Abstract
Objective
The human cytidine deaminase APOBEC3G (A3G) potently restricts HIV-1 but the virus, in turn, expresses a Vif protein which degrades A3G. A natural A3G-H186R variant, common in African populations, has been associated with a more rapid AIDS disease progression, but the underlying mechanism remains unknown. We hypothesized that differences in HIV-1 Vif activity towards A3G wild-type (WT) and A3G-H186R contribute to the distinct clinical AIDS manifestation.
Methods
Vif variants were cloned from plasma samples of 26 South African HIV-1 subtype C infected patients that either express WT A3G or A3G-H186R. The Vif alleles were assessed for their ability to counteract A3G variants using Western blot and single-cycle infectivity assays.
Results
We obtained a total of 392 Vif sequences which displayed an amino acid sequence difference of 6.2–19.2% between patients. The intra-patient Vif diversity from patient groups A3GWT/WT, A3GWT/H186R and A3GH186R/H186R was similar. Vif variants obtained from patients expressing A3GWT/WT and A3GH186R/H186R were capable of counteracting both A3G variants with similar efficiency. However, the antiviral activity of A3G-H186R was significantly reduced in both the presence and absence of Vif, indicating that the A3G-H186R variant intrinsically exerts less antiviral activity.
Conclusion
A3G WT and A3G-H186R are equally susceptible to counteraction by Vif, regardless of whether the Vif variant was obtained from A3GWT/WT and A3GH186R/H186R patients. However, the A3G-H186R variant intrinsically displayed lower antiviral activity, which could explain the higher plasma viral loads and accelerated disease progression reported for patients expressing A3GH186R/H186R.
Keywords: HIV-1, HIV, Vif, subtype C, APOBEC3, APOBEC3G, Restriction factor
Introduction
Successful HIV-1 replication in the host depends on its ability to evade a myriad of innate and adaptive immune defences [1–7]. Among the innate immune factors that exert pressure on HIV-1 are host restriction factors such as the apolipoprotein B mRNA-editing enzyme catalytic polypeptide-like 3G protein (APOBEC3G, A3G), which belongs to the family of cytidine deaminases [8, 9]. A3G inhibits HIV-1 replication by deaminating single stranded viral DNA during reverse transcription resulting in guanine-to-adenine (G-to-A) mutations across the proviral genome [8, 10, 11]. Deaminase-independent mechanisms of A3G restriction have also been described [12–15]. HIV-1 viral infectivity factor protein Vif, promotes the proteasomal degradation of A3G allowing productive viral replication [16–18].
A natural A3G polymorphism H186R is frequent in African populations with minor allele frequencies ranging from 30% in South Africans [19], and between 25% to 51% in other African populations [20]. The polymorphism is rare in Caucasian (2% to 3%) and Asian (0% to 10%) populations [20]. This A3G-H186R variant has been associated with higher viral loads, decreased CD4+ T cell counts and a more rapid progression to AIDS in patients homozygous for A3G-H186R [19, 21–23]. However, other studies which mainly analysed the effect of the heterozygous A3GWT/H186R variant found no association with disease progression [24–27]. Despite the clear correlation of A3G-H186R in vivo, the mechanism remains unknown [21, 28].
We recently showed that HIV-1 Vif adapts to different APOBEC3H (A3H) haplotypes in HIV-1-infected patients [29]. However, it remains unknown whether A3G haplotypes similarly select for specific Vif variants. If a given A3G variant is more or less susceptible to HIV-1 Vif mediated degradation it is conceivable that this altered viral host interaction could result in an altered HIV/AIDS disease presentation.
Of note, most Vif-A3G studies have focused on subtype B Vif variants, which only represent ~10% of all global infections [30]. However, the greatest burden of infections is in sub-Saharan Africa where HIV-1 subtype C predominates [30]. Only two studies, including one from our group, functionally analysed a limited number of subtype C variants. Both studies concluded that subtype C Vif had similar or enhanced activity against WT A3G [31, 32].
To our knowledge, the current study is the first to investigate the anti-A3G phenotype of subtype C Vif alleles obtained from patients with distinct A3G haplotypes.
Materials and Methods
Study participants
We selected 26 women from the “Centre for the AIDS Programme of Research in South Africa” acute infection study (CAPRISA 002) in Durban, South Africa [33] based on the A3G genotype information that was previously determined [19]. Plasma viremia and CD4+ T cell counts are regularly documented and samples are stored for future research. Participants provided written informed consent and ethical approval was obtained from the Biomedical Research Ethics Committee of the University of KwaZulu-Natal.
We obtained plasma samples from 11 A3GWT/WT, 10 A3GWT/H186R and 5 A3GH186R/H186R chronically infected, antiretroviral therapy-naïve study participants. The plasma viremia and absolute CD4+ T cell counts were obtained approximately 36 months post-infection and were comparable between patients with different A3G haplotypes.
HIV-1 Vif Amplification and Cloning
Viral RNA was extracted from plasma using QIAamp Viral RNA Mini Kit (Qiagen, Hilden, Germany) and cDNA synthesized using Thermoscript RT PCR System (Invitrogen, ThermoFisher Scientific, Massachusetts, USA). The HIV-1 Vif coding region was amplified by nested PCR with the Expand High Fidelity PCR System (Roche, Penzberg, Germany) using the primers: Vif1-forward 5’AAAATTAGCAGGAAGATGGCCAGT3’ and Vif1-reverse 5’CTCCGCTTCTTCCTGCCATAGGAGAT3’, and Vif2-forward 5’TACTCTGGAAAGGTGAAGG3’ and Vif2-reverse 5’ CTTCCTGCCATAGGAGATGCCTAA3’. Five separate PCR reactions were performed per patient and gel purified amplicons were cloned into the pCR2.1 TOPO cloning vector (Invitrogen). Two to four clones of each PCR were sequenced resulting in 10 to 20 sequences for each patient. The GenBank accession numbers for the vif sequences generated in this study are KT881902 - KT882293.
Vif and A3G Expression Plasmids
One representative HIV-1 Vif variant from each of the 11 A3GWT/WT and 5 A3GH186R/H186R donors was selected for functional characterization. The Vif ORF was carboxy-terminal FLAG tagged and cloned into the mammalian expression plasmid pCRV1 as previously described [34, 35]. Carboxy-terminal Hemagglutinin (HA)-tagged WT A3G and A3G-H186R were cloned into the mammalian expression plasmid PTR600 as previously described [36].
Cell Culture
TZM-bl cells were provided by J. C. Kappes and X. Wu through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, National Institutes of Health, NIH Reagent Program. HEK-293T and TZM-bl were maintained at 37°C in a humidified atmosphere of 5% CO2 in Dulbecco's high-glucose modified Eagle's medium (CellGro, Corning, New York, USA), supplemented with 10% fetal bovine serum (FBS) and Penicillin/Streptomycin.
A3G Degradation and Single-Cycle Viral Infectivity Assays
HEK-293T cells were co-transfected with 500 ng of HIV pNL4-3Δvif, 50 ng of each Vif expression plasmid and 20 ng of WT or A3G-H186R with 4 mg/ml of polyethylenimine as previously described [36]. The replication-competent molecular clone NL4-3 ΔVif was provided by the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, National Institutes of Health. After 48 hours, viral supernatants were collected and the cells were lysed and analysed by western blot as previously described [36]. Viral supernatants were used to infect TZM-bl cells and β-galactosidase activity was measured 48 hours post-infection as previously described [29].
Statistical Analysis
GraphPad Prism version 5.01 was used for statistical analyses (paired and unpaired t tests). P-values less than 0.05 were considered significant. Average relative infectivity values and their standard deviations were calculated from representative triplicate transfections.
Results
Phylogenetic Analysis of Vif sequences
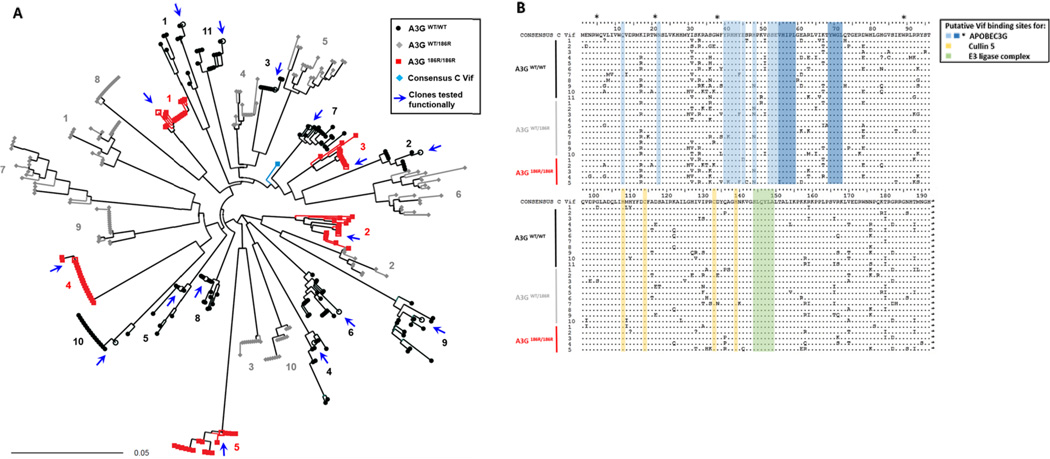
Despite the availability of HIV-1 subtype C vif sequences [37–39], functional data regarding their anti-A3G activity remains very limited [31, 32]. We therefore cloned, sequenced and analysed HIV-1 subtype C vif alleles from patients homozygous for WT A3G, homozygous for A3G-H186R and from heterozygous patients [19]. We generated 392 full length HIV-1 subtype C vif clonal sequences. Phylogenetic analysis confirmed that all sequences were subtype C (data not shown) and vif clonal sequences from each patient clustered independently (Figure 1A). Intra-patient sequences differed between 0.1% and 4.9% and inter-patient diversity ranged from 6.2% to 19.2% at the protein level. We observed no significant correlations between intra-patient sequence diversity and viral loads or CD4+ counts (data not shown).
Figure 1. Sequence analysis of patient-derived HIV-1 Vif sequences.
(A) Neighbour joining phylogenetic tree of 392 full length HIV-1 vif clonal sequences shows HIV-1 vif clonal sequences from each of 26 participants forming independent clusters. The patient’s A3G genotype from which Vif clones were derived are represented by the indicated symbols and colours. Vif clones that were functionally tested are represented by open symbols and arrows. Patient samples were assigned numbers that correspond in later figures. (B) Alignment of Vif amino acid consensus sequences of 26 study samples. Sequences are compared to a consensus subtype C reference sequence (obtained from the Los Alamos National Laboratory HIV database (http://www.hiv.lanl.gov). Protein domains putatively involved in interactions that lead to proteasomal degradation of A3G are indicated; blue indicates I9, N22, E45 and N48, YRHHY (40–44), amino acids 52 to 72 including the highlighted VHIPLx4-5Lx2YWGI motif which are important for binding to A3G. * indicates tryptophan residues important for A3G binding; yellow indicates the HCCH motif important for binding to Cullin 5 and green shows SLQYLA motif important for recruitment of ubiquitin-ligase (E3) complex containing elongin –B and –C, cullin-5 and Rbx.
An alignment of each patient’s consensus sequences is shown in Figure 1B. Putative sites of interaction with A3G or Cullin 5 and the E3 ubiquitin ligase complex are indicated by different colors (Figure 1B). The sites of potential interaction with A3G include amino acids at position 9, 22, 45 and 48 [34, 40]; the YRHHY motif (40–44) [41]; the VHIPLx4-5Lx2YWGL motif (positions 55–59, 64, 69–72) [42]; tryptophans at positions 5, 21, 38 and 89 [43], were not different between sequences. Similarly the HCCH and SLQYLA motifs, which are binding sites of Cullin 5 [44, 45] and Elongin C (144–149) were conserved [16, 46, 47].
Phenotypic characterization of HIV subtype C Vif variants
There is evidence that natural Vif variants differ in their ability to neutralize A3G [31, 32, 34] and we speculate that Vif diversity may emerge as it adapts to an individual’s A3G repertoire. We functionally characterized the patient-derived subtype C Vif panel for A3G degradation and counteraction in single-cycle infectivity assays. We co-transfected HIVΔVif with individual Vif expression plasmids, WT A3G or A3G-H186R. HIV-1 infectivity was subsequently analysed by infecting TZM-bl reporter cells with viral supernatants collected two days post-transfection. Infectivity values were plotted relative to HIV-1 in the absence of A3G, which was set to 100%.
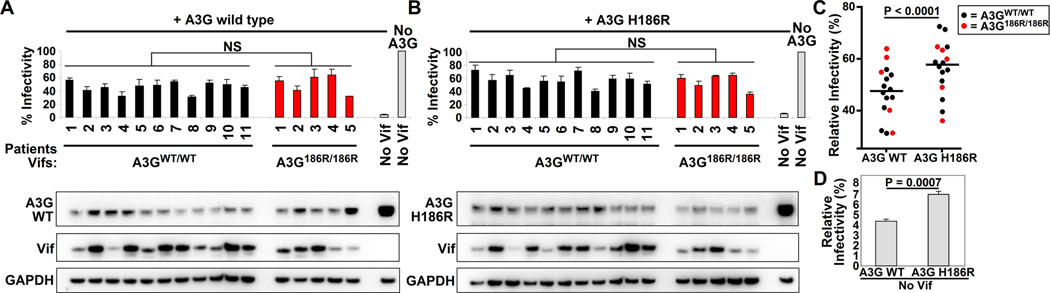
We first looked at whether Vifs obtained from A3GWT/WT or A3GH186R/H186R patients would differ in their ability to counteract A3G. All patient-derived Vif variants counteracted WT A3G and A3G-H186R to similar levels, irrespective of the patient’s A3G variant. (Figure 2, compare within A and B, NS; not significant, unpaired t test). This indicates that HIV-1 Vif does not adapt to the different A3G variants in vivo. Western blot analysis showed that the level of A3G degradation (compare to the No Vif control) was similar among Vifs (compare between 2A and 2B), which is in agreement with the infectivity data. Additionally, we observed that the expression levels of the individual Vif variants was highly variable, but was independent of both the patient’s A3G genotype from which they were derived as well as of their activity to degrade and counteract A3G (Figure 2A and B).
Figure 2. Activity of patient-derived subtype C Vifs against the WT A3G and A3G-H186R variants.
Patient-derived Vifs were cloned into an expression vector and co-transfected with NL4-3ΔVif and WT A3G (A) or A3G-H186R (B) expression plasmids and infectivity of the produced viruses was tested on TZM-bl reporter cells. NS; not significant, unpaired t test. 293T cell lysates were analysed by western blot and probed for HA, Vif and GAPDH serves as a loading control. Vif variants derived from homozygotes WT A3G are represented by black bars while Vifs derived from A3G-H186R homozygous carriers are represented by red bars. Error bars represent standard deviations from triplicate transfections. (C) Dot plot comparing the infectivity levels in the presence of WT A3G or A3G-H186R, P < 0001, paired t test. (D) Comparison of infectivities in the presence of WT A3G and A3G-H186R in the absence of Vif (also shown in 2A). P = 0.0007, unpaired t test. Error bars represent standard deviations from triplicate transfections.
Intrinsic antiviral activity of WT A3G and A3G-H186R
To determine whether the A3G variants differ in the restriction activity, we compared viral infectivity in the presence of WT A3G or A3G-H186H (Figure 2, compare between A and B). The infectivity values in the presence of A3G-H186R were always significantly higher compared to WT A3G (p<0.0001, paired t test), indicating that A3G-H186R restricts HIV less efficiently than WT A3G (Figure 2C). This difference is likely attributable to the lower basal antiviral activity of A3G-H186R, because the same result is also apparent in the absence of Vif (Figure 2D, 4.32 +/− 0.21 for WT A3G versus 6.9 +/− 0.28 for A3G-H186R, p = 0.0007, unpaired t test). This small but significant difference suggests that A3G-H186R intrinsically has reduced antiviral activity.
Taken together, we observed no evidence for functional adaptation of Vif to the different A3G variants in vivo. Importantly, our data show that the WT A3G was more efficient at restricting HIV compared to A3G-H186R variant, which may explain the accelerated HIV-1 disease progression in A3GH186R/ H186R patients.
Discussion
Several studies showed that A3GH186R/ H186R patients experience accelerated AIDS disease progression compared to individuals expressing A3GWT/WT or A3GWT/H186R [19, 21–23]. We observed that A3G-H186R restricts HIV-1 less potently than WT A3G both in the presence as well as in the absence of Vif. This novel finding indicates that A3G-H186R intrinsically has less antiviral activity, which is in agreement with a previous study which used cell free biochemical approaches which shows that A3G-H186R has reduced deaminase activity [48]. It is also possible that the H186R substitution may affect its packaging into virions, RNA binding or protein oligomerization and further mechanistic studies are required to elucidate the reduced antiviral activity of A3G-H186R. We speculate that A3G is not always fully counteracted by Vif in vivo and that the reduced restriction of A3G-H186R leads to higher viral loads and a more rapid disease progression in patients expressing A3GWT/H186R.
Studies show that SIV Vif adapts to polymorphisms in A3G [4, 49, 50]. We therefore anticipated that Vif would reduce its activity to degrade A3G-H186R because it poses less of a threat than A3G WT. However, Vif variants from A3GWT/WT or A3GH186R/ H186R showed no differences in activity, indicating that Vif does not adapt to A3G-H186R. It is conceivable that the difference in anti-HIV activity of the A3G variants is too small to create sufficient selective pressure for Vif adaptation given that A3G-H186R has less antiviral activity than the WT counterpart.
Together, polymorphisms in multiple APOBEC3 proteins such as A3G and A3H in combination with a HIV strain encoding a specific Vif variant could have profound effects on HIV replication and HIV-disease progression in patients [19, 21–23, 28, 29, 51]. Indeed, the time to progression to AIDS varies widely in patients of which only a small percentage could be attributed to natural polymorphisms in genes commonly associated with differential outcome such as CCR5 or HLA [52–58].
Acknowledgments
We thank all the CAPRISA 002 Acute Infection Study participants who are continuing to make an important contribution to HIV research. The scientific and supportive role of the whole CAPRISA 002 study and protocol teams are gratefully acknowledged. This study was funded in part by grants from South African Department of Science and Technology and National Research Foundation Research Chairs Initiative (grant #64809) (TN), the Victor Daitz Chair Foundation (TN), an International Early Career Scientist Award from the Howard Hughes Medical Institute (grant #55007427) (TN) and the NIH grants R01AI064001 (VS), R01 AI089246 (VS). The CAPRISA 002 study has received support from the National Institute of Allergy and infectious Disease (NIAID), National Institutes of Health (NIH) (grant # AI51794), from CONRAD (USAID co-operative grant #GP00-08-00005-00, subproject agreement # PPA-09-046), from the National Research Foundation (grant # 67385), the Technology Innovation Agency, and the Columbia University-Southern African Fogarty AIDS International Training and Research Programme (AITRP) funded by the Fogarty International Center, NIH (grant # D43TW00231).
Footnotes
Author Contributions
KR, MO, VS, TN conceived and designed the experiments. KR, MO and ML performed the experiments. KR and MO analysed the data. NG heads and manages the study cohort and provided the samples. All the authors contributed to the manuscript writing and reviewed the final version.
References
- 1.Harris RS, Hultquist JF, Evans DT. The restriction factors of human immunodeficiency virus. J Biol Chem. 2012;287:40875–40883. doi: 10.1074/jbc.R112.416925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kirchhoff F. Immune evasion and counteraction of restriction factors by HIV-1 and other primate lentiviruses. Cell Host Microbe. 2010;8:55–67. doi: 10.1016/j.chom.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 3.Malim MH, Bieniasz PD. HIV Restriction Factors and Mechanisms of Evasion. Cold Spring Harb Perspect Med. 2012;2:a006940. doi: 10.1101/cshperspect.a006940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Malim MH, Emerman M. HIV-1 accessory proteins--ensuring viral survival in a hostile environment. Cell Host Microbe. 2008;3:388–398. doi: 10.1016/j.chom.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 5.O'Brien SJ, Nelson GW. Human genes that limit AIDS. Nature Genetics. 2004;36:565–574. doi: 10.1038/ng1369. [DOI] [PubMed] [Google Scholar]
- 6.Sharp PM, Hahn BH. Origins of HIV and the AIDS Pandemic. Cold Spring Harb Perspect Med. 2011;1:a006841. doi: 10.1101/cshperspect.a006841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Simon V, Bloch N, Landau NR. Intrinsic host restrictions to HIV-1 and mechanisms of viral escape. Nat Immunol. 2015;16:546–553. doi: 10.1038/ni.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mangeat B, Turelli P, Caron G, Friedli M, Perrin L, Trono D. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature. 2003;424:99–103. doi: 10.1038/nature01709. [DOI] [PubMed] [Google Scholar]
- 9.Sheehy AM, Gaddis NC, Choi JD, Malim MH. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418:646–650. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- 10.Harris RS, Bishop KN, Sheehy AM, Craig HM, Petersen-Mahrt SK, Watt IN, et al. DNA deamination mediates innate immunity to retroviral infection. Cell. 2003;113:803–809. doi: 10.1016/s0092-8674(03)00423-9. [DOI] [PubMed] [Google Scholar]
- 11.Zhang H, Yang B, Pomerantz RJ, Zhang C, Arunachalam SC, Gao L. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature. 2003;424:94–98. doi: 10.1038/nature01707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo F, Cen S, Niu M, Yang Y, Gorelick RJ, Kleiman L. The interaction of APOBEC3G with human immunodeficiency virus type 1 nucleocapsid inhibits tRNA3Lys annealing to viral RNA. J Virol. 2007;81:11322–11331. doi: 10.1128/JVI.00162-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klarmann GJ, Chen X, North TW, Preston BD. Incorporation of uracil into minus strand DNA affects the specificity of plus strand synthesis initiation during lentiviral reverse transcription. J Biol Chem. 2003;278:7902–7909. doi: 10.1074/jbc.M207223200. [DOI] [PubMed] [Google Scholar]
- 14.Newman EN, Holmes RK, Craig HM, Klein KC, Lingappa JR, Malim MH, et al. Antiviral function of APOBEC3G can be dissociated from cytidine deaminase activity. Curr Biol. 2005;15:166–170. doi: 10.1016/j.cub.2004.12.068. [DOI] [PubMed] [Google Scholar]
- 15.Shindo K, Takaori-Kondo A, Kobayashi M, Abudu A, Fukunaga K, Uchiyama T. The enzymatic activity of CEM15/Apobec-3G is essential for the regulation of the infectivity of HIV-1 virion but not a sole determinant of its antiviral activity. J Biol Chem. 2003;278:44412–44416. doi: 10.1074/jbc.C300376200. [DOI] [PubMed] [Google Scholar]
- 16.Marin M, Rose KM, Kozak SL, Kabat D. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat Med. 2003;9:1398–1403. doi: 10.1038/nm946. [DOI] [PubMed] [Google Scholar]
- 17.Sheehy AM, Gaddis NC, Malim MH. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat Med. 2003;9:1404–1407. doi: 10.1038/nm945. [DOI] [PubMed] [Google Scholar]
- 18.Stopak K, de Noronha C, Yonemoto W, Greene WC. HIV-1 Vif blocks the antiviral activity of APOBEC3G by impairing both its translation and intracellular stability. Mol Cell. 2003;12:591–601. doi: 10.1016/s1097-2765(03)00353-8. [DOI] [PubMed] [Google Scholar]
- 19.Reddy K, Winkler CA, Werner L, Mlisana K, Abdool Karim SS, Ndung'u T. APOBEC3G expression is dysregulated in primary HIV-1 infection and polymorphic variants influence CD4+ T-cell counts and plasma viral load. AIDS. 2010;24:195–204. doi: 10.1097/QAD.0b013e3283353bba. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Genomes Project C. Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.An P, Bleiber G, Duggal P, Nelson G, May M, Mangeat B, et al. APOBEC3G genetic variants and their influence on the progression to AIDS. J Virol. 2004;78:11070–11076. doi: 10.1128/JVI.78.20.11070-11076.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bunupuradah T, Imahashi M, Iampornsin T, Matsuoka K, Iwatani Y, Puthanakit T, et al. Association of APOBEC3G genotypes and CD4 decline in Thai and Cambodian HIV-infected children with moderate immune deficiency. AIDS Res Ther. 2012;9:34. doi: 10.1186/1742-6405-9-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Singh KK, Wang Y, Gray KP, Farhad M, Brummel S, Fenton T, et al. Genetic variants in the host restriction factor APOBEC3G are associated with HIV-1-related disease progression and central nervous system impairment in children. J Acquir Immune Defic Syndr. 2013;62:197–203. doi: 10.1097/QAI.0b013e31827ab612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bizinoto MC, Leal E, Diaz RS, Janini LM. Loci polymorphisms of the APOBEC3G gene in HIV type 1-infected Brazilians. AIDS Res Hum Retroviruses. 2011;27:137–141. doi: 10.1089/aid.2010.0146. [DOI] [PubMed] [Google Scholar]
- 25.De Maio FA, Rocco CA, Aulicino PC, Bologna R, Mangano A, Sen L. Effect of HIV-1 Vif variability on progression to pediatric AIDS and its association with APOBEC3G and CUL5 polymorphisms. Infect Genet Evol. 2011;11:1256–1262. doi: 10.1016/j.meegid.2011.04.020. [DOI] [PubMed] [Google Scholar]
- 26.De Maio FA, Rocco CA, Aulicino PC, Bologna R, Mangano A, Sen L. APOBEC3-mediated editing in HIV type 1 from pediatric patients and its association with APOBEC3G/CUL5 polymorphisms and Vif variability. AIDS Res Hum Retroviruses. 2012;28:619–627. doi: 10.1089/AID.2011.0291. [DOI] [PubMed] [Google Scholar]
- 27.Do H, Vasilescu A, Diop G, Hirtzig T, Heath SC, Coulonges C, et al. Exhaustive genotyping of the CEM15 (APOBEC3G) gene and absence of association with AIDS progression in a French cohort. J Infect Dis. 2005;191:159–163. doi: 10.1086/426826. [DOI] [PubMed] [Google Scholar]
- 28.Duggal NK, Fu W, Akey JM, Emerman M. Identification and antiviral activity of common polymorphisms in the APOBEC3 locus in human populations. Virology. 2013;443:329–337. doi: 10.1016/j.virol.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ooms M, Brayton B, Letko M, Maio SM, Pilcher CD, Hecht FM, et al. HIV-1 Vif adaptation to human APOBEC3H haplotypes. Cell Host Microbe. 2013;14:411–421. doi: 10.1016/j.chom.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 30.Hemelaar J, Gouws E, Ghys PD, Osmanov S. Isolation W-UNfH, Characterisation. Global trends in molecular epidemiology of HIV-1 during 2000–2007. AIDS. 2011;25:679–689. doi: 10.1097/QAD.0b013e328342ff93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Binka M, Ooms M, Steward M, Simon V. The activity spectrum of Vif from multiple HIV-1 subtypes against APOBEC3G, APOBEC3F, and APOBEC3H. J Virol. 2012;86:49–59. doi: 10.1128/JVI.06082-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iwabu Y, Kinomoto M, Tatsumi M, Fujita H, Shimura M, Tanaka Y, et al. Differential anti-APOBEC3G activity of HIV-1 Vif proteins derived from different subtypes. J Biol Chem. 2010;285:35350–35358. doi: 10.1074/jbc.M110.173286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Loggerenberg F, Mlisana K, Williamson C, Auld SC, Morris L, Gray CM, et al. Establishing a cohort at high risk of HIV infection in South Africa: challenges and experiences of the CAPRISA 002 acute infection study. PLoS ONE. 2008;3:e1954. doi: 10.1371/journal.pone.0001954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Simon V, Zennou V, Murray D, Huang Y, Ho DD, Bieniasz PD. Natural variation in Vif: differential impact on APOBEC3G/3F and a potential role in HIV-1 diversification. PLoS Pathog. 2005;1:e6. doi: 10.1371/journal.ppat.0010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zennou V, Perez-Caballero D, Gottlinger H, Bieniasz PD. APOBEC3G incorporation into human immunodeficiency virus type 1 particles. J Virol. 2004;78:12058–12061. doi: 10.1128/JVI.78.21.12058-12061.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Letko M, Silvestri G, Hahn BH, Bibollet-Ruche F, Gokcumen O, Simon V, et al. Vif proteins from diverse primate lentiviral lineages use the same binding site in APOBEC3G. J Virol. 2013;87:11861–11871. doi: 10.1128/JVI.01944-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jacobs GB, Nistal M, Laten A, van Rensburg EJ, Rethwilm A, Preiser W, et al. Molecular analysis of HIV type 1 vif sequences from Cape Town, South Africa. AIDS Res Hum Retroviruses. 2008;24:991–994. doi: 10.1089/aid.2008.0077. [DOI] [PubMed] [Google Scholar]
- 38.Scriba TJ, Treurnicht FK, Zeier M, Engelbrecht S, van Rensburg EJ. Characterization and phylogenetic analysis of South African HIV-1 subtype C accessory genes. AIDS Res Hum Retroviruses. 2001;17:775–781. doi: 10.1089/088922201750237059. [DOI] [PubMed] [Google Scholar]
- 39.Bell CM, Connell BJ, Capovilla A, Venter WD, Stevens WS, Papathanasopoulos MA. Molecular characterization of the HIV type 1 subtype C accessory genes vif, vpr, and vpu. AIDS Res Hum Retroviruses. 2007;23:322–330. doi: 10.1089/aid.2006.0181. [DOI] [PubMed] [Google Scholar]
- 40.Wichroski MJ, Ichiyama K, Rana TM. Analysis of HIV-1 viral infectivity factor-mediated proteasome-dependent depletion of APOBEC3G: correlating function and subcellular localization. J Biol Chem. 2005;280:8387–8396. doi: 10.1074/jbc.M408048200. [DOI] [PubMed] [Google Scholar]
- 41.Russell RA, Pathak VK. Identification of two distinct human immunodeficiency virus type 1 Vif determinants critical for interactions with human APOBEC3G and APOBEC3F. J Virol. 2007;81:8201–8210. doi: 10.1128/JVI.00395-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.He Z, Zhang W, Chen G, Xu R, Yu XF. Characterization of conserved motifs in HIV-1 Vif required for APOBEC3G and APOBEC3F interaction. J Mol Biol. 2008;381:1000–1011. doi: 10.1016/j.jmb.2008.06.061. [DOI] [PubMed] [Google Scholar]
- 43.Tian C, Yu X, Zhang W, Wang T, Xu R, Yu XF. Differential requirement for conserved tryptophans in human immunodeficiency virus type 1 Vif for the selective suppression of APOBEC3G and APOBEC3F. J Virol. 2006;80:3112–3115. doi: 10.1128/JVI.80.6.3112-3115.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Luo K, Xiao Z, Ehrlich E, Yu Y, Liu B, Zheng S, et al. Primate lentiviral virion infectivity factors are substrate receptors that assemble with cullin 5-E3 ligase through a HCCH motif to suppress APOBEC3G. Proc Natl Acad Sci U S A. 2005;102:11444–11449. doi: 10.1073/pnas.0502440102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xiao Z, Xiong Y, Zhang W, Tan L, Ehrlich E, Guo D, et al. Characterization of a novel Cullin5 binding domain in HIV-1 Vif. J Mol Biol. 2007;373:541–550. doi: 10.1016/j.jmb.2007.07.029. [DOI] [PubMed] [Google Scholar]
- 46.Mehle A, Strack B, Ancuta P, Zhang C, McPike M, Gabuzda D. Vif overcomes the innate antiviral activity of APOBEC3G by promoting its degradation in the ubiquitin-proteasome pathway. J Biol Chem. 2004;279:7792–7798. doi: 10.1074/jbc.M313093200. [DOI] [PubMed] [Google Scholar]
- 47.Yu X, Yu Y, Liu B, Luo K, Kong W, Mao P, et al. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science. 2003;302:1056–1060. doi: 10.1126/science.1089591. [DOI] [PubMed] [Google Scholar]
- 48.Feng Y, Chelico L. Intensity of deoxycytidine deamination of HIV-1 proviral DNA by the retroviral restriction factor APOBEC3G is mediated by the noncatalytic domain. J Biol Chem. 2011;286:11415–11426. doi: 10.1074/jbc.M110.199604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Compton AA, Hirsch VM, Emerman M. The host restriction factor APOBEC3G and retroviral Vif protein coevolve due to ongoing genetic conflict. Cell Host Microbe. 2012;11:91–98. doi: 10.1016/j.chom.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Krupp A, McCarthy KR, Ooms M, Letko M, Morgan JS, Simon V, et al. APOBEC3G polymorphism as a selective barrier to cross-species transmission and emergence of pathogenic SIV and AIDS in a primate host. PLoS Pathog. 2013;9:e1003641. doi: 10.1371/journal.ppat.1003641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Refsland EW, Hultquist JF, Luengas EM, Ikeda T, Shaban NM, Law EK, et al. Natural polymorphisms in human APOBEC3H and HIV-1 Vif combine in primary T lymphocytes to affect viral G-to-A mutation levels and infectivity. PLoS Genet. 2014;10:e1004761. doi: 10.1371/journal.pgen.1004761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Carrington M, O'Brien SJ. The influence of HLA genotype on AIDS. Annu Rev Med. 2003;54:535–551. doi: 10.1146/annurev.med.54.101601.152346. [DOI] [PubMed] [Google Scholar]
- 53.Dean M, Carrington M, Winkler C, Huttley GA, Smith MW, Allikmets R, et al. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Hemophilia Growth and Development Study, Multicenter AIDS Cohort Study, Multicenter Hemophilia Cohort Study, San Francisco City Cohort, ALIVE Study. Science. 1996;273:1856–1862. doi: 10.1126/science.273.5283.1856. [DOI] [PubMed] [Google Scholar]
- 54.Kawashima Y, Pfafferott K, Frater J, Matthews P, Payne R, Addo M, et al. Adaptation of HIV-1 to human leukocyte antigen class I. Nature. 2009;458:641–645. doi: 10.1038/nature07746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Leslie AJ, Pfafferott KJ, Chetty P, Draenert R, Addo MM, Feeney M, et al. HIV evolution: CTL escape mutation and reversion after transmission. Nat Med. 2004;10:282–289. doi: 10.1038/nm992. [DOI] [PubMed] [Google Scholar]
- 56.Liu R, Paxton WA, Choe S, Ceradini D, Martin SR, Horuk R, et al. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell. 1996;86:367–377. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- 57.Rousseau CM, Daniels MG, Carlson JM, Kadie C, Crawford H, Prendergast A, et al. HLA class I-driven evolution of human immunodeficiency virus type 1 subtype c proteome: immune escape and viral load. J Virol. 2008;82:6434–6446. doi: 10.1128/JVI.02455-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stephens HA. HIV-1 diversity versus HLA class I polymorphism. Trends Immunol. 2005;26:41–47. doi: 10.1016/j.it.2004.11.001. [DOI] [PubMed] [Google Scholar]