Abstract
After postoperative deaths in children who were prescribed codeine, several pediatric hospitals have removed it from their formularies. These deaths were attributed to atypical cytochrome P450 2D6 (CYP2D6) pharmacogenetics, which is also implicated in poor analgesic response. Because codeine is often prescribed to patients with sickle cell disease and is now the only Schedule III opioid analgesic in the United States, we implemented a precision medicine approach to safely maintain codeine as an option for pain control. Here we describe the implementation of pharmacogenetics-based codeine prescribing that accounts for CYP2D6 metabolizer status. Clinical decision support was implemented within the electronic health record to guide prescribing of codeine with the goal of preventing its use after tonsillectomy or adenoidectomy and in CYP2D6 ultra-rapid and poor metabolizer (high-risk) genotypes. As of June 2015, CYP2D6 genotype results had been reported for 2468 unique patients. Of the 830 patients with sickle cell disease, 621 (75%) had a CYP2D6 genotype result; 7.1% were ultra-rapid or possible ultra-rapid metabolizers, and 1.4% were poor metabolizers. Interruptive alerts recommended against codeine for patients with high-risk CYP2D6 status. None of the patients with an ultra-rapid or poor metabolizer genotype were prescribed codeine. Using genetics to tailor analgesic prescribing retained an important therapeutic option by limiting codeine use to patients who could safely receive and benefit from it. Our efforts represent an evidence-based, innovative medication safety strategy to prevent adverse drug events, which is a model for the use of pharmacogenetics to optimize drug therapy in specialized pediatric populations.
The use of codeine in pediatric medicine has been questioned following reports of postoperative deaths in children who were prescribed codeine and the subsequent US Food and Drug Administration (FDA) boxed warning.1,2 Citing its unpredictable pharmacokinetics and pharmacodynamics that could lead to deleterious outcomes, some have suggested ending all codeine use in children.3,4 In response, several pediatric hospitals have removed codeine from their formularies.5–7 Although these efforts may be a justifiable use of the formulary system to improve patient safety, it is a blunt approach that limits therapeutic options. Since the recent rescheduling of hydrocodone-containing analgesics from the Drug Enforcement Administration (DEA) Schedule III to the more tightly regulated Schedule II, codeine coformulated with acetaminophen is now the only opioid analgesic that is classified as a Schedule III controlled substance under federal law.8 Schedule III regulations, unlike their Schedule II counterparts, allow for verbal and facsimile prescribing to pharmacies as well as refills with the original prescription. Furthermore, codeine remains a safe and effective analgesic for the majority of patients who are not cytochrome P450 2D6 (CYP2D6) ultra-rapid metabolizers or poor metabolizers compared with alternatives.9
Codeine with acetaminophen has been prescribed to children for many years, is relatively inexpensive, and is available in both liquid and tablet form. Although codeine is commonly prescribed (>18 million US outpatient prescriptions in 2013)10 and largely well tolerated, the reports of severe adverse events and death in children ignited discussion about its risks and benefits.3,4,11–13 It is well established that some individuals may not experience any pain relief from codeine, likely a result of its poor metabolism, but both lack of efficacy and safety concerns may be reduced through preemptive CYP2D6 genotype testing.9 Using this approach, clinicians know in advance of prescribing which patients may be at risk for a poor outcome with codeine and conversely, which patients are most likely to benefit from it.
Codeine is a prodrug that requires hepatic biotransformation to morphine via CYP2D6 to yield analgesic effects (Fig 1).14 CYP2D6 is a highly polymorphic gene with numerous allelic variants that differ in drug metabolizing potential. Moreover, the CYP2D6 gene is subject to copy number variations that can also significantly increase an individual’s overall CYP2D6 enzyme activity. It is well established that variability in CYP2D6 enzyme activity can affect the efficacy and toxicity of codeine in patients.15–20 Notably, CYP2D6 ultra-rapid metabolizers are at risk for toxic systemic concentrations of morphine with label-recommended dosages of codeine, and CYP2D6 poor metabolizers are unable to achieve adequate systemic concentrations of morphine to experience a therapeutic benefit from codeine. CYP2D6 genotyping options are available from several clinical laboratories.21
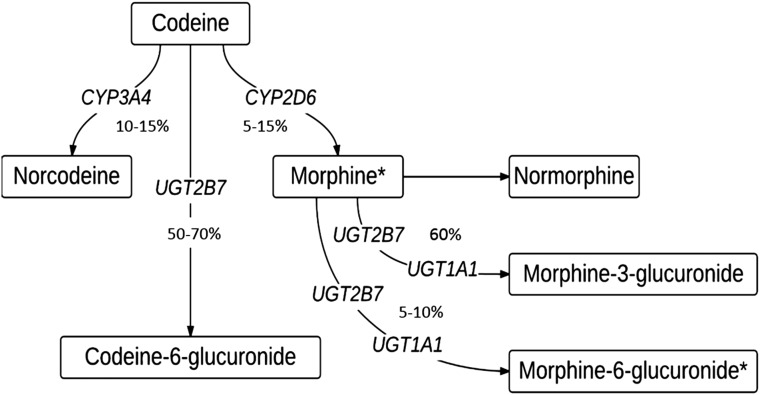
FIGURE 1.
Pathway of codeine metabolism in a CYP2D6 extensive metabolizer. Asterisks (*) denote active metabolite.
In environments without the routine use of CYP2D6 genotyping, several cases of fatal and severe, nonfatal respiratory depression were reported in children receiving seemingly appropriate weight-based dosing of codeine for postoperative pain management after elective adenotonsillectomies.22,23 The common thread that emerged from these reports was clear; children with functional CYP2D6 gene duplications are at high risk for codeine toxicity because of its excessive conversion to morphine. Given these reports, the FDA issued a Drug Safety Communication in 2012 that warned clinicians to exercise caution when prescribing codeine to children after tonsillectomy and/or adenoidectomy and alerting parents/caregivers to the signs of morphine toxicity.1 In 2013, the FDA added a new boxed warning, the FDA’s strongest warning, to the drug label of codeine-containing products: “Respiratory depression and death have occurred in children who received codeine following tonsillectomy and/or adenoidectomy and had evidence of being ultra-rapid metabolizers of codeine due to a CYP2D6 polymorphism.”2,24 The FDA also added a specific contraindication regarding the use of codeine-containing products in children for postoperative pain management after tonsillectomy and/or adenoidectomy. Most recently, in late 2015, an FDA Advisory Committee recommended that codeine not be used to treat children or the majority of teenagers suffering from pain or a cough.25
The Clinical Pharmacogenetics Implementation Consortium (CPIC) provides actionable, peer-reviewed, evidence-based guidelines for pharmacogenetic testing to optimize drug therapy, thereby facilitating the uptake of precision medicine in clinical practice.26 CPIC offers codeine prescribing recommendations based on CYP2D6 metabolizer status (eg, ultra-rapid, extensive, intermediate, and poor metabolizers).9 The CPIC guideline advises that CYP2D6 ultra-rapid metabolizers (∼2% of the general population) should avoid codeine due to the potential for toxicity (eg, sedation, respiratory depression) as a result of morphine overdose with normal doses of codeine. Furthermore, the guideline advises that CYP2D6 poor metabolizers (∼10% of the general population) should avoid codeine because of the lack of efficacy as a result of little to no conversion of codeine to morphine. The frequency of poor and ultra-rapid metabolizers in a given population varies based on ethnicity, with CYP2D6 ultra-rapid metabolizers occurring in as many as 20% to 30% of some African and Arab populations.27–29 The guideline specifies that it is safe to prescribe the label-recommended dose of codeine to CYP2D6 extensive metabolizers and intermediate metabolizers, noting that intermediate metabolizers may not achieve adequate pain control, and a change in analgesic may be warranted per patient response.
Although we acknowledge that multiple enzymes are involved in the metabolism of codeine (Fig 1), including CYP3A4 and the glucuronidating enzymes UGT1A1 and UGT2B7, the only pharmacogene currently rated by CPIC as actionable for codeine prescribing is CYP2D6, a rating based on a high standard of evidence in peer-reviewed literature.26
After the FDA boxed warning was added to codeine products in 2013, a controversial regulatory development changed the landscape of opioid prescribing. In 2014, the US DEA reclassified hydrocodone-containing analgesics from Schedule III to the more restrictive Schedule II of the Controlled Substances Act.8 Medications in Schedule II have the highest potential for abuse and dependence and are subject to stricter requirements, including DEA registration, labeling, inventory, recordkeeping, and reporting, which has wide implications for prescribing and dispensing. These additional regulations governing Schedule II opioid analgesics complicate outpatient access to these medications because refills are not allowed, and additional prescribing steps are required.8,30–32 A recent survey indicated that only 10.7% of hospitals with e-prescribing systems electronically prescribed Schedule II medications in 2013, so most patients must still bring a paper prescription to the pharmacy.33 Although opioid misuse is a national problem, legitimate access to opioids can also be challenging, particularly for individuals living in predominantly minority areas30 and for those with chronic pain for whom prescribing of opioid analgesics is frequently necessary. The rescheduling debate for hydrocodone/acetaminophen lasted for years and even prompted congressional action.31 Eventual regulatory action made the change, but the FDA advisory committee vote was divided. Advocates against rescheduling these products cited access as one of their primary concerns.32
Patients with sickle cell disease (SCD) experience recurrent and unexpected episodes of vaso-occlusive pain crises throughout their lives.34 Codeine is used extensively for SCD-related vaso-occlusive pain, usually in combination with nonsteroidal anti-inflammatory agents.35 Some patients with SCD report suboptimal analgesic effects from codeine, possibly a result of its poor conversion to morphine. National guidelines recommend codeine as a front-line drug for the management of SCD-associated pain, and this practice is adopted by hematologists at our institution and many others.35 Recent data show comparable efficacy between codeine/acetaminophen (Schedule III) and oxycodone/acetaminophen (Schedule II) as well as codeine/acetaminophen and hydrocodone/acetaminophen (Schedule II) for the treatment of acute pain.36,37 The rescheduling of hydrocodone-containing analgesics and codeine’s ability to adequately provide analgesia in comparison with Schedule II analgesics has reinforced our preference to retain codeine on the hospital formulary and to initiate measures that ensure its safe and effective use.
We chose a precision medicine approach to codeine prescribing by implementing a pharmacogenetics-based prescribing strategy to guide the rational use of codeine across the institution. Such an approach could optimize the use of codeine in SCD and other chronic, recurrent pain disorders by reducing the incidence of excessive toxicity and poor analgesic effect.
We describe the development and implementation of a pharmacogenetics-based strategy for codeine prescribing that accounts for CYP2D6 metabolizer status. We report the prescribing patterns for a subset of our population, those with SCD, who most frequently require codeine-containing analgesics at our specialized pediatric institution.
Methods
Preemptive Pharmacogenetic Testing at St. Jude Children’s Research Hospital
St. Jude Children’s Research Hospital provides comprehensive inpatient and outpatient care, including the provision of all medications, to children with catastrophic illnesses. Each year, ∼8000 patients are treated at St. Jude, including 830 active patients with SCD. St. Jude is the only referral center for SCD in the region, which encompasses a geographic radius of ∼300 miles. Most St. Jude patients are eligible to enroll on an ongoing clinical trial called PG4KDS, a research protocol designed to implement preemptive pharmacogenetic test results into routine clinical care (www.stjude.org/pg4kds).38 Approximately 97% of approached patients consent to the protocol.38 PG4KDS was approved by the hospital’s institutional review board in 2011. Genomic DNA from consented patients is genotyped in a Clinical Laboratory Improvement Amendments-certified laboratory for 230 pharmacogenes, including CYP2D6, by using the Affymetrix DMET Plus array supplemented with a CYP2D6 copy number assay.39 PG4KDS uses a rational, stepwise process to integrate select pharmacogenetic test results (eg, CYP2D6 genotype) into the electronic health record (EHR) to guide prescribing of relevant medications (eg, codeine). Each result is coupled with an interpretive consultation note (Fig 2) and clinical decision support (CDS) alerts that are presented via the EHR.40,41 For those patients who do not have a CYP2D6 result available through PG4KDS, a single gene test for CYP2D6 may be ordered as a routine clinical laboratory test to guide codeine prescribing, an option that has a turn-around time of ∼5 to 7 days.42 St. Jude hematologists often order a CYP2D6 genotype before codeine is prescribed, anticipating the future use of codeine as a potential therapeutic option for patients with SCD.
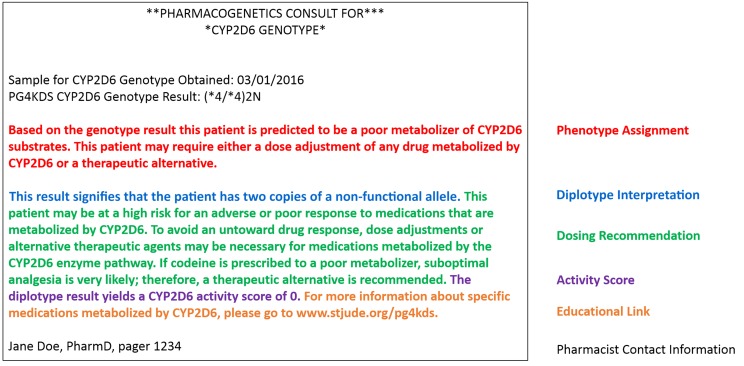
FIGURE 2.
Elements of a CYP2D6 pharmacogenetic consultation note posted to the EHR.
Assigning CYP2D6 Phenotype Based on Genotype
CYP2D6 phenotypes are assigned to diplotypes based on assessments of functional allele activity from previous studies, generally as summarized in the CYP2D6/codeine CPIC guideline.9 The 4 phenotype categories are extensive (normal) metabolizer, intermediate metabolizer, poor metabolizer, and ultra-rapid metabolizer. Ultra-rapid metabolizers express more functional CYP2D6 enzyme than normal, whereas poor metabolizers express little to no functional CYP2D6 enzyme. Intermediate metabolizers express lower than normal amounts of functional CYP2D6 enzyme but still have some CYP2D6 activity. Poor metabolizers and ultra-rapid metabolizers are considered high-priority (high-risk) phenotypes for codeine because they require a change in typical prescribing. Cases with an observed CYP2D6 gene duplication in which we cannot determine which allele is duplicated may be categorized as “possible ultra-rapid metabolizers” or “possible intermediate metabolizers,” depending on the functional status of the identified alleles. It is recommended that patients with a “possible” phenotype status that could be high risk be treated the same as those with a definitive high-risk result (eg, avoid codeine in patients who are “possible CYP2D6 ultra-rapid metabolizers”). Cases for which the genotyping assay detected a complex CYP2D6 hybrid structure of indeterminate function43 do not have an assigned phenotype. These results are placed in the EHR and assigned a CYP2D6 phenotype of “indeterminate.” Clinicians are informed that additional CYP2D6 testing may be available, and they are given the option of pursuing more definitive CYP2D6 copy number testing if it is desired to know copy number/hybrid status.
Pharmacogenetics-Based Strategy for Codeine Prescribing
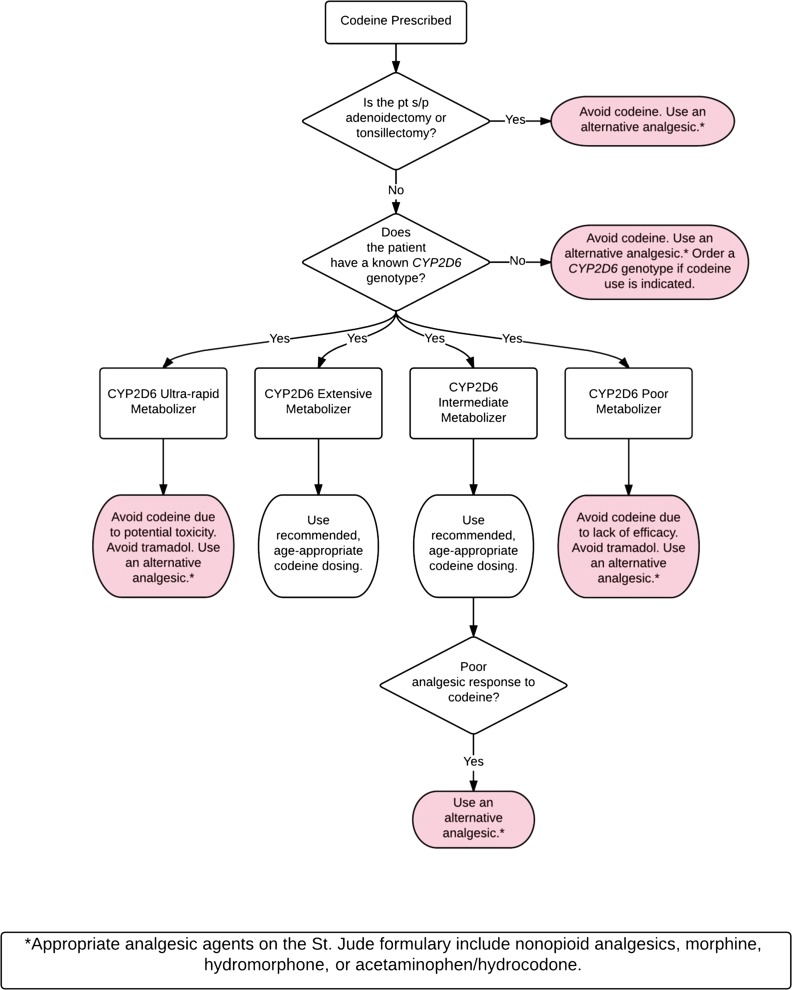
Our pharmacogenetics-based codeine prescribing strategy is consistent with both the FDA boxed warning for codeine-containing products and the CPIC guideline for CYP2D6/codeine (Fig 3); it was approved by the St. Jude Pharmacy and Therapeutics Committee in 2013. Per this strategy, codeine should be avoided in the postoperative period for patients who underwent tonsillectomy and/or adenoidectomy. In these cases, an alternative oral analgesic agent on the hospital formulary (eg, nonopioid analgesic, morphine, hydrocodone/acetaminophen) that is not significantly metabolized by CYP2D6 is to be prescribed. To ensure compliance with this practice, we removed codeine from relevant postoperative EHR order sets and created a CDS alert that is presented in the rare case codeine is ordered for a patient who is status-post tonsillectomy and/or adenoidectomy, regardless of whether CYP2D6 genotype is known. Codeine should also be avoided in patients with an unknown CYP2D6 genotype and in patients whose genotype result yields an indeterminate phenotype assignment because their level of risk for toxicity or therapeutic failure is not known. Tramadol is not an acceptable alternative because its pharmacogenetics mirrors that of codeine.9,44 In the rare event of tramadol use, CDS alerts for tramadol remain in place for CYP2D6 poor and ultra-rapid metabolizers as an additional safety measure. In addition, tramadol is not listed as an acceptable alternative in CDS alerts for codeine. Codeine should only be prescribed to patients with a known CYP2D6 genotype to ensure that it is not used in the minority of patients who would likely experience an unfavorable response. This strategy preserves appropriate codeine use in the majority of patients expected to respond favorably.
FIGURE 3.
The pharmacogenetics-based codeine prescribing strategy used across all services at St. Jude Children’s Research Hospital. pt, patient; s/p, status-post.
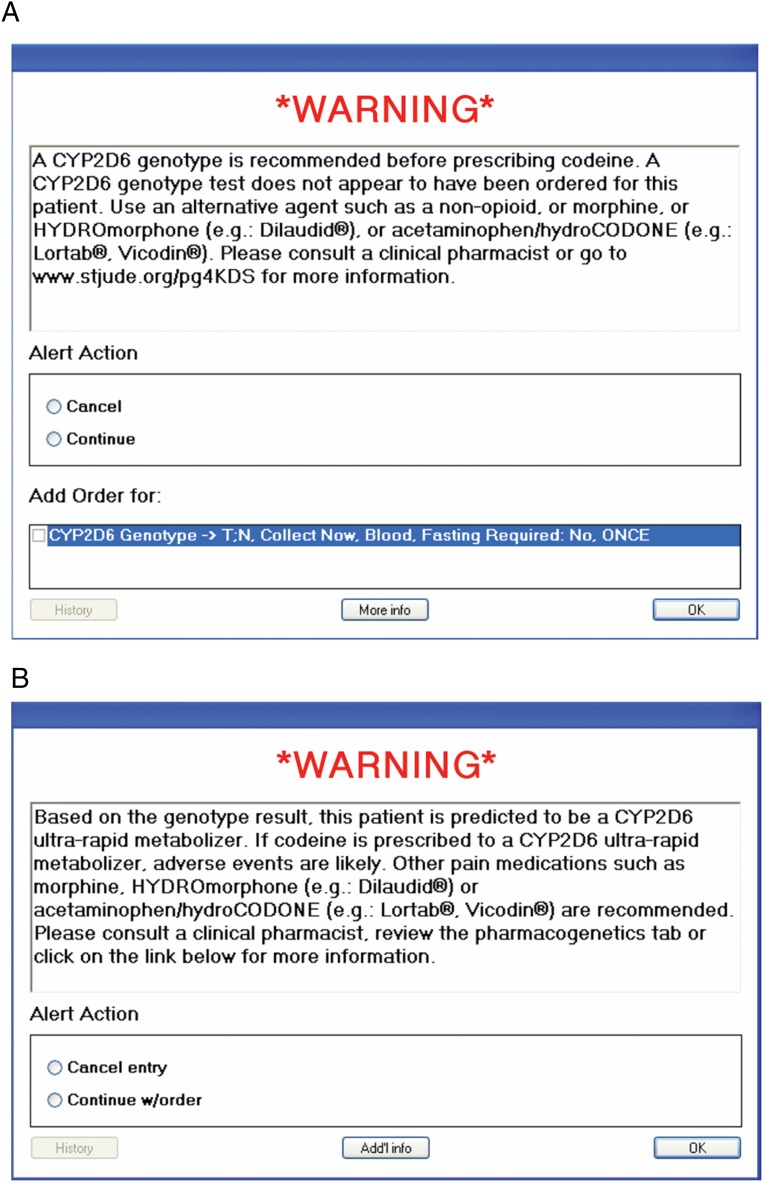
Active CDS implemented into the St. Jude EHR (Cerner, Kansas City, MO) was essential to support the successful implementation of our CYP2D6 genotype-based codeine prescribing strategy.41 Pretest (pregenotype) alerts are presented when a prescriber orders codeine for a patient for whom there is no CYP2D6 test result in the EHR (Fig 4A). This alert communicates that a CYP2D6 genotype should be obtained before prescribing codeine, and a CYP2D6 test may be ordered directly from the alert screen. The prescriber may override the alert and continue with the order; in this instance, a second screen prompts the prescriber to enter an override reason. Preset override reasons include (1) patient has previously tolerated codeine or had efficacy with codeine therapy; (2) patient has undergone an allogeneic bone marrow transplant (and therefore his or her genetic blood test results would not reflect hepatic CYP2D6 activity, but rather that of their bone marrow donor); (3) a CYP2D6 genotype was just ordered; and (4) other (with required free-text reason).
FIGURE 4.
CDS alerts for CYP2D6/codeine. A, A pretest alert is presented when a prescriber orders codeine for a patient for whom there is no CYP2D6 test result. B, A posttest alert is presented when a prescriber orders codeine for a patient who has a high-risk CYP2D6 genotype result (eg, ultra-rapid metabolizer).
Posttest alerts are presented when a prescriber orders codeine for a patient who has a high-risk (actionable) CYP2D6 result (eg, ultra-rapid, possible ultra-rapid, or poor metabolizer) in the EHR (Fig 4B). This alert communicates that increased toxicity or lack of efficacy may occur in this patient if codeine is prescribed (to an ultra-rapid metabolizer or a poor metabolizer, respectively). Appropriate oral alternative agents on our hospital’s formulary unaffected by CYP2D6 status are specifically recommended in the alert. Prescribers who order codeine for known intermediate metabolizers of CYP2D6 will not be presented with a posttest alert because this is not considered a high-risk result; per the CPIC recommendation, these patients may receive label-recommended doses of codeine. However, all results that confer CYP2D6 intermediate metabolizer status are accompanied by a written consultation in the EHR that includes a recommendation for an alternative to codeine (unaffected by CYP2D6 status) should these patients experience a poor analgesic response (Fig 3).
Through our CDS system, we retrieved CYP2D6/codeine alert data including alert type (pretest versus posttest), date and time of the alert, CYP2D6 phenotype, prescriber’s discipline (eg, physician, pharmacist, nurse practitioner), patient’s medical service, and general codeine prescription data, including whether the prescription was dispensed to the patient. Regression analysis was used to assess trends in the number of CYP2D6/codeine pretest alerts presented to clinicians and the number of St. Jude patients genotyped for CYP2D6 over time.
Results
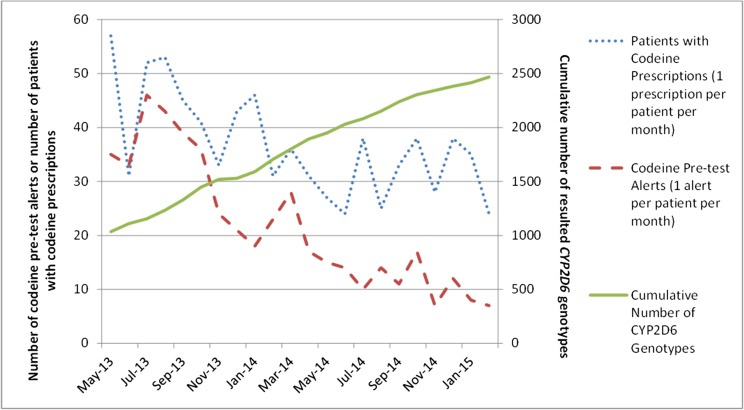
Since May 2013 when CYP2D6/codeine pretest alerts were initiated, the number of pretest alerts presented when a codeine order was entered for a patient with no CYP2D6 genotype result has been steadily declining (P < .001 for trend) as the number of patients across the institution genotyped in our preemptive pharmacogenetics model has increased (P < .001 for trend) (Fig 5). Our goal is to have minimal CYP2D6/codeine pretest alerts (ideally none), which indicates that we are obtaining preemptive CYP2D6 genotypes for all patients to optimize the prescribing of codeine.
FIGURE 5.
Inverse relationship over time between utilization of CYP2D6 genotyping (right y axis) and the number of patients for whom CYP2D6/codeine pretest alerts occurred upon codeine orders at St. Jude (left y axis). This trend (P < .001) indicates increased utilization of genetic data within the EHR to guide codeine prescribing.
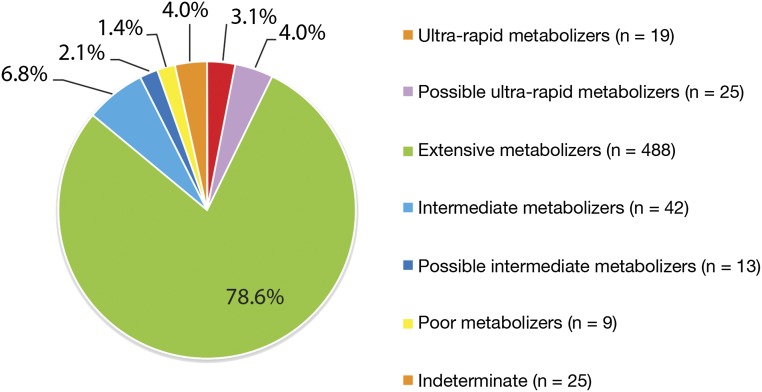
Approximately 30% (n = 757) of the enrolled PG4KDS patient population have nonmalignant blood disorders and are treated by the St. Jude Hematology service. Most of these patients have SCD, and they represent our principal users of codeine-containing analgesics. Approximately 97% of our patients with SCD are African American (other races include white, Hispanic, and mixed race). Nearly 70% of patients for whom a CYP2D6/codeine pretest or posttest alert have been presented to the prescriber are treated by the Hematology service. As of June 2015, 621 (75%) of the 830 active patients with SCD at our hospital had a CYP2D6 genotype result posted to their EHR. Their SCD hemoglobin genotypes included HbSS, HbSC, HbSβ0 thalassemia, HbSβ+ thalassemia, and HbSD, and ages ranged from 9 months to 18 years. The distribution of CYP2D6 phenotypes in these patients reveals 1.4% of patients categorized as poor metabolizers and 7% of patients categorized as ultra-rapid or possible ultra-rapid metabolizers (Fig 6). These data represent the largest cohort of SCD patients who have been genotyped for CYP2D6 thus far. With respect to the percentages of poor metabolizers and ultra-rapid metabolizers, our findings are consistent with other CYP2D6 genotype analyses in African American patients with or without SCD and differ from the distribution of CYP2D6 phenotypes in individuals of European ancestry.45–48 African American patients are more likely to be ultra-rapid metabolizers and less likely to be poor metabolizers compared with individuals of European ancestry. In our cohort, 53 patients with SCD (9% of patients with a known CYP2D6 genotype) have a high-risk CYP2D6 result (44 ultra-rapid metabolizers or possible ultra-rapid metabolizers and 9 poor metabolizers). Thus, excluding 25 patients with an indeterminate phenotype (Table 1), the large majority of patients with SCD at our institution (n = 543; 87%) can safely receive label-recommended doses of codeine based on their genotypes. Patients with an indeterminate phenotype were not counted as “high risk” because their phenotype status is not known with certainty.
FIGURE 6.
The distribution of CYP2D6 phenotypes among St. Jude patients with SCD (n = 621). “Indeterminate” implies that the genotyping assay detected a complex CYP2D6 hybrid structure of indeterminate function.
TABLE 1.
Codeine Prescribing Patterns Among Patients With SCD and Known CYP2D6 Phenotypes (n = 621)
| CYP2D6 phenotype | n | Patients for Whom a Codeine-Containing Analgesic Was Dispensed, n (%) |
|---|---|---|
| High-risk phenotypes | ||
| Ultra-rapid metabolizer | 19 | 0 |
| Possible ultra-rapid metabolizer | 25 | 1a (4) |
| Poor metabolizer | 9 | 0 |
| Non–high-risk phenotypes | ||
| Extensive metabolizer | 488 | 161 (33) |
| Intermediate metabolizer | 42 | 8 (19) |
| Possible intermediate metabolizer | 13 | 4 (31) |
| Unknown risk | ||
| Indeterminate | 25 | 1 (4) |
Patient had a documented history of tolerating codeine well in the past.
Of the 543 patients without high-risk CYP2D6 results (eg, CYP2D6 extensive, intermediate, or possible intermediate metabolizers), 173 patients (32%) received codeine (Table 1). Only 1 of the 53 patients with high-risk CYP2D6 results (eg, ultra-rapid metabolizers, possible ultra-rapid metabolizers, and poor metabolizers) received codeine. The most common alternative analgesic used in place of codeine was hydrocodone/acetaminophen. Tramadol was never used as an alternative analgesic in these patients. Six patients with high-risk CYP2D6 genotypes were initially prescribed codeine, but a posttest CDS alert prompted the prescriber to change the order to a recommended alternative analgesic. In 5 of these cases, the prescriber changed the order and the patient did not receive codeine. The only patient with a high-risk genotype who received codeine was a “possible CYP2D6 ultra-rapid metabolizer,” who had previously tolerated codeine. Importantly, none of the patients with SCD in this analysis who received codeine experienced severe adverse events from its use. We have educated our SCD care providers about using CYP2D6 genotypes to guide codeine prescribing, and the hematology clinical note template includes a section to document the patient’s CYP2D6 phenotype. Therefore, prescribers systematically consider genetic data along with other clinical factors when selecting the most appropriate analgesic for each patient.
Discussion
Pharmacogenetic testing was leveraged at our pediatric hospital to preserve the safe and effective use of codeine as an analgesic in our pediatric SCD population. Our efforts represent an evidence-based, innovative medication safety strategy to proactively prevent severe adverse drug events and avoid ineffective medications in children, both of which are pressing and important issues in pediatric medicine.6 Because we offer genotyping to all patients, there is no a priori selection of those who would be offered the study.
Application
Other pediatric hospitals have also adopted a pharmacogenetic approach to codeine use.49 Pharmacogenetics-based prescribing strategies for medications other than codeine may similarly benefit other special needs pediatric populations. Although our institution-wide strategy to implement preemptive pharmacogenetic testing may not be immediately feasible elsewhere, focusing on high-risk, pharmacogenetically relevant medications frequently used among a specialized patient population (such as SCD) is rational and generalizable. The frequency of actionable genotypes in specialized patient populations may differ from the general population, as was the case for CYP2D6 genotypes in our patients with SCD. This difference may make pharmacogenetic testing more compelling and useful in certain groups.
Pharmacogenetic testing allows for the continued use of medications that may otherwise be deemed unsafe from a population perspective, which broadens and personalizes therapeutic options. The 2013 American Academy of Pediatrics policy statement “Ethical and Policy Issues in Genetic Testing and Screening of Children” supports pharmacogenetic testing for therapeutic purposes, including drug targeting and dose-responsiveness.50 Moreover, genetics-guided precision medicine may reduce health care burdens and costs. For example, mild to moderately severe SCD-associated pain crises may be managed if a prescriber can call in a codeine prescription for home administration, thereby circumventing emergency department visits and hospitalizations. As pharmacogenetic testing becomes more economical and accessible, opportunities and new strategies for enhancing the safe use of pharmaceuticals in children will become more widespread. Future economic studies may facilitate the adoption of a pharmacogenetic strategy for codeine prescribing.
Potential Impact of Precision Medicine on Prescribing Practices
Weighing the possibility of drug-seeking behavior against the provision of adequate pain control remains a challenge, particularly for specialized patient populations such as SCD that legitimately require recurrent opioids.51,52 Understanding drug response patterns in certain populations may reduce the misinterpretation of symptoms. For instance, patients may refuse codeine from perceived “lack of effect,” leading SCD providers to incorrectly label them as “drug seekers.” However, patients with SCD who have failed codeine therapy for a pain crisis are likely to have reduced-functioning CYP2D6 variants.53 Patients at risk for failing codeine therapy include CYP2D6 poor metabolizers and CYP2D6 intermediate metabolizers, which constitute ∼10% of patients in our cohort. Although accurately identifying drug-seeking behavior is wrought with complexity, our experience shows genotyping may facilitate appropriate opioid prescribing and help reduce the mislabeling of patients.
Limitations
There are several limitations to the widespread adoption of this pharmacogenetically guided approach to codeine use at other institutions. Although the cost of genotyping continues to decrease, the cost of preemptively genotyping all patients with SCD remains a limitation.54 It is generally less costly on a per gene basis to genotype preemptively for a panel of genes compared with single gene testing; the cost of a single gene test for CYP2D6 is several hundred dollars, whereas the costs for any 1 gene (including CYP2D6) is considerably lower when averaged out among a panel of 230 genes, as we do preemptively for PG4KDS samples. All relevant costs to health systems and patients should be studied, such as costs associated with inadequate pain control due to the use of less effective analgesics, the costs associated with using more tightly regulated Schedule II narcotics, emergency department visits that led to admissions (due to insufficient pain control), additional acute care clinic visits, quality of life, and family member time related to extra time spent in the health care system seeking more appropriate pain relief. Another limitation is the lack of broad pharmacogenetic expertise, which is needed to efficiently interpret a patient’s CYP2D6 genotype. Multiple laboratories offer clinical CYP2D6 genotyping21; however, CYP2D6 gene test results are some of the most complex to interpret, given the large number of variants, copy number considerations, and the possibility of hybrid rearrangements with the nonfunctional, neighboring CYP2D7 and CYP2D8 genes.55 Although CPIC’s Informatics Working Group and other researchers have emphasized that these technical problems exist for CYP2D6, complex results on copy number variants and hybrid genes may preclude definitive interpretation of CYP2D6 phenotype for a small percent of tested patients.56,57 Lastly, our current study illustrates the implementation process to incorporate CYP2D6 genotyping into routine clinical practice; however, future studies are needed to show the effect of this pharmacogenetic algorithm on clinical end points such as adverse effects and pain control.
Conclusions
Although some advocate for the cessation of codeine use in children, codeine can remain an important option to treat pediatric pain if its prescribing is guided by pharmacogenetics to ensure safety and efficacy. We demonstrated that a rational approach informed by the patient’s individual metabolism of codeine can be implemented and widely used in children with SCD. This approach may serve as a model for the implementation of pharmacogenetic testing to optimize drug therapy in other specialized pediatric populations. Future studies should focus on how CYP2D6 genotyping affects the frequency of adequate pain control and toxicity with codeine and how this approach to optimize codeine use for SCD affects quality of life and overall health care costs and utilization.
Acknowledgments
We thank the following individuals from St. Jude Children’s Research Hospital: Melinda Wood, RN, for her assistance with obtaining consent from patients for the clinical trial; and Nancy Kornegay, MBA, Mark Wilkinson, BS, Wenjian Yang, PhD, and Colton Smith, PhD, for their expertise and assistance in specific components of data analysis.
Glossary
- CDS
clinical decision support
- CPIC
Clinical Pharmacogenetics Implementation Consortium
- CYP2D6
cytochrome P450 2D6
- DEA
Drug Enforcement Administration
- EHR
electronic health record
- FDA
Food and Drug Administration
- SCD
sickle cell disease
Footnotes
Dr Gammal collected data, carried out the initial analyses, and drafted the initial manuscript; Drs Crews, Haidar, and Hoffman are coinvestigators on the PG4KDS study; they supervised the collection and analysis of data and critically reviewed and revised the manuscript; Dr Baker created the clinical decision support alerts, collected data, and critically reviewed the manuscript; Drs Barker, Estepp, Wang, and Weiss critically reviewed and revised the manuscript; Ms Pei designed and conducted the statistical analysis; Dr Broeckel supervises all PG4KDS-related genotyping; Dr Relling conceptualized and designed the PG4KDS study, supervised the collection and analysis of data, and critically reviewed and revised the manuscript; Dr Hankins conceptualized and designed the study and critically reviewed and revised the manuscript; and all authors approved the final manuscript as submitted.
FINANCIAL DISCLOSURE: The authors have indicated they have no financial relationships relevant to this article to disclose.
FUNDING: Supported by National Cancer Institute grants CA 36401, CA 21765; National Institute of Health/National Institute of General Medical Sciences Pharmacogenomics Research Network (grants U01 GM92666, U01 HL105918); and by the American Lebanese Syrian Associated Charities. Funded by the National Institutes of Health (NIH).
POTENTIAL CONFLICT OF INTEREST: The authors have indicated they have no potential conflicts of interest to disclose.
COMPANION PAPER: A companion to this article can be found online at www.pediatrics.org/cgi/doi/10.1542/peds.2016.1359.
References
- 1.US Food and Drug Administration FDA Drug Safety Communication: Codeine use in certain children after tonsillectomy and/or adenoidectomy may lead to rare, but life-threatening adverse events or death. 2012. Available at: www.fda.gov/Drugs/DrugSafety/ucm313631.htm. Accessed June 15, 2015
- 2.US Food and Drug Administration FDA Drug Safety Communication: Safety review update of codeine use in children; new boxed warning and contraindication on use after tonsillectomy and/or adenoidectomy. 2013. Available at: www.fda.gov/Drugs/DrugSafety/ucm339112.htm. Accessed June 15, 2015
- 3.Woolf AD, Greco C. Why can’t we retire codeine? Pediatrics. 2014;133(5). Available at: www.pediatrics.org/cgi/content/full/133/5/e1354 [DOI] [PubMed] [Google Scholar]
- 4.Kaiser SV, Asteria-Penaloza R, Vittinghoff E, Rosenbluth G, Cabana MD, Bardach NS. National patterns of codeine prescriptions for children in the emergency department. Pediatrics. 2014;133(5). Available at: www.pediatrics.org/cgi/content/full/133/5/e1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jerome J, Solodiuk JC, Sethna N, McHale J, Berde C. A single institution’s effort to translate codeine knowledge into specific clinical practice. J Pain Symptom Manage. 2014;48(1):119–126 [DOI] [PubMed] [Google Scholar]
- 6.Rieder MJ, Carleton B. Pharmacogenomics and adverse drug reactions in children. Front Genet. 2014;5:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cartabuke RS, Tobias JD, Taghon T, Rice J. Current practices regarding codeine administration among pediatricians and pediatric subspecialists. Clin Pediatr (Phila). 2014;53(1):26–30 [DOI] [PubMed] [Google Scholar]
- 8.Drug Enforcement Administration. Schedules of controlled substances: rescheduling of hydrocodone combination products from Schedule III to Schedule II. Available at: www.deadiversion.usdoj.gov/fed_regs/rules/2014/fr0822.htm. Accessed June 15, 2015 [PubMed]
- 9.Crews KR, Gaedigk A, Dunnenberger HM, et al. ; Clinical Pharmacogenetics Implementation Consortium . Clinical Pharmacogenetics Implementation Consortium guidelines for cytochrome P450 2D6 genotype and codeine therapy: 2014 update. Clin Pharmacol Ther. 2014;95(4):376–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dunnenberger HM, Crews KR, Hoffman JM, et al. Preemptive clinical pharmacogenetics implementation: current programs in five US medical centers. Annu Rev Pharmacol Toxicol. 2015;55:89–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tremlett M, Anderson BJ, Wolf A. Pro-con debate: is codeine a drug that still has a useful role in pediatric practice? Paediatr Anaesth. 2010;20(2):183–194 [DOI] [PubMed] [Google Scholar]
- 12.Lazaryan M, Shasha-Zigelman C, Dagan Z, Berkovitch M. Codeine should not be prescribed for breastfeeding mothers or children under the age of 12. Acta Paediatr. 2015;104(6):550–556 [DOI] [PubMed] [Google Scholar]
- 13.Fleming ML, Wanat MA. To prescribe codeine or not to prescribe codeine? J Pain Palliat Care Pharmacother. 2014;28(3):251–254 [DOI] [PubMed] [Google Scholar]
- 14.Thorn CF, Klein TE, Altman RB. Codeine and morphine pathway. Pharmacogenet Genomics. 2009;19(7):556–558 [DOI] [PubMed] [Google Scholar]
- 15.Eckhardt K, Li S, Ammon S, Schänzle G, Mikus G, Eichelbaum M. Same incidence of adverse drug events after codeine administration irrespective of the genetically determined differences in morphine formation. Pain. 1998;76(1-2):27–33 [DOI] [PubMed] [Google Scholar]
- 16.Lötsch J, Rohrbacher M, Schmidt H, Doehring A, Brockmöller J, Geisslinger G. Can extremely low or high morphine formation from codeine be predicted prior to therapy initiation? Pain. 2009;144(1–2):119–124 [DOI] [PubMed] [Google Scholar]
- 17.Kirchheiner J, Schmidt H, Tzvetkov M, et al. Pharmacokinetics of codeine and its metabolite morphine in ultra-rapid metabolizers due to CYP2D6 duplication. Pharmacogenomics J. 2007;7(4):257–265 [DOI] [PubMed] [Google Scholar]
- 18.Gasche Y, Daali Y, Fathi M, et al. Codeine intoxication associated with ultrarapid CYP2D6 metabolism. N Engl J Med. 2004;351(27):2827–2831 [DOI] [PubMed] [Google Scholar]
- 19.Caraco Y, Sheller J, Wood AJ. Pharmacogenetic determination of the effects of codeine and prediction of drug interactions. J Pharmacol Exp Ther. 1996;278(3):1165–1174 [PubMed] [Google Scholar]
- 20.Koren G, Cairns J, Chitayat D, Gaedigk A, Leeder SJ. Pharmacogenetics of morphine poisoning in a breastfed neonate of a codeine-prescribed mother. Lancet. 2006;368(9536):704. [DOI] [PubMed] [Google Scholar]
- 21.Genetic Testing Registry CYP2D6 cytochrome P450, family 2, subfamily D, polypeptide 6. Available at: www.ncbi.nlm.nih.gov/gtr/genes/1565. Accessed June 18, 2015
- 22.Ciszkowski C, Madadi P, Phillips MS, Lauwers AE, Koren G. Codeine, ultrarapid-metabolism genotype, and postoperative death. N Engl J Med. 2009;361(8):827–828 [DOI] [PubMed] [Google Scholar]
- 23.Kelly LE, Rieder M, van den Anker J, et al. More codeine fatalities after tonsillectomy in North American children. Pediatrics. 2012;129(5). Available at: www.pediatrics.org/cgi/content/full/129/5/e1343 [DOI] [PubMed] [Google Scholar]
- 24.Acetaminophen and codeine [package insert]. Available at: https://dailymed.nlm.nih.gov. Accessed June 9, 2015
- 25.National Public Radio. Limits urged on the use of codeine to stop kids’ coughs and pain. Available at: www.npr.org/sections/health-shots/2015/12/10/459224775/limits-urged-on-the-use-of-codeine-to-stop-kids-coughs-and-pain. Accessed December 12, 2015
- 26.Caudle KE, Klein TE, Hoffman JM, et al. Incorporation of pharmacogenomics into routine clinical practice: the Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline development process. Curr Drug Metab. 2014;15(2):209–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.LLerena A, Naranjo ME, Rodrigues-Soares F, Penas-LLedó EM, Fariñas H, Tarazona-Santos E. Interethnic variability of CYP2D6 alleles and of predicted and measured metabolic phenotypes across world populations. Expert Opin Drug Metab Toxicol. 2014;10(11):1569–1583 [DOI] [PubMed] [Google Scholar]
- 28.Aklillu E, Persson I, Bertilsson L, Johansson I, Rodrigues F, Ingelman-Sundberg M. Frequent distribution of ultrarapid metabolizers of debrisoquine in an ethiopian population carrying duplicated and multiduplicated functional CYP2D6 alleles. J Pharmacol Exp Ther. 1996;278(1):441–446 [PubMed] [Google Scholar]
- 29.McLellan RA, Oscarson M, Seidegård J, Evans DA, Ingelman-Sundberg M. Frequent occurrence of CYP2D6 gene duplication in Saudi Arabians. Pharmacogenetics. 1997;7(3):187–191 [DOI] [PubMed] [Google Scholar]
- 30.Green CR, Ndao-Brumblay SK, West B, Washington T. Differences in prescription opioid analgesic availability: comparing minority and white pharmacies across Michigan. J Pain. 2005;6(10):689–699 [DOI] [PubMed] [Google Scholar]
- 31.Gershman JA, Fass AD. Hydrocodone rescheduling amendment and pipeline products on the horizon. P&T. 2012;37(7):399–404 [PMC free article] [PubMed] [Google Scholar]
- 32.Kuehn BM. FDA committee . FDA committee: More restrictions needed on hydrocodone combination products. JAMA. 2013;309(9):862. [DOI] [PubMed] [Google Scholar]
- 33.Fox BI, Pedersen CA, Gumpper KF. ASHP national survey on informatics: assessment of the adoption and use of pharmacy informatics in U.S. hospitals—2013. Am J Health Syst Pharm. 2015;72(8):636–655 [DOI] [PubMed] [Google Scholar]
- 34.McCavit TL. Sickle cell disease. Pediatr Rev. 2012;33(5):195–204, quiz 205–206 [DOI] [PubMed] [Google Scholar]
- 35.Yawn BP, Buchanan GR, Afenyi-Annan AN, et al. Management of sickle cell disease: summary of the 2014 evidence-based report by expert panel members. JAMA. 2014;312(10):1033–1048 [DOI] [PubMed] [Google Scholar]
- 36.Chang AK, Bijur PE, Lupow JB, Gallagher EJ. Comparative analgesic efficacy of oxycodone/acetaminophen vs. codeine/acetaminophen for short-term pain management following ED discharge. Pain Med. 2015;16(12):2397–2404 [DOI] [PubMed] [Google Scholar]
- 37.Chang AK, Bijur PE, Munjal KG, John Gallagher E. Randomized clinical trial of hydrocodone/acetaminophen versus codeine/acetaminophen in the treatment of acute extremity pain after emergency department discharge. Acad Emerg Med. 2014;21(3):227–235 [DOI] [PubMed] [Google Scholar]
- 38.Hoffman JM, Haidar CE, Wilkinson MR, et al. PG4KDS: a model for the clinical implementation of pre-emptive pharmacogenetics. Am J Med Genet C Semin Med Genet. 2014;166C(1):45–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fernandez CA, Smith C, Yang W, et al. Concordance of DMET plus genotyping results with those of orthogonal genotyping methods. Clin Pharmacol Ther. 2012;92(3):360–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hicks JK, Crews KR, Hoffman JM, et al. A clinician-driven automated system for integration of pharmacogenetic interpretations into an electronic medical record. Clin Pharmacol Ther. 2012;92(5):563–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bell GC, Crews KR, Wilkinson MR, et al. Development and use of active clinical decision support for preemptive pharmacogenomics. J Am Med Inform Assoc. 2014;21(e1):e93–e99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Crews KR, Cross SJ, McCormick JN, et al. Development and implementation of a pharmacist-managed clinical pharmacogenetics service. Am J Health Syst Pharm. 2011;68(2):143–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Black JL III, Walker DL, O’Kane DJ, Harmandayan M. Frequency of undetected CYP2D6 hybrid genes in clinical samples: impact on phenotype prediction. Drug Metab Dispos. 2012;40(1):111–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Orliaguet G, Hamza J, Couloigner V, et al. A case of respiratory depression in a child with ultrarapid CYP2D6 metabolism after tramadol. Pediatrics. 2015;135(3). Available at: www.pediatrics.org/cgi/content/full/135/3/e753 [DOI] [PubMed] [Google Scholar]
- 45.Joly P, Gagnieu MC, Bardel C, Francina A, Pondarre C, Martin C. Genotypic screening of the main opiate-related polymorphisms in a cohort of 139 sickle cell disease patients. Am J Hematol. 2012;87(5):534–536 [DOI] [PubMed] [Google Scholar]
- 46.Yee MM, Josephson C, Hill CE, et al. Cytochrome P450 2D6 polymorphisms and predicted opioid metabolism in African American children with sickle cell disease. J Pediatr Hematol Oncol. 2013;35(7):e301–e305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Relling MV, Cherrie J, Schell MJ, Petros WP, Meyer WH, Evans WE. Lower prevalence of the debrisoquin oxidative poor metabolizer phenotype in American black versus white subjects. Clin Pharmacol Ther. 1991;50(3):308–313 [DOI] [PubMed] [Google Scholar]
- 48.Gaedigk A, Simon SD, Pearce RE, Bradford LD, Kennedy MJ, Leeder JS. The CYP2D6 activity score: translating genotype information into a qualitative measure of phenotype. Clin Pharmacol Ther. 2008;83(2):234–242 [DOI] [PubMed] [Google Scholar]
- 49.Rasmussen-Torvik LJ, Stallings SC, Gordon AS, et al. Design and anticipated outcomes of the eMERGE-PGx project: a multicenter pilot for preemptive pharmacogenomics in electronic health record systems. Clin Pharmacol Ther. 2014;96(4):482–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.American Academy of Pediatrics Committee on Bioethics and Committee on Genetics; and the American College of Medical Genetics and Genomics Social, Ethical, and Legal Issues Committee . Ethical and policy issues in genetic testing and screening of children. Pediatrics. 2013;131(3):620–622 [DOI] [PubMed] [Google Scholar]
- 51.Elander J, Lusher J, Bevan D, Telfer P, Burton B. Understanding the causes of problematic pain management in sickle cell disease: evidence that pseudoaddiction plays a more important role than genuine analgesic dependence. J Pain Symptom Manage. 2004;27(2):156–169 [DOI] [PubMed] [Google Scholar]
- 52.Brown SE, Weisberg DF, Balf-Soran G, Sledge WH. Sickle cell disease patients with and without extremely high hospital use: pain, opioids, and coping. J Pain Symptom Manage. 2015;49(3):539–547 [DOI] [PubMed] [Google Scholar]
- 53.Brousseau DC, McCarver DG, Drendel AL, Divakaran K, Panepinto JA. The effect of CYP2D6 polymorphisms on the response to pain treatment for pediatric sickle cell pain crisis. J Pediatr. 2007;150(6):623–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sequencing Costs DNA. National Human Genome Research Institute. Available at: www.genome.gov/sequencingcosts/. Accessed June 15, 2015
- 55.Hicks JK, Swen JJ, Gaedigk A. Challenges in CYP2D6 phenotype assignment from genotype data: a critical assessment and call for standardization. Curr Drug Metab. 2014;15(2):218–232 [DOI] [PubMed] [Google Scholar]
- 56.CPIC Informatics Working Group PharmGKB. Available at: https://www.pharmgkb.org/page/cpicInformatics. Accessed November 29, 2015
- 57.Twist GP, Gaedigk A, Miller NA, et al. Constellation: a tool for rapid, automated phenotype assignment of a highly polymorphic pharmacogene, CYP2D6, from whole-genome sequences. NPJ Genomic Med. 2016;1:15007. [DOI] [PMC free article] [PubMed] [Google Scholar]