Abstract
The mosquito, Aedes aegypti (L.) originated in Sub-Saharan Africa as a dark form sylvan species (A. aegypti formosus). Evolution of A. aegypti aegypti type form as a human commensal facilitated its colonization of most semitropical and tropical areas. We investigated the genetic basis for abdominal white scale presence that represents the diagnostic for sylvan A. aegypti formosus (scales absent), from type form (scales present) and A. aegypti queenslandensis form (dense scaling). We performed quantitative trait locus (QTL) mapping using 3 criteria for scale patterns among 192 F1 intercross progeny from matings between a queenslandensis type and an aegypti type form. Results identified 3 QTL determining scale patterns and indicated that classification criteria impact robustness of QTL LOD support. Dark- and light-colored forms exist in sympatry, but vary in multiple phenotypic characteristics, including preferences for vertebrate host, oviposition container, house-entering behavior, and dengue vector competence. Markers associated with 2 QTL regions reflected major reductions in recombination frequencies compared with the standard type form linkage map, suggestive of inversion polymorphisms associated with observed linkage disequilibrium between type-specific characteristics. Understanding the genic basis for differences in A. aegypti forms could inform efforts to develop new mosquito and arboviral disease control strategies.
Key words: arbovirus, evolution, linkage analysis, quantitative genetics, quantitative trait locus, speciation
Aedes aegypti (L.) is the principal mosquito vector for transmission of dengue, chikungunya, Zika, and urban yellow fever. It has successfully established populations across much of the tropical and subtropical regions of the planet, and this range is expanding in concert with climate change (Capinha et al. 2014; Campbell et al. 2015; Kraemer et al. 2015). The species has evolved from a primarily dark-colored sylvan form with little human interaction in Sub-Saharan Africa, to include a light-colored domestic form well adapted to cohabitation with humans in urban environments (Powell and Tabachnick 2013). Early efforts to describe this variation distinguished 3 forms or subspecies that generally reflected the frequency of light-colored scales across the dorsal abdomen: 1) A. aegypti aegypti, the type form with a relatively small number of scales located mainly on anterior tergites; 2) A. aegypti formosus, the dark form with no dorsal scales; and 3) A. aegypti queenslandensis, the pale form with more extensive scaling across multiple tergites (Mattingly 1957; McClelland 1974). A review of A. aegypti domestication and population expansion with human activities does validate the concept of distinct sylvan and domestic forms (Powell and Tabachnick 2013), and recent genetic analyses confirmed an African ancestry and provided evidence for a single subspeciation event that produced the domestic form (Moore et al. 2013; Brown et al. 2014).
Evolution of the A. aegypti domestic form from a sylvan ancestor likely included simultaneous natural selection on multiple traits as evidenced by the observed genetic correlations between seemingly unrelated phenotypes that distinguish the 2 forms. Adaptation to human habitats included anthropophilic blood feeding preferences, a propensity to enter human domiciles, oviposition in human derived containers, competence to transmit dengue virus, as well as the forementioned variability in scale patterns (Mattingly 1967; Tabachnick and Powell 1979; Wallis et al. 1983; Failloux et al. 2002). Although the 2 forms continue to exist in sympatry in Sub-Saharan Africa, they can readily mate (Moore 1979). Of interest too, are multiple reports of phenotypic changes in the domestic form associated with wet versus dry season including scale patterns and breeding site choice (Tsuda et al. 2003; Huber et al. 2008; Sylla et al. 2009; Paupy et al. 2010).
Here, we performed genome-wide quantitative trait locus (QTL) mapping of dorsal abdominal tergite scale patterns among F1 intercross progeny from matings between laboratory colonies representing a queenslandensis type originating from Indonesia and an aegypti type form originating from Tanzania. We compared QTL results for abdominal scale patterns using 3 criteria modified around the CKM system of classification (Hartberg et al. 1986). Our results indicate the influence of at least 3 QTL determining scale patterns in A. aegypti, and that classification criteria have an impact on robustness of LOD support for individual QTL.
Materials and Methods
Mosquito Strains and Crosses
The A. aegypti aegypti type form strain (Tanz) was established from stocks collected in Tanzania. The A. aegypti queenslandensis form strain (Queen) was derived following 7 generations of selection for dense tergal abdominal scaling from stocks collected in Surabaya, Indonesia, as described elsewhere (Tsuda et al. 2003). Typical scale patterns for individuals in each strain are shown in Figure 1. Mosquitoes were reared and maintained in an environmental chamber at 27 °C and 80% relative humidity under a 16-h light:8-h dark cycle that included a 1h crepuscular period at the beginning and end of each cycle. Adults were provided a 2% sucrose solution ad libitum. Adult females 5 days post-eclosion were blood fed on anesthetized mice to stimulate oviposition. Genetic and phenotypic data were derived from an F1 intercross population prepared by pairwise matings, where a single Tanz strain male and 5 virgin Queen strain females were placed in a 30×30×30cm cheesecloth cage. Individual females were subsequently transferred to 10mL glass vials and provided a strip of paper towel as an oviposition substrate. F1 progeny from individual females were reared in 40×25×8cm plastic cheesecloth covered containers and thereafter subjected to pairwise matings as described above. F2 adults were scored for phenotype as described below and then preserved in 95% ethanol.
Figure 1.
Examples of Aedes aegypti aegypti type form (left panels) and A. aegypti queenslandensis form (right panels).
Phenotype Analysis
We employed 3 methods for classification of abdominal tergite scaling patterns. For scale phenotype A (spA), we followed the CKM method as outlined by Hartberg et al. (1986) that rates individual tergites for abdominal segments I–VII as no white scales present (score = 0) or some white scale(s) present (score = 1), wherein total scores per individual ranged from 0 to 7. For phenotype B (spB), we rated individual tergites with white scales present that defined a median anterior-posterior line across the entire tergite as score = 1; if not, the score = 0. For phenotype C (spC), we rated individual tergites as being covered by >50% white scales as score = 1, otherwise the score = 0. Again, for spB and spC total scores per individual ranged from 0 to 7.
DNA Extractions and Genotyping
In preparation for DNA extractions, individuals were removed from ethanol, placed on filter paper, and allowed to dry at room temperature overnight. Thereafter, DNA extractions were performed as described previously (Severson et al. 1993). Purified DNA from each mosquito was resuspended in 15 µL of TE (10mM Tris–HCl, 1mM EDTA, pH 8.0). Individuals were genotyped at loci based on restriction fragment length polymorphism markers following Mori et al. (2008). PCR amplifications for loci based on comparative anchor-tagged sequence analyses were performed as described previously (Chambers et al. 2003) using 1 µL of a 1:1200 dilution of genomic DNA as template. Microsatellite locus genotyping followed standard protocols (Chambers et al. 2007; Lovin et al. 2009), with loci reported from multiple sources (Ravel et al. 2002; Chambers et al. 2007; Slotman et al. 2007; Lovin et al. 2009).
QTL Analysis
Multipoint linkage analysis was conducted using the MAPMAKER computer program (Lander et al. 1987) with a LOD of 3.0 as the threshold for significance. QTL controlling abdominal scale patterns were identified using the R/qtl computer package (Broman et al. 2003). QTL detection was performed by interval mapping using a nonparametric model. LOD significance thresholds for QTL were determined using 1000 permutations of the data. Confidence intervals for individual QTL were estimated as Bayes 95% credible intervals. The estimated phenotypic variance (EV) explained (i.e., QTL heritability H 2) was estimated from the difference in LOD scores as: EV = 1–10−(2/n)LOD (Broman and Sen 2009). Additive (a) and dominance (d) effects of individual QTL were calculated relative to alleles at the marker locus closest to the predicted QTL location relative to the Queen strain parent allele as outlined in Edwards et al. (1987). Mode of gene action was determined as the absolute value of d/a, where additive = 0–0.20, partial dominance = 0.21–0.80, dominance = 0.81–1.20, and overdominance >1.20 (Stuber et al. 1987; Babu et al. 2006). Graphical presentation of individual linkage groups was performed using the MapChart program (Voorrips 2002).
Results
Linkage Analysis
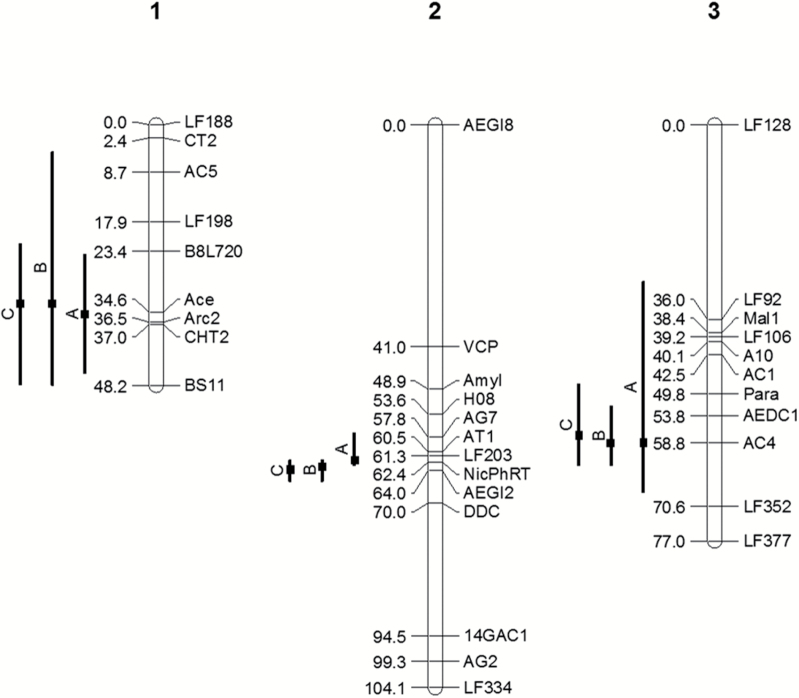
Linkage analysis was performed on 192 progeny from a Tanz × Queen F1 intercross population and included 33 marker loci distributed broadly across the majority of the 3 linkage groups (Severson et al. 2002): 9 loci on chromosome 1 spanning 48.2 cM (Haldane units) for a mean interval size of 6.0 cM; 13 loci on chromosome 2 spanning 104.1 cM for a mean interval size of 8.7 cM; and 11 loci on chromosome 3 spanning 77.0 cM for a mean interval size of 7.7 cM (Figure 2). Individual marker details are provided in Supplementary Table S1 online.
Figure 2.
Linkage map (Haldane cM) for F1 intercross progeny from matings between Aedes aegypti aegypti type form and A. aegypti queenslandensis form laboratory strains. Horizontal bars show predicted QTL positions and 95% Bayes confidence intervals for abdominal scale pattern phenotype classifications A, B, and C, respectively. QTL for phenotype B on chromosome 1 was not significant but is included for reference with the other phenotypes.
Phenotype Distribution
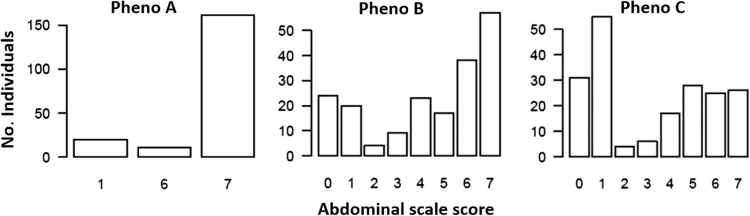
Histograms representing observed distributions of the F2 progeny observed using scale phenotype A, B, and C scoring criteria as outlined in Materials and Methods are shown in Figure 3. The spA data reflected a limited distribution where relatively small numbers of individuals received scores of 1 or 6, whereas the majority of individuals received the maximum score of 7. None of the progeny received a score of 0, reflecting the type form genetic backgrounds of the strains used for the crosses. Distributions observed for spB and spC spanned the entire scoring range but reflected opposite biases. For spB, a majority of the progeny received scores of 6 or 7. Conversely, with spC, a majority received scores of 0 or 1. All 3 phenotypes showed highly nonnormal distributions.
Figure 3.
Distribution of abdominal scale pattern classes observed for 192 F1 intercross progeny from matings between Aedes aegypti aegypti type form and A. aegypti queenslandensis form laboratory strains. See Materials and Methods for phenotype scoring criteria.
QTL Analysis
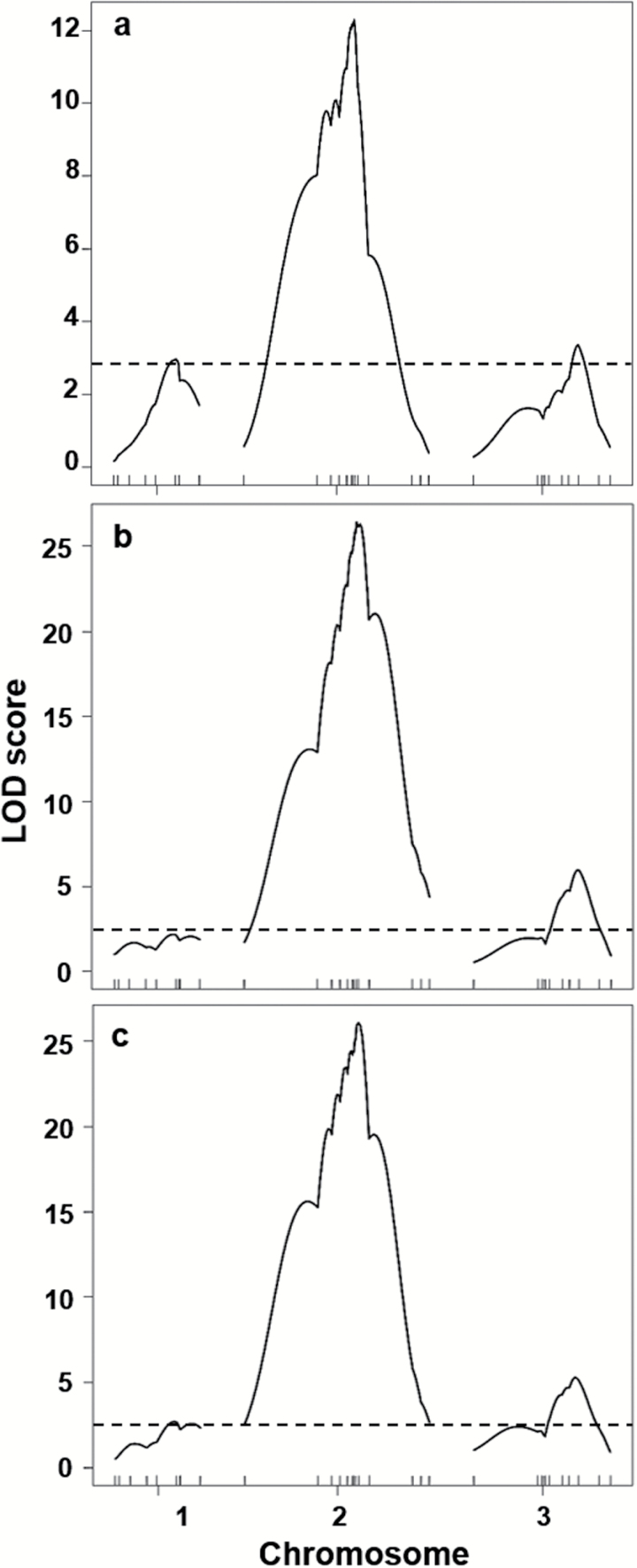
Simple interval mapping LOD results for each of the 3 phenotypes are shown in Figure 4. Three QTL genome regions, associated with each of the 3 A. aegypti linkage groups, were significantly linked to tergal abdominal scale patterns (Table 1, Figure 2). Results for chromosomes 2 and 3 were generally consistent across all 3 phenotypic classifications, whereas only spA and spC identified a genome region on chromosome 1 that exceeded the LOD threshold (P < 0.05). All 3 phenotypic classifications identified a major QTL in the same genome region on chromosome 2 that accounted for 26.5–46.9% of the estimated phenotypic variance (EV) explained, wherein spB and spC reflected the greatest EV. The 2 minor QTL on chromosomes 1 and 3 accounted for EV values of ~7–12%. Of note, however, the specific interval defining individual QTL positions varied across phenotype classifications, with spB and spC QTL each defined by the same flanking markers, whereas with spA QTL positions on chromosomes 1 and 2 were placed within an adjacent interval.
Figure 4.
LOD score profiles identifying QTL for abdominal scale phenotype classifications A, B, and C, respectively. The horizontal line represents the experimentwise threshold value (P = 0.05) for identifying a QTL based on 1000 permutations. See Figure 2 and Supplementary Table S1 online for information on marker loci.
Table 1.
QTL for abdominal scale patterns in Aedes aegypti
| Phenotype | Chrom | Position | LOD (P) | BCI (95%) | Closest marker | Flanking markers | EV | a | d | Action | Degree |
|---|---|---|---|---|---|---|---|---|---|---|---|
| A | 1 | 35.0 | 2.97 (0.019) | (24; 46) | Ace | Ace-ARC2 | 6.9 | 0.56 | −0.29 | PD | 0.52 |
| 2 | 62.0 | 12.29 (0.0001) | (57; 63) | NicPhRT | LF203-NicPhRT | 25.5 | 1.49 | 1.4 | D | 0.94 | |
| 3 | 58.8 | 3.37 (0.006) | (29; 68) | AC4 | AEDC1-AC4 | 7.8 | 0.65 | 0.4 | PD | 0.62 | |
| B | 1 | 33.0 | 2.16 (0.126) | (5; 48) | Ace | B8L720-Ace | na | na | na | na | na |
| 2 | 63.0 | 26.43 (0.0001) | (62; 66) | NicPhRT | NicPhRT-AEG12 | 46.9 | 3.13 | 1.6 | PD | 0.51 | |
| 3 | 58.8 | 5.96 (0.0001) | (52; 63) | AC4 | AEDC1-AC4 | 13.3 | 1.39 | 0.93 | PD | 0.67 | |
| C | 1 | 33.0 | 2.69 (0.046) | (22; 48) | Ace | B8L720-Ace | 6.2 | 0.83 | 0.65 | PD | 0.78 |
| 2 | 64.0 | 26.09 (0.0001) | (62; 66) | AEGI2 | NicPhRT-AEG12 | 46.5 | 2.86 | −0.02 | A | 0.01 | |
| 3 | 57.0 | 5.28 (0.0001) | (48; 63) | AC4 | AEDC1-AC4 | 11.9 | 1.39 | 0.76 | PD | 0.55 |
BCI, Bayes confidence interval; EV, estimated phenotypic variance explained; a, additive effect; d, dominance effect; Action, mode of action determined as d/a where additive = 0.0–0.20, partial dominance = 0.21–0.80, dominance = 0.81–1.20, and overdominance = >1.20 (after Stuber et al. 1987; Babu et al. 2006); Degree, d/a.
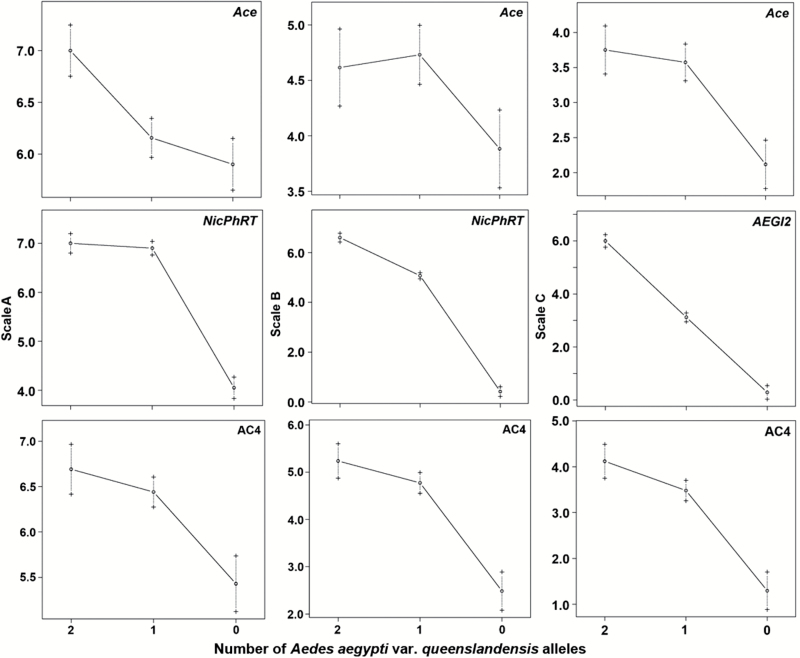
Estimates of additive (a) and dominance (d) effects allowed us to estimate degree of dominance (d/a) for each QTL. Gene effects were calculated relative to the Queen allele at the marker closest to the predicted QTL position (Table 1, Figure 5). For the QTL on chromosome 1, both spA and spC showed a partial dominance (PD) effect of the Queen allele (d/a for spB was not considered as the QTL region was not significant). For the QTL on chromosome 3, all 3 phenotype classifications showed a PD effect. Interestingly, for the QTL on chromosome 2, the observed QTL effect varied dependent on phenotype classification. The Queen allele effect for spA was dominant (D), for spB it was PD, and for spC it was additive (A).
Figure 5.
Means and standard deviations of abdominal scale classifications for genotypes at marker locus nearest QTL positions of 192 F1 intercross progeny from matings between Aedes aegypti aegypti type form and A. aegypti queenslandensis form laboratory strains. QTL for phenotype B on chromosome 1 was not significant, but data for Ace locus are included for reference with the other phenotypes.
Discussion
The evolution of A. aegypti aegypti type form as a human commensal has facilitated its remarkable success in colonizing a majority of the semitropical and tropical areas of the planet (Powell and Tabachnick 2013). Here, we investigated the genetic basis for white scale presence on abdominal tergites that represents the diagnostic for distinguishing the type form (with scales present) from its sylvan ancestor, A. aegypti formosus (scales absent), as well as the A. aegypti queenslandensis form (abdomen with dense scaling). We specifically performed QTL analyses among F2 progeny from crosses between a type form strain selected for an A. aegypti queenslandensis scale pattern phenotype characterized by dense white scale presence (Tsuda et al. 2003) and a type form strain originating from Tanzania. We elected to compare results from 3 different criteria for scoring the scale pattern type to determine if outcomes would be consistent or whether one criteria might prove more effective for defining the phenotype.
Our results validate a multigenic mode of inheritance for abdominal scale pattern among type form populations, wherein we identified 3 QTL located on the 3 A. aegypti chromosomes. These consist of a major QTL on chromosome 2, and 2 minor QTLs on chromosomes 1 and 3, respectively. Interestingly, classical inheritance studies by Hartberg et al. (1986) similarly suggested that abdominal scaling patterns were determined by 3 independently segregating loci. Here, alleles from the Queen genotype generally showed a partial dominant mode of inheritance, although mode of inheritance predicted for the major QTL on chromosome 2 was dependent on the choice of phenotype characterization and ranged from dominant (spA) to partial dominant (PD), to additive (spC) effect. Of note, multiple single locus scale pattern and color mutations are known to be located on chromosome 2 (Craig and Vandehey 1962; Munstermann and Craig 1979; Verna and Munstermann 2011), although none of these loci appear to be located within or even near the 95% Bayes confidence interval relative to the QTL genome position (Munstermann 1993; Severson et al. 2002, 2004).
Although evidence for gene introgression between A. aegypti forms is clear (Huber et al. 2008; Sylla et al. 2009), persistent evidence for linkage disequilibrium among genes determining the individual phenotypes suggests some level of integration of these as adaptive trait complexes. That is, for example, the finding of significant correlations between dry versus wet seasonal changes in abdominal scale patterns and oviposition in natural, peridomestic, or domestic containers (Tsuda et al. 2003; Paupy et al. 2010) as well as area of collection and preference for natural, peridomestic, or domestic habitats (Trpis and Hausermann 1975) strongly supports the case for linkage disequilibrium. Of note, the correlation between scale patterns and breeding site choice (Tsuda et al. 2003; Paupy et al. 2010) was not strictly defined by presence or absence of pale scales on the first abdominal tergite to infer A. aegypti type form versus sylvan (McClelland 1974). Instead, the significant correlations were defined by evaluating the scale patterns across abdominal tergites, which interestingly span the range of phenotypes we observed in our F2 segregant population using the spB or spC phenotype descriptions. These results suggest that correlations between phenotypes may be maintained by tight physical linkage of the underlying genes, existence of pleiotrophic gene effects, or strong selection for unlinked gene effects inherent to the 2 forms.
Intriguing questions remain as to the apparent disconnect between reported genetic introgression among A. aegypti forms in West Africa (Huber et al. 2008; Sylla et al. 2009), but not so in East Africa (Tabachnick et al. 1979), and particularly important to this discussion, the strong persistent correlations observed among phenotypes (Tsuda et al. 2003; Paupy et al. 2010) in both West Africa and Indonesia. A hypothesis that could explain these dichotomies would be that genes determining phenotypes characteristic of the different A. aegypti forms are located within chromosomal inversions. It is well established in other mosquito species, especially, for example, the Anopheles gambiae complex wherein sympatric speciation has occurred in association with well-defined inversion polymorphisms (Coluzzi et al. 1979). However, documenting the presence of inversions in A. aegypti is problematic as quality polytene chromosome preparations remain a challenge because of the highly repetitive nature of its genome (Brown et al. 1995). Results from classical linkage analysis of segregating progeny from crosses between A. aegypti aegypti type form and A. aegypti formosus suggested the presence of multiple inversions, that tentatively included up to 4 inversions on chromosome 1, one on chromosome 2, and up to 3 on chromosome 3 (Bernhardt et al. 2009). This conclusion was deduced from their finding that these genome regions reflected severe reductions in recombination frequencies as well as some evidence for changes in linear orders when compared with the standard A. aegypti aegypti type form map (Severson et al. 2002, 2004). Here, we found that markers associated with individual QTL regions for chromosomes 1 and 2 do reflect reductions in recombination frequencies as well. The map distances observed in the present study between B8L720-Arc2 (Chr 1) and between LF203-DDC (Chr 2) show ~59% and 57% reduction in map distance, respectively, compared with the standard map (Severson et al. 2002, 2004). However, we did not find any evidence of reduced recombination or change in linear orders among markers on chromosome 3.
Understanding the genic basis for the observed differences in A. aegypti forms remains an area of great interest, as this knowledge could inform efforts to develop new mosquito control and arboviral disease prevention strategies. The draft A. aegypti genome assembly (Nene et al. 2007) in its present fragmented form provides limited utility for detailed evaluation of the QTL regions identified here. The obvious need to better understand these and other important phenotypes, therefore, awaits availability of a finished assembly and that effort should be a research community priority.
Supplementary Material
Supplementary material can be found at http://www.jhered.oxfordjournals.org/.
Funding
National Institutes of Health, National Institute of Allergy and Infectious Diseases (RO1-AI059342, R56-AI110721-A1) to D.W.S.
Supplementary Material
References
- Babu R, Nair SK, Kumar A, Rao HS, Verma P, Gahalain A, Singh IS, Gupta HS. 2006. Mapping QTLs for popping ability in a popcorn x flint corn cross. Theor Appl Genet. 112:1392–1399. [DOI] [PubMed] [Google Scholar]
- Bernhardt SA, Blair C, Sylla M, Bosio C, Black WC IV. 2009. Evidence of multiple chromosomal inversions in Aedes aegypti formosus from Senegal. Insect Mol Biol. 18:557–569. [DOI] [PubMed] [Google Scholar]
- Broman KW, Sen S. 2009. A guide to QTL mapping with R/qtl. New York: Springer. [Google Scholar]
- Broman KW, Wu H, Sen S, Churchill GA. 2003. R/qtl: QTL mapping in experimental crosses. Bioinformatics. 19:889–890. [DOI] [PubMed] [Google Scholar]
- Brown JE, Evans BR, Zheng W, Obas V, Barrera-Martinez L, Egizi A, Zhao H, Caccone A, Powell JR. 2014. Human impacts have shaped historical and recent evolution in Aedes aegypti, the dengue and yellow fever mosquito. Evolution. 68:514–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SE, Menninger J, Difillipantonio M, Beaty BJ, Ward DC, Knudson DL. 1995. Toward a physical map of Aedes aegypti . Insect Mol Biol. 4:161–167. [DOI] [PubMed] [Google Scholar]
- Campbell LP, Luther C, Moo-Llanes D, Ramsey JM, Danis-Lozano R, Peterson AT. 2015. Climate change influences on global distributions of dengue and chikungunya virus vectors. Philos Trans R Soc Lond B Biol Sci. 370:20140135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capinha C, Rocha J, Sousa CA. 2014. Macroclimate determines the global range limit of Aedes aegypti . Ecohealth. 11:420–428. [DOI] [PubMed] [Google Scholar]
- Chambers EW, Lovin DD, Severson DW. 2003. Utility of comparative anchor-tagged sequences as physical anchors for comparative genome analysis among the Culicidae. Am J Trop Med Hyg. 69:98–104. [PubMed] [Google Scholar]
- Chambers EW, Meece JK, McGowan JA, Lovin DD, Hemme RR, Chadee DD, McAbee K, Brown SE, Knudson DL, Severson DW. 2007. Microsatellite isolation and linkage group identification in the yellow fever mosquito Aedes aegypti . J Hered. 98:202–210. [DOI] [PubMed] [Google Scholar]
- Coluzzi M, Sabatini A, Petrarca V, Di Deco MA. 1979. Chromosomal differentiation and adaptation to human environments in the Anopheles gambiae complex. Trans R Soc Trop Med Hyg. 73:483–497. [DOI] [PubMed] [Google Scholar]
- Craig GB, Vandehey RC. 1962. Genetic variability in Aedes aegypti (Diptera: Culicidae) I. Mutations affecting color pattern. Annals Entomol Soc Am 55:47–58. [Google Scholar]
- Edwards MD, Stuber CW, Wendel JF. 1987. Molecular-marker-facilitated investigations of quantitative-trait loci in maize. I. Numbers, genomic distribution and types of gene action. Genetics. 116:113–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Failloux AB, Vazeille M, Rodhain F. 2002. Geographic genetic variation in populations of the dengue virus vector Aedes aegypti . J Mol Evol. 55:653–663. [DOI] [PubMed] [Google Scholar]
- Hartberg WK, Meeks CK, Williams KR. 1986. A model for polygenic inheritance of abdominal tergal scale pattern in Aedes aegypti . J Am Mosq Control Assoc. 2:490–502. [PubMed] [Google Scholar]
- Huber K, Ba Y, Dia I, Mathiot C, Sall AA, Diallo M. 2008. Aedes aegypti in Senegal: genetic diversity and genetic structure of domestic and sylvatic populations. Am J Trop Med Hyg. 79:218–229. [PubMed] [Google Scholar]
- Kraemer MU, Sinka ME, Duda KA, Mylne AQ, Shearer FM, Barker CM, Moore CG, Carvalho RG, Coelho GE, Van Bortel W, et al. 2015. The global distribution of the arbovirus vectors Aedes aegypti and Ae. albopictus . Elife. 4:e08347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Green P, Abrahamson J, Barlow A, Daly M, Lincoln S, Newberg L. 1987. MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1:174–181. [DOI] [PubMed] [Google Scholar]
- Lovin DD, Washington KO, deBruyn B, Hemme RR, Mori A, Epstein SR, Harker BW, Streit TG, Severson DW. 2009. Genome-based polymorphic microsatellite development and validation in the mosquito Aedes aegypti and application to population genetics in Haiti. BMC Genomics. 10:590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattingly PF. 1957. Genetical aspects of the Aedes aegypti problem. I. Taxonom: and bionomics. Ann Trop Med Parasitol. 51:392–408. [PubMed] [Google Scholar]
- Mattingly PF. 1967. Taxonomy of Aedes aegypti and related species. Bull World Health Organ. 36:552–554. [PMC free article] [PubMed] [Google Scholar]
- McClelland GAH. 1974. A worldwide survey of variation in scale pattern of the abdominal tergum of Aedes aegypti (L.) (Dipera: Culicidae). Trans R Ent Soc Lond 126:239–259. [Google Scholar]
- Moore DF. 1979. Hybridization and mating behavior in Aedes aegypti (Diptera: Culicidae). J Med Entomol. 16:223–226. [DOI] [PubMed] [Google Scholar]
- Moore M, Sylla M, Goss L, Burugu MW, Sang R, Kamau LW, Kenya EU, Bosio C, de Lourdes Munoz M, Sharakova M, et al. 2013. Dual African origins of global Aedes aegypti s.l. populations revealed by mitochondrial DNA. PLoS Negl Trop Dis. 7:e2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori A, Romero-Severson J, Black WC IV, Severson DW. 2008. Quantitative trait loci determining autogeny and body size in the Asian tiger mosquito (Aedes albopictus). Heredity 101:75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munstermann LE. 1993. Gene map of the yellow fever mosquito, Aedes aegypti (Stegomyia) aegypti (2N=6). In: O’Brien SJ, editor. Genetic maps: locus maps of complex genomes, vol. 6. New York: Cold Spring Harbor; p. 3264–3268. [Google Scholar]
- Munstermann LE, Craig GB. 1979. Genetics of Aedes aegypti: updating the linkage map. J Hered 70:291–296. [Google Scholar]
- Nene V, Wortman JR, Lawson D, Haas B, Kodira C, Tu ZJ, Loftus B, Xi Z, Megy K, Grabherr M, et al. 2007. Genome sequence of Aedes aegypti, a major arbovirus vector. Science. 316:1718–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paupy C, Brengues C, Ndiath O, Toty C, Hervé JP, Simard F. 2010. Morphological and genetic variability within Aedes aegypti in Niakhar, Senegal. Infect Genet Evol. 10:473–480. [DOI] [PubMed] [Google Scholar]
- Powell JR, Tabachnick WJ. 2013. History of domestication and spread of Aedes aegypti–a review. Mem Inst Oswaldo Cruz. 108(Suppl 1):11–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravel S, Hervé JP, Diarrassouba S, Kone A, Cuny G. 2002. Microsatellite markers for population genetic studies in Aedes aegypti (Diptera: Culicidae) from Côte d’Ivoire: evidence for a microgeographic genetic differentiation of mosquitoes from Bouaké. Acta Trop. 82:39–49. [DOI] [PubMed] [Google Scholar]
- Severson DW, DeBruyn B, Lovin DD, Brown SE, Knudson DL, Morlais I. 2004. Comparative genome analysis of the yellow fever mosquito Aedes aegypti with Drosophila melanogaster and the malaria vector mosquito Anopheles gambiae . J Hered. 95:103–113. [DOI] [PubMed] [Google Scholar]
- Severson DW, Meece JK, Lovin DD, Saha G, Morlais I. 2002. Linkage map organization of expressed sequence tags and sequence tagged sites in the mosquito, Aedes aegypti . Insect Mol Biol. 11:371–378. [DOI] [PubMed] [Google Scholar]
- Severson DW, Mori A, Zhang Y, Christensen BM. 1993. Linkage map for Aedes aegypti using restriction fragment length polymorphisms. J Hered. 84:241–247. [DOI] [PubMed] [Google Scholar]
- Slotman MA, Kelly NB, Harrington LC, Kitthawee S, Jones JW, Scott TW, Caccone A, Powell JR. 2007. Polymorphic microsatellite markers for studies of Aedes aegypti (Diptera: Culicidae), the vector of dengue and yellow fever. Mol Ecol Notes 7:168–171. [Google Scholar]
- Stuber CW, Edwards MD, Wendel JF. 1987. Molecular marker facilitated investigations of quantitative trait loci in maize. II. Factors influencing yield and its component traits. Crop Sci 27:639–648. [Google Scholar]
- Sylla M, Bosio C, Urdaneta-Marquez L, Ndiaye M, Black WC., 4th 2009. Gene flow, subspecies composition, and dengue virus-2 susceptibility among Aedes aegypti collections in Senegal. PLoS Negl Trop Dis. 3:e408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabachnick WJ, Munstermann LE, Powell JR. 1979. Genetic distinctness of sympatric forms of Aedes aegypti in East Africa. Evolution 33:287–296. [DOI] [PubMed] [Google Scholar]
- Tabachnick WJ, Powell JR. 1979. A world-wide survey of genetic variation in the yellow fever mosquito, Aedes aegypti . Genet Res. 34:215–229. [DOI] [PubMed] [Google Scholar]
- Trpis M, Hausermann W. 1975. Demonstration of differential domesticity of Aedes aegypti (L.) (Diptera, Culicidae) in Africa by mark release recapture. Bull Entomol Res 65:199–208. [Google Scholar]
- Tsuda Y, Yotopranoto S, Bendryman SS, Rosmanida R, Dachlan YP, Takagi M. 2003. Seasonal changes in variation of dorsal scale pattern of Aedes aegypti (L.) (Diptera: Culicidae) in Surabaya, Indonesia. Jpn Soc Med Entomol Zool 54:73–80. [Google Scholar]
- Verna TN, Munstermann LE. 2011. Morphological variants of Aedes aegypti collected from the Leeward Island of Antigua. J Am Mosq Control Assoc. 27:308–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorrips RE. 2002. MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered. 93:77–78. [DOI] [PubMed] [Google Scholar]
- Wallis GP, Tabachnick WJ, Powell JR. 1983. Macrogeographic genetic variation in a human commensal: Aedes aegypti, the yellow fever mosquito. Genet Res. 41:241–258. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.