Abstract
Hydrophobic nanoparticles have shown substantial potential for bioanalysis and biomedical applications. However, their use is hindered by complex phase transfer and inefficient surface modification. This paper reports a facile and universal strategy for phase transfer and surface biofunctionalization of hydrophobic nanomaterials using aptamer-pendant DNA tetrahedron nanostructures (Apt-tet). The Janus DNA tetrahedron nanostructures are constructed by three carboxyl group modified DNA strands and one aptamer sequence. Each tetrahedron edge is an 18-base-pair double helix, making the tetrahedral edges about 5.8 nm in length. The pendant linear sequence is an aptamer, in this case AS1411, known to specifically bind nucleolin, typically overexpressed on the plasma membranes of tumor cells. The incorporation of the aptamers adds targeting ability and also enhances intracellular uptake. Phase-transfer efficiency using Apt-tet is much higher than that achieved using single-stranded DNA. In addition, the DNA tetrahedron nanostructures can be programmed to permit the incorporation of other functional nucleic acids, such as DNAzymes, siRNA, or antisense DNA, allowing, in turn, the construction of promising theranostic nanoagents for bioanalysis and biomedical applications. Given these unique features, we believe that our strategy of surface modification and functionalization may become a new paradigm in phase-transfer-agent design and further expand biomedical applications of hydrophobic nanomaterials.
Owing to their unique physical and chemical properties, hydrophobic nanoparticles have attracted considerable attention for their potential biomedical applications in, for example, biosensing, cell imaging, and cancer therapy.1–3 In particular, iron oxide nanoparticles (IONPs), because of their strong magnetic susceptibility, chemical stability, and low toxicity, have been intensively investigated as T2-negative contrast agents for magnetic resonance imaging (MRI), drug delivery, cell tracking, and magnetic hyperthermia.4–6 Currently, commercially available IONPs are usually synthesized by the coprecipitation method in aqueous media. However, IONPs synthesized under these conditions present a wide size distribution and poor crystallinity as a consequence of the low boiling point of water. Therefore, in order to prepare high-quality and uniform IONPs, many high boiling point nonpolar solvents have been used as reaction media. Unfortunately, IONPs synthesized in nonpolar solvents are often coated with hydrophobic ligands, making them water insoluble with correspondingly limited biological applications. Recently, many phase-transfer agents have been exploited to transfer hydrophobic IONPs to an aqueous phase via ligand exchange routes, using 2,3-dimercaptosuccinic acid (DMSA),7 3,4-dihydroxyhydrocinnamic acid (DHAC),8 silanes,9 etc. In particular, dopamine-based agents can form a stable, robust anchor on the surface of hydrophobic IONPs without regard to surface functional groups, rendering them hydrophilic.10–13 The process of phase transfer is simple and high efficiency. While substantial progress has been made, some flaws remain. Specifically, the above-mentioned phase-transfer agents are commonly monofunctional, making it necessary to incorporate biological moieties for specific molecular recognition through further surface functionalization steps. This makes the entire process complicated, time- and labor-consuming, and energy intensive. Moreover, some of them suffer from safety issues.13 Therefore, the development of a facile and universal method for phase transfer of hydrophobic nanoparticles, as well as readily attainable surface biofunctionalization, remains a desirable goal.
Since the pioneering work of Seeman and Mirkin et al. in the 1990s,14,15 interest in functional DNA nanostructures has increased rapidly. It is now possible to assemble many different molecular shapes and structures from DNA.16–18 Among them, the three-dimensional (3D) DNA tetrahedron nanostructure has excellent mechanical rigidity and structural stability.19 The DNA tetrahedron is a pyramid-like structure with four triangular faces and six double-stranded (dsDNA) edges. It can be rapidly assembled by annealing four designed single-stranded DNA, and each vertex can be readily functionalized with different chemical moieties and biomolecules. By exploiting these properties, the DNA tetrahedron has found a broad spectrum of applications in sensor construction, drug delivery, and molecular logic gates.20–23 To the best of our knowledge, using the DNA tetrahedron nanostructure as a phase-transfer agent has not been previously reported.
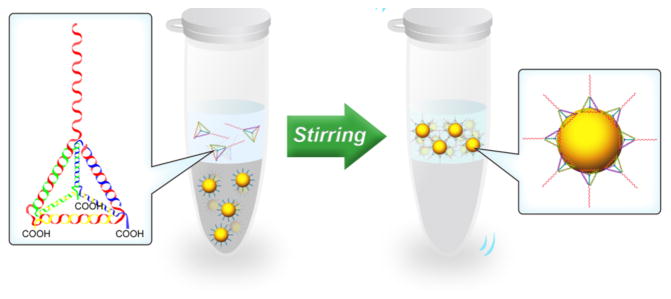
Herein, we describe a facile and universal method for phase transfer and surface biofunctionalization of hydrophobic nanoparticles using Janus DNA tetrahedron nanostructures (Scheme 1). To demonstrate the utility of our design, we employed hydrophobic IONPs as the first model. The Janus DNA tetrahedron nanostructures were first constructed by three carboxyl group modified DNA strands (strand A, B, C, 55-nt) and one aptamer sequence containing DNA strand (strand D, 87-nt) (see Table S1 for detailed sequences). Each edge of a tetrahedron is an 18-base-pair double helix, making the tetrahedral edges 5.8 nm in length.24 The presence of carboxyl groups on three tetrahedron vertices may favor the attachment of IONPs, since carboxyl groups have a strong affinity for Fe3+ ions on the surface of IONPs.25,26 Therefore, the DNA tetrahedron nanostructures could firmly adhere to oleic acid coated IONPs by ligand exchange, rendering the particles water soluble. The pendant linear sequence of strand D is aptamer AS1411, known to specifically bind nucleolin, which is typically overexpressed on the plasma membrane of tumor cells,27 thus enhancing intracellular uptake. As such, this Janus DNA tetrahedron functionalized with IONPs possesses excellent selectivity to cancer cells and could, moreover, serve as a contrast agent for T2-weighted MRI.
Scheme 1.
Phase Transfer and Surface Biofunctionalization of Hydrophobic Nanoparticles through a Facile Ligand Exchange Strategy
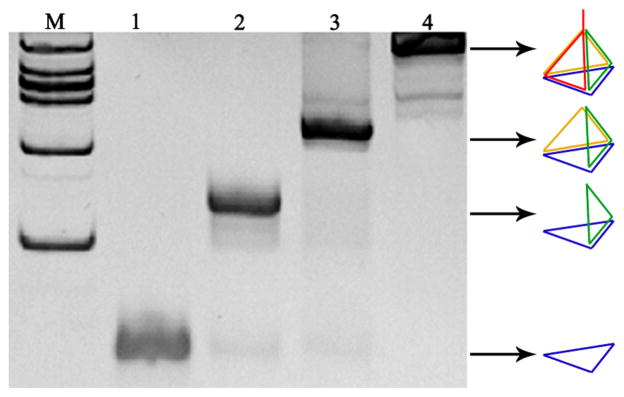
To self-assemble the Janus DNA tetrahedron nanostructures, strands A, B, C, and D were mixed in stoichiometric equivalents in buffer, heated, and then rapidly cooled to 4 °C. Formation of the aptamer-pendant DNA tetrahedron nanostructures (Apt-tet) was verified by native polyacrylamide gel electrophoresis (PAGE) (Figure 1). As strands were added from lane 1 to lane 4, the DNA tetrahedron migrated more slowly than the ssDNA and other DNA combinations lacking one or two strands. The results demonstrated that the DNA tetrahedron did, indeed, form as designed and that the self-assembly process was carried out efficiently by the presence of clean single bands.
Figure 1.
Analysis by 12.5% native polyacrylamide gel electrophoresis. Lane 1 is strand A, lane 2 is strand A + strand B, lane 3 is strand A + strand B + strand C, and lane 4 is strand A + strand B + strand C + strand D.
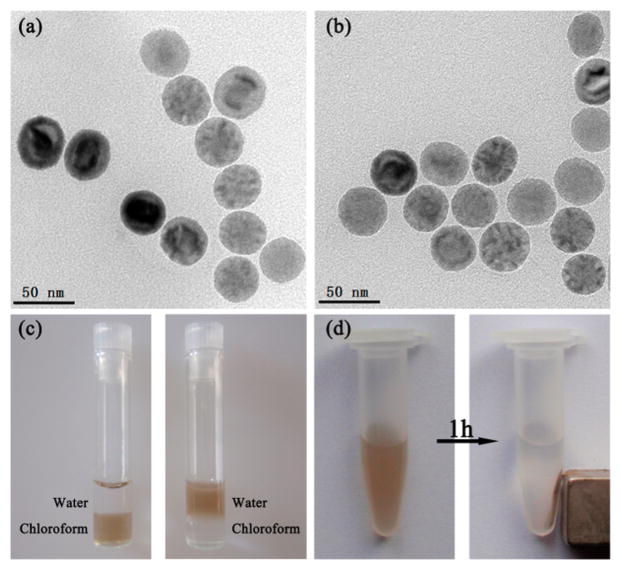
To test the phase-transfer ability of Apt-tet, we first applied oleic acid capped IONPs as a model. The hydrophobic IONPs were spherical and fairly monodisperse, as shown by transmission electron microscopy (TEM) (Figure 2a). To initiate phase transfer, IONPs in chloroform were mixed with Apt-tet in water, and the solution was vigorously stirred at 4 °C. During the mixing process, the brown color gradually disappeared from the chloroform phase, and the aqueous layer became brown with a colorless chloroform phase on the underside (Figure 2c). In addition, images of Apt-tet-IONPs in aqueous solution with and without a magnet clearly demonstrated its excellent magnetic properties (Figure 2d). After ligand exchange, excess Apt-tet was removed by washing, and the redispersed aptamer-pendant DNA tetrahedron nanostructure-functionalized IONPs (Apt-tet-IONPs) were inspected using TEM (Figure 2b). In addition to maintaining their spherical morphology, dynamic light scattering (DLS) data indicated that Apt-tet-IONPs (about 58 nm) showed larger size than that of IONPs (about 43 nm) (Figure S1). These results suggested that hydrophobic IONPs were stabilized by DNA tetrahedrons in water and formed uniformly hydrophilic nanoparticles. Additionally, the UV–vis spectrum of Apt-tet-IONPs showed a characteristic peak of DNA at 260 nm after DNA tetrahedron functionalization (red line), suggesting successful attachment of DNA tetrahedron to IONPs (Figure S2).
Figure 2.
(a) TEM images of IONPs before DNA tetrahedron functionalization in chloroform. (b) TEM images of IONPs after DNA tetrahedron functionalization in water. (c) Solvent dispersity of IONPs (left) before and (right) after DNA tetrahedron functionalization. (d) Magnetic separation of DNA tetrahedron nanostructure-functionalized IONPs.
To test the functionality of the DNA tetrahedron nanostructures attached on IONPs, Apt-tet-IONPs were assembled with 5 nm AuNPs modified with a complementary DNA strand (cDNA-AuNPs). As shown in Figure S3, Apt-tet-IONPs were surrounded by many cDNA-AuNPs, forming a satellite structure. As a comparison, when AuNPs were modified with a noncomplementary DNA strand (ncDNA-AuNPs), no assembly of the AuNPs was observed. These results were consistent with DLS analysis (Figure S3D) and confirmed that the DNA tetrahedrons not only were functionalized to IONPs but also retained their molecule recognition properties. Next, we investigated the solubility and stability of the Apt-tet-IONPs in aqueous solution with pH values ranging from 2 to 11. As shown in Figure S4, Apt-tet-IONPs demonstrated excellent dispersion and solubility over a wide range of pH (pH = 3–10), with aggregation only at extremely acidic (pH 2.0) and basic (pH 11.0) conditions. Additionally, Apt-tet-IONPs also exhibited good dispersion and stability in either phosphate buffered saline (PBS) or cell culture medium for over a period of two months (Figure S5). Thus, with long-term stability having been demonstrated, the Apt-tet-IONPs were found to be suitable for biomedical applications.
Since carboxyl groups have a strong affinity for Fe3+ ions, we surmised that the three carboxyl groups in the designed DNA tetrahedron (Apt-tet) may have a strong ability to drag IONPs from the organic phase into aqueous media. To confirm our hypothesis, we compared the phase-transfer ability of the carboxyl group-modified DNA tetrahedron nanostructures with two other DNA molecules: nonmodified DNA tetrahedron nanostructures (Nmo-tet, self-assembled by strands D, F, G, and H) and carboxyl-terminated ssDNA (Ct-ssDNA, strand I). Three kinds of DNA solution were mixed with hydrophobic IONPs under the same conditions, respectively. As expected, only the carboxyl group modified DNA tetrahedron nanostruc-tures could efficiently drag IONPs from the organic phase into aqueous media (Figure S6). Consequently, Apt-tet showed higher phase-transfer efficiency than either Nmo-tet or Ct-ssDNA. These results demonstrated that the strong affinity between carboxyl groups and Fe3+ ions is critically important for phase transfer of IONPs from chloroform to water. We presume that multivalent interactions between three carboxyl groups with IONPs have substantially contributed to the observed higher phase-transfer efficiency. Furthermore, the excellent structural stability and mechanical rigidity of the DNA tetrahedron nanostructures may also facilitate favorable interaction between Apt-tet and IONPs, resulting in the acceleration of phase transfer.
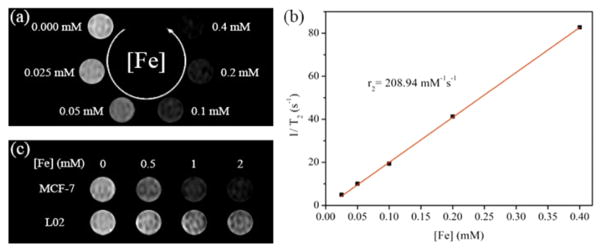
Having demonstrated a facile method for preparing hydrophilic Apt-tet-IONPs, we further explored their potential biomedical applications. IONPs are known to shorten the transverse relaxation time of water protons, and, as such, they have been used as negative contrast agents in MRI.4–6 To investigate the MRI performance of Apt-tet-IONPs, we conducted relaxivity and phantom MRI measurements on a 0.5 T MRI system. As shown in Figure 3, both T2 relaxation time and the corresponding signal intensity of the T2-weighted MR images of the Apt-tet-IONPs decreased as the iron concentration was increased. The T2-weighted MR images of the Apt-tet-IONPs exhibited a concentration-dependent darkening phenomenon, demonstrating the negative contrast enhancement by the Apt-tet-IONPs (Figure 3a). As expected, the transverse relaxation rates (1/T2) varied linearly with the iron concentration (Figure 3b). The weighted transverse-relaxivity value (r2) for Apt-tet-IONPs was calculated as 206.93 mM−1 s−1. This value shows the promise of Apt-tet-IONPs as T2 contrast agents. After demonstrating the good MRI performance of the transferred IONPs in aqueous media, we next established their utility in living cells. To accomplish this, Apt-tet-IONPs were incubated with MCF-7 and L02 cells for 1 h, and excess IONPs were removed by washing. MCF-7 cells treated with Apt-tet-IONPs presented higher T2-weighted MRI contrast than L02 cells treated with Apt-tet-IONPs (Figure 3c). These results indicated that the Apt-tet-IONPs retained their magnetic characteristics and that the aptamer-pendant DNA tetrahedron nanostructures could still recognize specific cancer cells, suggesting that the Apt-tet-IONP can be considered as a potential candidate for targeted MRI.
Figure 3.
(a) T2-weighted MR images of the Apt-tet-IONPs in aqueous solution at different iron concentrations. (b) Corresponding T2 relaxation rate of the Apt-tet-IONPs as a function of iron concentrations. (c) T2-weighted MR images of MCF-7 and L02 cells (5 × 105 cells) incubated with Apt-tet-IONPs at different iron concentrations.
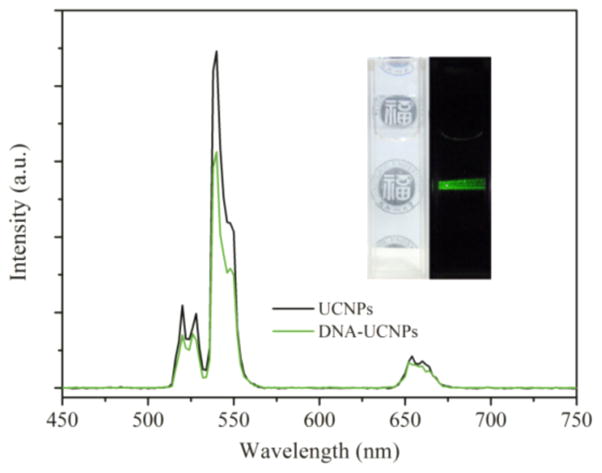
To explore the universality of this method, we applied upconversion nanoparticles (UCNPs) as another model. UCNPs in cyclohexane were mixed with Apt-tet in water, and the solution was vigorously stirred at 4 °C. Water-soluble UCNPs could be achieved in this process. The TEM image revealed that the prepared UCNPs were roughly monodisperse without aggregation (Figure S7). The DNA tetrahedron attachment on UCNPs was confirmed by the observation of a characteristic peak of DNA at 260 nm after DNA tetrahedron functionalization (Figure S8). Upconverting emission spectra of UCNPs in cyclohexane and aptamer-pendant DNA tetrahedron nanostructure-functionalized UCNPs (Apt-tet-UCNPs) in water are shown in Figure 4. Upon continuous excitation at 980 nm, the luminescence of the Apt-tet-UCNPs in water appeared predominantly green (Figure 4 inset and Figure S9). The emission peaks that appeared at 520, 539, and 654 nm were due to the transitions from the energy levels 4H11/2, 4S3/2, and 4F9/2 to 4I15/2 of Er3+. Furthermore, the corresponding upconversion luminescence spectrum of the Apt-tet-UCNPs in water was similar to that of the UCNPs in cyclohexane. These results revealed that the upconversion luminescent property of the UCNPs was unaffected by Apt-tet functionalization. In addition, Apt-tet-UCNPs also exhibited good dispersion and stability in a range of physiological solutions (Figure S10).
Figure 4.
Room-temperature upconversion luminescence spectrum of UCNPs in cyclohexane (black) and Apt-tet-UCNPs in water (green) under excitation at 980 nm. Inset: a photograph of Apt-tet-UCNPs in water taken under ambient light (left) and corresponding luminescence photograph in the dark under excitation by a 980 nm laser (right).
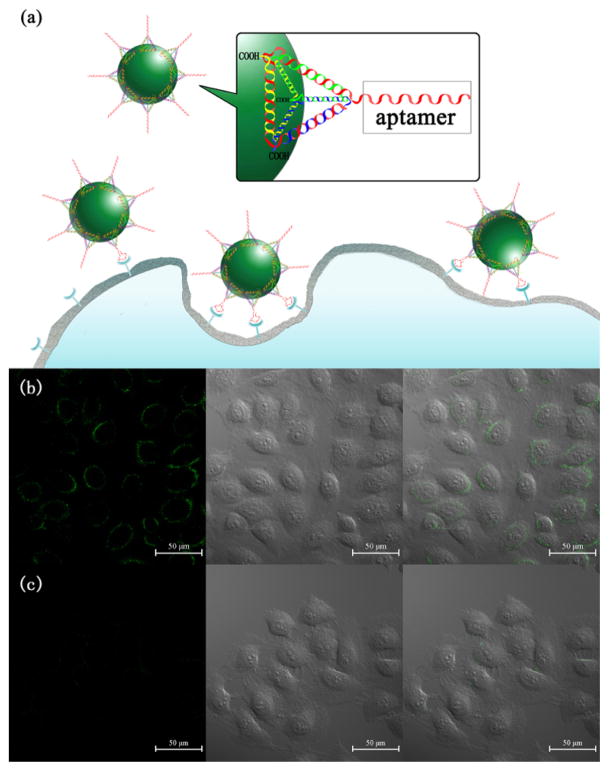
Finally, we tested the potential of the transferred UCNPs for cellular imaging (Figure 5a). The upconversion luminescence images of MCF-7 cells incubated with Apt-tet-UCNPs and random sequence pendant DNA tetrahedron nanostructure-functionalized UCNPs (Rdm-tet-UCNPs) were taken by a confocal microscope equipped with a 980 nm NIR laser. Upconversion luminescent microscopy (UCLM) results showed that the MCF-7 cells treated with Apt-tet-UCNPs displayed a strong green fluorescence (Figure 5b). In contrast, only negligible fluorescence was observed after MCF-7 cells were treated with Rdm-tet-UCNPs (Figure 5c). Furthermore, both the Apt-tet-UCNPs and Rdm-tet-UCNPs exhibited negligible binding with a nucleolin-negative cell line (L02 cells) (Figure S11), further validating aptamer-specific targeting.
Figure 5.
(a) Targeted imaging of cancer cells with DNA tetrahedron nanostructure-functionalized UCNPs. Confocal microscopy images of MCF-7 cells treated with (b) Apt-tet-UCNPs and (c) Rdm-tet-UCNPs. Each series can be classified as the upconversion luminescent images (left), bright-field (middle), and overlay of both (right), respectively.
In summary, we developed a facile and universal method for phase transfer and surface biofunctionalization of hydrophobic nanoparticles using the Janus DNA tetrahedron nanostructures. The designed DNA tetrahedron nanostructures not only drag IONPs from an organic phase into aqueous media but also give them target-specific properties. Significantly, the method is generally applicable to different types of hydrophobic nanoparticles, such as UCNPs. Moreover, functional nucleic acids, such as aptamer, DNAzymes, siRNA, or antisense DNA, can be readily incorporated into the desired tetrahedral DNA nanostructures to construct multifunctional phase-transfer agents. With these remarkable advantages, we believe that the proposed phase-transfer and surface functionalization method will provide new opportunities to extend the use of hydrophobic nanoparticles in biomedical applications.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge the financial support from the National Natural Science Foundation of China (No. 21125524, 21221003, 21327009, 21475026, 21305016), the National Institutes of Health (GM079359 and CA133086), the National Key Scientific Program of China (2011CB911000), and China Nat ional Instrumentat ion Program 2011YQ03012412.
Footnotes
Notes
The authors declare no competing financial interest.
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.5b05650.
Experimental details and additional characterization data (PDF)
References
- 1.Villaraza L, AJ, Bumb A, Brechbiel MW. Chem Rev. 2010;110:2921–2959. doi: 10.1021/cr900232t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee H, Yoon TJ, Figueiredo JL, Swirski FK, Weissleder R. Proc Natl Acad Sci U S A. 2009;106:12459–12464. doi: 10.1073/pnas.0902365106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Namiki Y, Namiki T, Yoshida H, Ishii Y, Tsubota A, Koido S, Nariai K, Mitsunaga M, Yanagisawa S, Kashiwagi H, Mabashi Y, Yumoto Y, Hoshina S, Fujise K, Tada N. Nat Nanotechnol. 2009;4:598–606. doi: 10.1038/nnano.2009.202. [DOI] [PubMed] [Google Scholar]
- 4.Gao J, Gu H, Xu B. Acc Chem Res. 2009;42:1097–1107. doi: 10.1021/ar9000026. [DOI] [PubMed] [Google Scholar]
- 5.Lee DE, Koo H, Sun IC, Ryu JH, Kim K, Kwon IC. Chem Soc Rev. 2012;41:2656–2672. doi: 10.1039/c2cs15261d. [DOI] [PubMed] [Google Scholar]
- 6.Lin LS, Cong ZX, Cao JB, Ke KM, Peng QL, Gao J, Yang HH, Liu G, Chen X. ACS Nano. 2014;8:3876–3883. doi: 10.1021/nn500722y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jun YW, Huh YM, Choi JS, Lee JH, Song HT, Kim S, Yoon S, Kim KS, Shin JS, Suh JS, Cheon J. J Am Chem Soc. 2005;127:5732–5733. doi: 10.1021/ja0422155. [DOI] [PubMed] [Google Scholar]
- 8.Liu Y, Chen T, Wu C, Qiu L, Hu R, Li J, Cansiz S, Zhang L, Cui C, Zhu G, You M, Zhang T, Tan W. J Am Chem Soc. 2014;136:12552–12555. doi: 10.1021/ja5060324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Palma R, Peeters S, Van Bael MJ, Van den Rul H, Bonroy K, Laureyn W, Mullens J, Borghs G, Maes G. Chem Mater. 2007;19:1821–1831. [Google Scholar]
- 10.Xu CJ, Xu KM, Gu HW, Zheng RK, Liu H, Zhang XX, Guo ZH, Xu B. J Am Chem Soc. 2004;126:9938–9939. doi: 10.1021/ja0464802. [DOI] [PubMed] [Google Scholar]
- 11.Cheng K, Peng S, Xu C, Sun S. J Am Chem Soc. 2009;131:10637–10644. doi: 10.1021/ja903300f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ling D, Park W, Park YI, Lee N, Li F, Song C, Yang SG, Choi SH, Na K, Hyeon T. Angew Chem, Int Ed. 2011;50:11360–11365. doi: 10.1002/anie.201101521. [DOI] [PubMed] [Google Scholar]
- 13.Shultz MD, Reveles JU, Khanna SN, Carpenter EE. J Am Chem Soc. 2007;129:2482–2487. doi: 10.1021/ja0651963. [DOI] [PubMed] [Google Scholar]
- 14.Kallenbach NR, Ma RI, Seeman NC. Nature. 1983;305:829–831. [Google Scholar]
- 15.Jones MR, Seeman NC, Mirkin CA. Science. 2015;347:1260901. doi: 10.1126/science.1260901. [DOI] [PubMed] [Google Scholar]
- 16.Tao J, Zheng J, Li JS, Zhao P, Li JP, Ma C, Yi M, Yang RH. Sci China: Chem. 2014;57:453–458. [Google Scholar]
- 17.Li J, Zheng C, Cansiz S, Wu C, Xu J, Cui C, Liu Y, Hou W, Wang Y, Zhang L, Teng IT, Yang HH, Tan W. J Am Chem Soc. 2015;137:1412–1415. doi: 10.1021/ja512293f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen X, Hong CY, Lin YH, Chen JH, Chen GN, Yang HH. Anal Chem. 2012;84:8277–8283. doi: 10.1021/ac3017828. [DOI] [PubMed] [Google Scholar]
- 19.Shih WM, Quispe JD, Joyce GF. Nature. 2004;427:618–621. doi: 10.1038/nature02307. [DOI] [PubMed] [Google Scholar]
- 20.Wen Y, Pei H, Wan Y, Su Y, Huang Q, Song S, Fan C. Anal Chem. 2011;83:7418–7423. doi: 10.1021/ac201491p. [DOI] [PubMed] [Google Scholar]
- 21.Lee H, Lytton-Jean AKR, Chen Y, Love KT, Park AI, Karagiannis ED, Sehgal A, Querbes W, Zurenko CS, Jayaraman M, Peng CG, Charisse K, Borodovsky A, Manoharan M, Donahoe JS, Truelove J, Nahrendorf M, Langer R, Anderson DG. Nat Nanotechnol. 2012;7:389–393. doi: 10.1038/nnano.2012.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li J, Pei H, Zhu B, Liang L, Wei M, He Y, Chen N, Li D, Huang Q, Fan C. ACS Nano. 2011;5:8783–8789. doi: 10.1021/nn202774x. [DOI] [PubMed] [Google Scholar]
- 23.Pei H, Liang L, Yao G, Li J, Huang Q, Fan C. Angew Chem, Int Ed. 2012;51:9020–9024. doi: 10.1002/anie.201202356. [DOI] [PubMed] [Google Scholar]
- 24.Flory JD, Shinde S, Lin S, Liu Y, Yan H, Ghirlanda G, Fromme P. J Am Chem Soc. 2013;135:6985–6993. doi: 10.1021/ja400762c. [DOI] [PubMed] [Google Scholar]
- 25.Liu J, Sun ZK, Deng YH, Zou Y, Li CY, Guo XH, Xiong LQ, Gao Y, Li FY, Zhao DY. Angew Chem, Int Ed. 2009;48:5875–5879. doi: 10.1002/anie.200901566. [DOI] [PubMed] [Google Scholar]
- 26.Huang G, Zhu X, Li H, Wang L, Chi X, Chen J, Wang X, Chen Z, Gao J. Nanoscale. 2015;7:2667–2675. doi: 10.1039/c4nr06616b. [DOI] [PubMed] [Google Scholar]
- 27.Qiu L, Wu C, You M, Han D, Chen T, Zhu G, Jiang J, Yu R, Tan W. J Am Chem Soc. 2013;135:12952–12955. doi: 10.1021/ja406252w. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.