical analysis
Statistical comparisons were performed using GraphPad Prism v6.0 software. For two group comparisons, the Student’s t test was used. For multi-group comparisons, one-way ANOVA followed by Bonferroni’s posttest for multiple comparisons adjustment. The applied statistical analyses and the numbers of independent replicates (n) are reported in the figure legends.
Online supplemental material
Video 1 shows in vivo behavior of KCs. Video 2 shows catching of MRSA by KCs. Video 3 shows intracellular accumulation of MRSA in KCs. Video 4 shows staphylococcal-induced acidification of phagolysosomes of KCs. Video 5 shows in vivo ROS production induced by MRSA in KCs. Video 6 shows neutrophils are not attracted to intracellular proliferating MRSA. Video 7 shows that KC lyses induces release of MRSA and attracts neutrophils in swarming behavior. Video 8 shows rapid binding of fluorescent vancomycin to MRSA in circulation. Video 9 shows that fluorescent vancomycin does not access the intracellular MRSA reservoir. Video 10 shows that vancosomes are taken up by MRSA infected KCs. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20160334/DC1.
Acknowledgments
We thank Trecia Nussbaumer for the breeding of mice.
P. Kubes is supported by Alberta Innovates Health Solutions (AIHS), the Canadian Institutes of Health Research, and the Canada Research Chairs Program. B.G.J. Surewaard is partially funded by Marie Currie actions FP7-PEOPLE-2013-IOF (grant no. 627575) and AIHS. J.F. Deniset and F.J. Zemp are financially supported by AIHS.
The authors declare no competing financial interests.
Author contributions: B.G.J. Surewaard, J.D. Deniset, and F.J. Zemp conceived the study and performed experiments, analyzed data, and wrote the manuscript. M. Amrein performed electron microscopy and analyzed data. M. Otto, J. Conly, A. Omri, and R. Yates provided valuable material and critically evaluated the manuscript. P. Kubes wrote the manuscript and directed the study.
Footnotes
Abbreviations used:
- Cat C
- cathepsin C
- iNOS
- inducible nitric oxide synthase
- KC
- Kupffer cell
- MPO
- myeloperoxidase
- MRSA
- methicillin-resistant Staphylococcus aureus
- ROS
- reactive oxygen species
- SD-IVM
- spinning-disk intravital microscopy
References
- Altman D.R., Sebra R., Hand J., Attie O., Deikus G., Carpini K.W., Patel G., Rana M., Arvelakis A., Grewal P., et al. . 2014. Transmission of methicillin-resistant Staphylococcus aureus via deceased donor liver transplantation confirmed by whole genome sequencing. Am. J. Transplant. 14:2640–2644. 10.1111/ajt.12897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flannagan R.S., Heit B., and Heinrichs D.E.. 2016. Intracellular replication of Staphylococcus aureus in mature phagolysosomes in macrophages precedes host cell death, and bacterial escape and dissemination. Cell. Microbiol. 18:514–535. 10.1111/cmi.12527 [DOI] [PubMed] [Google Scholar]
- Fowler V.G. Jr., and Proctor R.A.. 2014. Where does a Staphylococcus aureus vaccine stand? Clin. Microbiol. Infect. 20(Suppl 5):66–75. 10.1111/1469-0691.12570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gresham H.D., Lowrance J.H., Caver T.E., Wilson B.S., Cheung A.L., and Lindberg F.P.. 2000. Survival of Staphylococcus aureus inside neutrophils contributes to infection. J. Immunol. 164:3713–3722. 10.4049/jimmunol.164.7.3713 [DOI] [PubMed] [Google Scholar]
- Grosz M., Kolter J., Paprotka K., Winkler A.C., Schäfer D., Chatterjee S.S., Geiger T., Wolz C., Ohlsen K., Otto M., et al. . 2014. Cytoplasmic replication of Staphylococcus aureus upon phagosomal escape triggered by phenol-soluble modulin α. Cell. Microbiol. 16:451–465. 10.1111/cmi.12233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmy K.Y., Katschke K.J. Jr., Gorgani N.N., Kljavin N.M., Elliott J.M., Diehl L., Scales S.J., Ghilardi N., and van Lookeren Campagne M.. 2006. CRIg: a macrophage complement receptor required for phagocytosis of circulating pathogens. Cell. 124:915–927. 10.1016/j.cell.2005.12.039 [DOI] [PubMed] [Google Scholar]
- Jensen L.J., Aagaard M.T., and Schifter S.. 1985. Prophylactic vancomycin versus placebo in arterial prosthetic reconstructions. Thorac. Cardiovasc. Surg. 33:300–303. 10.1055/s-2007-1014145 [DOI] [PubMed] [Google Scholar]
- Kinoshita M., Uchida T., Sato A., Nakashima M., Nakashima H., Shono S., Habu Y., Miyazaki H., Hiroi S., and Seki S.. 2010. Characterization of two F4/80-positive Kupffer cell subsets by their function and phenotype in mice. J. Hepatol. 53:903–910. 10.1016/j.jhep.2010.04.037 [DOI] [PubMed] [Google Scholar]
- Klevens R.M., Edwards J.R., Tenover F.C., McDonald L.C., Horan T., and Gaynes R.. National Nosocomial Infections Surveillance System . 2006. Changes in the epidemiology of methicillin-resistant Staphylococcus aureus in intensive care units in US hospitals, 1992-2003. Clin. Infect. Dis. 42:389–391. 10.1086/499367 [DOI] [PubMed] [Google Scholar]
- Klevens R.M., Morrison M.A., Nadle J., Petit S., Gershman K., Ray S., Harrison L.H., Lynfield R., Dumyati G., Townes J.M., et al. Active Bacterial Core surveillance (ABCs) MRSA Investigators . 2007. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA. 298:1763–1771. 10.1001/jama.298.15.1763 [DOI] [PubMed] [Google Scholar]
- Kolaczkowska E., Jenne C.N., Surewaard B.G., Thanabalasuriar A., Lee W.Y., Sanz M.J., Mowen K., Opdenakker G., and Kubes P.. 2015. Molecular mechanisms of NET formation and degradation revealed by intravital imaging in the liver vasculature. Nat. Commun. 6:6673 10.1038/ncomms7673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koziel J., Maciag-Gudowska A., Mikolajczyk T., Bzowska M., Sturdevant D.E., Whitney A.R., Shaw L.N., DeLeo F.R., and Potempa J.. 2009. Phagocytosis of Staphylococcus aureus by macrophages exerts cytoprotective effects manifested by the upregulation of antiapoptotic factors. PLoS One. 4:e5210 10.1371/journal.pone.0005210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubica M., Guzik K., Koziel J., Zarebski M., Richter W., Gajkowska B., Golda A., Maciag-Gudowska A., Brix K., Shaw L., et al. . 2008. A potential new pathway for Staphylococcus aureus dissemination: the silent survival of S. aureus phagocytosed by human monocyte-derived macrophages. PLoS One. 3:e1409 10.1371/journal.pone.0001409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kullar R., Davis S.L., Levine D.P., and Rybak M.J.. 2011. Impact of vancomycin exposure on outcomes in patients with methicillin-resistant Staphylococcus aureus bacteremia: support for consensus guidelines suggested targets. Clin. Infect. Dis. 52:975–981. 10.1093/cid/cir124 [DOI] [PubMed] [Google Scholar]
- Lehar S.M., Pillow T., Xu M., Staben L., Kajihara K.K., Vandlen R., DePalatis L., Raab H., Hazenbos W.L., Morisaki J.H., et al. . 2015. Novel antibody-antibiotic conjugate eliminates intracellular S. aureus. Nature. 527:323–328. 10.1038/nature16057 [DOI] [PubMed] [Google Scholar]
- Lowy F.D. 1998. Staphylococcus aureus infections. N. Engl. J. Med. 339:520–532. 10.1056/NEJM199808203390806 [DOI] [PubMed] [Google Scholar]
- Lublin M., Bartlett D.L., Danforth D.N., Kauffman H., Gallin J.I., Malech H.L., Shawker T., Choyke P., Kleiner D.E., Schwartzentruber D.J., et al. . 2002. Hepatic abscess in patients with chronic granulomatous disease. Ann. Surg. 235:383–391. 10.1097/00000658-200203000-00010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magill S.S., Edwards J.R., Bamberg W., Beldavs Z.G., Dumyati G., Kainer M.A., Lynfield R., Maloney M., McAllister-Hollod L., Nadle J., et al. Emerging Infections Program Healthcare-Associated Infections and Antimicrobial Use Prevalence Survey Team . 2014. Multistate point-prevalence survey of health care-associated infections. N. Engl. J. Med. 370:1198–1208. 10.1056/NEJMoa1306801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVicker G., Prajsnar T.K., Williams A., Wagner N.L., Boots M., Renshaw S.A., and Foster S.J.. 2014. Clonal expansion during Staphylococcus aureus infection dynamics reveals the effect of antibiotic intervention. PLoS Pathog. 10:e1003959 10.1371/journal.ppat.1003959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mermel L.A., Allon M., Bouza E., Craven D.E., Flynn P., O’Grady N.P., Raad I.I. II, Rijnders B.J., Sherertz R.J., and Warren D.K.. 2009. Clinical practice guidelines for the diagnosis and management of intravascular catheter-related infection: 2009 Update by the Infectious Diseases Society of America. Clin. Infect. Dis. 49:1–45. 10.1086/599376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang Y.Y., Schwartz J., Thoendel M., Ackermann L.W., Horswill A.R., and Nauseef W.M.. 2010. agr-Dependent interactions of Staphylococcus aureus USA300 with human polymorphonuclear neutrophils. J. Innate Immun. 2:546–559. 10.1159/000319855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pumerantz A., Muppidi K., Agnihotri S., Guerra C., Venketaraman V., Wang J., and Betageri G.. 2011. Preparation of liposomal vancomycin and intracellular killing of meticillin-resistant Staphylococcus aureus (MRSA). Int. J. Antimicrob. Agents. 37:140–144. 10.1016/j.ijantimicag.2010.10.011 [DOI] [PubMed] [Google Scholar]
- Rogers D.E. 1956. Studies on bacteriemia. I. Mechanisms relating to the persistence of bacteriemia in rabbits following the intravenous injection of staphylococci. J. Exp. Med. 103:713–742. 10.1084/jem.103.6.713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saginur R., and Suh K.N.. 2008. Staphylococcus aureus bacteraemia of unknown primary source: where do we stand? Int. J. Antimicrob. Agents. 32(Suppl 1):S21–S25. 10.1016/j.ijantimicag.2008.06.008 [DOI] [PubMed] [Google Scholar]
- Sande L., Sanchez M., Montes J., Wolf A.J., Morgan M.A., Omri A., and Liu G.Y.. 2012. Liposomal encapsulation of vancomycin improves killing of methicillin-resistant Staphylococcus aureus in a murine infection model. J. Antimicrob. Chemother. 67:2191–2194. 10.1093/jac/dks212 [DOI] [PubMed] [Google Scholar]
- Schlam D., Bohdanowicz M., Chatgilialoglu A., Steinberg B.E., Ueyama T., Du G., Grinstein S., and Fairn G.D.. 2013. Diacylglycerol kinases terminate diacylglycerol signaling during the respiratory burst leading to heterogeneous phagosomal NADPH oxidase activation. J. Biol. Chem. 288:23090–23104. 10.1074/jbc.M113.457606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surewaard B.G., Nijland R., Spaan A.N., Kruijtzer J.A., de Haas C.J., and van Strijp J.A.. 2012. Inactivation of staphylococcal phenol soluble modulins by serum lipoprotein particles. PLoS Pathog. 8:e1002606 10.1371/journal.ppat.1002606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tacke F., and Zimmermann H.W.. 2014. Macrophage heterogeneity in liver injury and fibrosis. J. Hepatol. 60:1090–1096. 10.1016/j.jhep.2013.12.025 [DOI] [PubMed] [Google Scholar]
- Thwaites G.E., Edgeworth J.D., Gkrania-Klotsas E., Kirby A., Tilley R., Török M.E., Walker S., Wertheim H.F., Wilson P., and Llewelyn M.J.. UK Clinical Infection Research Group . 2011. Clinical management of Staphylococcus aureus bacteraemia. Lancet Infect. Dis. 11:208–222. 10.1016/S1473-3099(10)70285-1 [DOI] [PubMed] [Google Scholar]
- Tian W., Li X.J., Stull N.D., Ming W., Suh C.I., Bissonnette S.A., Yaffe M.B., Grinstein S., Atkinson S.J., and Dinauer M.C.. 2008. Fc gamma R-stimulated activation of the NADPH oxidase: phosphoinositide-binding protein p40phox regulates NADPH oxidase activity after enzyme assembly on the phagosome. Blood. 112:3867–3877. 10.1182/blood-2007-11-126029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong S.Y., Davis J.S., Eichenberger E., Holland T.L., and Fowler V.G. Jr. 2015. Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 28:603–661. 10.1128/CMR.00134-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuchscherr L., Medina E., Hussain M., Völker W., Heitmann V., Niemann S., Holzinger D., Roth J., Proctor R.A., Becker K., et al. . 2011. Staphylococcus aureus phenotype switching: an effective bacterial strategy to escape host immune response and establish a chronic infection. EMBO Mol. Med. 3:129–141. 10.1002/emmm.201000115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong C.H., Jenne C.N., Lee W.Y., Léger C., and Kubes P.. 2011. Functional innervation of hepatic iNKT cells is immunosuppressive following stroke. Science. 334:101–105. 10.1126/science.1210301 [DOI] [PubMed] [Google Scholar]